Abstract
The staphylococcal accessory regulator SarA and the alternative sigma factor σB have been previously identified as positive regulators, and IcaR as a negative regulator, of icaADBC expression. Here, we show that in Staphylococcus aureus SarA and σB are also required for icaR expression and that IcaR does not have a significant effect on its own expression.
The proteins encoded by the intercellular adhesin genes (icaADBC) synthesize the polysaccharide poly-N-acetylglucosamine (PNAG), which contributes to the formation of a biofilm by Staphylococcus aureus. In addition to the proteins that synthesize PNAG, the ica locus also encodes the TetR family transcriptional regulator IcaR (23). The icaR gene is transcribed divergently from icaADBC (10) and is a negative regulator of icaADBC expression (9, 14). Biofilm formation by S. aureus plays an important role in the pathogenesis of endocarditis, osteomyelitis, and corneal and medical device infections (18). Although the majority of clinical isolates of S. aureus contain the ica operon, in vitro expression is tightly controlled (19) and the regulation of ica has been shown to be a complex and multifactorial process, involving a variety of external environmental factors and internal regulators.
Rachid et al. demonstrated that σB was required for biofilm formation under environmental stress conditions in an S. aureus mucosal isolate (22). They suggested that the effect of σB on ica expression could be indirect, as the ica promoter does not appear to contain a consensus σB binding site. Later, Conlon et al. demonstrated that icaR encodes a repressor of icaADBC transcription in Staphylococcus epidermidis, and we found the same to be true in S. aureus (9, 14). Alleviation of IcaR-mediated repression occurs in response to certain icaADBC-inducing stimuli such as ethanol but not in response to others (e.g., NaCl) (8), suggesting a role for additional regulatory mechanisms. Valle et al. and Beenken et al. demonstrated a role for SarA in icaADBC expression and biofilm formation in S. aureus (2, 27), and shortly thereafter Tormo et al. demonstrated a similar role in S. epidermidis (26).
While negative regulation of icaR by σB in S. epidermidis has been shown previously (16), its regulation in S. aureus has not yet been studied. IcaR belongs to the tetracycline repressor family of proteins, which are involved in gene regulation, acting as either transcriptional activators or repressors. The aim of this work was to elucidate regulation of the icaR gene in S. aureus.
All strains used in this study were grown at 37°C and 200 rpm in Luria-Bertani (LB) broth or LB agar, except for biofilm formation assays, where tryptic soy broth supplemented with 1% glucose was used. When appropriate, LB broth or LB agar was supplemented with the antibiotics at the following concentrations: 10 μg chloramphenicol/ml, 5 μg tetracycline/ml, and 10 μg erythromycin/ml. After overnight growth, cultures were diluted 1:50 in 5 ml fresh medium and incubated for 6 h before cells were collected for expression studies. Using DNA from strain S. aureus MN8 as the template and the primers IcaRproEcoRI (5′-GAATTCTAGTATTTTAATTTGCAATAGATTGTTGTTATAATTAAACGG-3′) and IcaRproSma (5′-CCCGGGCTTATCCTTCAATTTTTATAACCCC-3′), we amplified the promoter region and the first four codons of the icaR gene by PCR, and using the primers icapro1932 (5′-GAATTCGATATAAAGCATCAATTGAATAGTTCG-3′) and icaproREV (5′-CCCGGGGTTAAAAAATTGCAATTTCTTTACCTTTCG-3′), we amplified the icaADBC promoter and the first six codons of icaA. The DNA fragments were digested with SmaI, fused to the β-galactosidase gene (bgaB) from Bacillus stearothermophilus, and cloned into the staphylococcal shuttle vector pRB473 (constructed by Reinhold Brückner, Universitat Tubingen, Germany) (4). The reporter construct was sequenced to verify the absence of mutations, electroporated into S. aureus RN4220, and transduced to other strains using phage 80 (15, 17). S. aureus strain Newman ΔrsbUVW, in which the σB operon has been replaced with an erythromycin resistance (Erm) cassette, was kindly provided by Kenneth Bayles, University of Nebraska (24). Strain ALC1342, a derivative of strain RN6390 in which the sarA gene was replaced with an Erm cassette, was kindly provided by Ambrose Cheung, Dartmouth Medical School (7). RN4220 ΔicaR::Erm was constructed previously in our lab (14). Mutations were transduced to strains SA113 and Newman using phage 80. To complement the deletion mutations, sarA and 1.2 kb of the region upstream from the gene, which contains the promoters and a 5′ untranslated region; the rsbUVWσB locus; or the icaR gene was amplified from genomic DNA from strain Newman using the primer pairs attB2SarAFWD (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTATATCATTGGTGTCCTAGTTGG-3′) and attB1SarAREV (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCATGGATTGGATGGTAATTTAGCTGG-3′); attB2SigBFWD (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGAATCAATTGGAGGTTCTCATATG-3′) and attB1SigBREV (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCCTTTACGTTTCGCCTCAGTTCG-3′); or attB2IcaRFWD (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGTTTCCTCCACATAATCAATCATTG-3′) and attB1IcaRREV (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTCTTTACCTACCTTTCGTTAGTTAGG-3′) and cloned into pKOR1 (1). The plasmids were electroporated into strain RN4220 and transduced to the respective deletion mutants of Newman and SA113 using phage 80 (15). The erythromycin resistance cassettes were then replaced with the intact genetic loci by inducible counterselection.
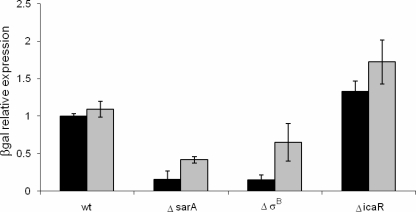
Regulation of the icaADBC promoter was assessed by 2-nitrophenyl β-d-galactopyranoside (ONPG) degradation (as a measurement of β-galactosidase activity) as previously described (6) with minor modifications: bacteria were lysed with lysostaphin, and 50 μl of cell extract and 50 μl of ONPG were combined and incubated for 2 h (for SA113) or 6 h (for Newman). β-Galactosidase activity was normalized to 1.0 (ONPG activity detected in the wild-type strain). Confirming several previous reports, our results showed that in both SA113 and Newman, IcaR repressed icaADBC expression, while SarA and σB activated this promoter (Fig. 1). Chromosomal complementation of the mutations confirmed that the effects were due to deletion of icaR, sarA, and the rsbUVWσB locus, respectively. Biofilms were formed in 96-well microtiter plates and quantified using the modified microtiter plate method developed by Stepanovic et al. (25). SA113 formed a stronger biofilm than Newman (optical density at 570 nm [OD570] = 1.74 ± 0.05 and 0.44 ± 0.03, respectively). In the absence of SarA or σB, the biofilm formation of both strains was impaired (OD570 = 0.39 and 0.54 for SA113 and OD570 = 0.23 ± 0.02 and 0.33 ± 0.03 for Newman, respectively), whereas chromosomal complementation of the sarA and rsbUVWσB loci restored biofilm formation (OD570 = 1.74 ± 0.02 and 1.75 ± 0.09 for SA113 and OD570 = 0.53 ± 0.02 and 0.54 ± 0.05 for Newman, respectively). Conversely, deletion of icaR induced a mild positive effect on biofilm formation (OD570 = 1.95 ± 0.01 and 0.62 ± 0.04, respectively). Again, complementation of icaR restored the biofilms (OD570 = 1.80 ± 0.01 for SA113 and 0.49 ± 0.02 for Newman). Previously, in S. epidermidis, although icaR was shown to repress icaADBC expression, biofilm formation was not significantly different in an icaR mutant and the wild type in brain heart infusion medium (9), demonstrating that additional factors are also important in the regulation of biofilm formation.
FIG. 1.
Effect of SarA, σB, and IcaR on the icaADBC promoter as determined by β-galactosidase activity in SA113 (black bars) and Newman (gray bars). wt, wild type. Error bars indicate standard deviations.
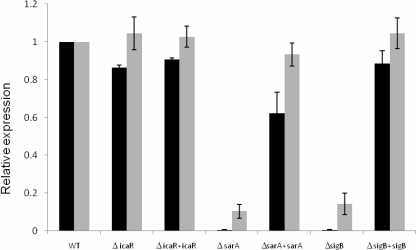
We then repeated the expression assays using the icaR promoter fusion. In a previous report it has been shown that in S. epidermidis, σB represses icaR (16). We were expecting a similar result with our constructs, but contrary to the report about S. epidermidis strain 1457, σB was an important positive regulator of expression of icaR in S. aureus strains SA113 and Newman (results for SA113 are shown in Fig. 2; results for Newman were similar and are not shown). The strains were fully complemented with an intact rsbuVWσB locus. Evidence that biofilm formation is regulated by somewhat different mechanisms within S. epidermidis and S. aureus comes from the fact that most icaADBC-positive strains of S. epidermidis produce biofilms in vitro, while most S. aureus strains do not (10, 19). While σB appears to have an indirect effect on the icaADBC promoter, the mechanism of action of its regulation is still unclear, and variability not only between different species of staphylococci but between different strains of S. aureus is apparent as well from reports regarding the effect of σB on biofilm formation. (22, 27). Furthermore, σB also appears to play an important role in the long-term stability of S. epidermidis biofilms (13) that is not necessarily related to the icaADBC, expression although no similar effect has been described for S. aureus biofilms.
FIG. 2.
Effect of SarA, σB, and IcaR on the icaR promoter on SA113 as determined by β-galactosidase activity (gray) and mRNA levels (black) determined by real-time PCR and normalized to the expression of 16S RNA. WT, wild type. Error bars indicate standard deviations.
Interestingly, SarA was also required for icaR expression in both strains (results for SA113 are shown in Fig. 2; results for Newman were similar and are not shown). Complementation of the sarA mutation by replacing the Erm cassette with an intact sarA gene restored the phenotype. It was previously reported that icaR was not repressed by SarA in S. epidermidis, but a positive regulatory effect was not noted (26). Positive regulation of both icaADBC and its repressor, icaR, by SarA could result from unwinding of the promoter and subsequent access for the RNA polymerase complex to initiate transcription in either direction and could serve as a mechanism to prevent overexpression of the polysaccharide.
Valle et al. demonstrated that both SarA and σB affected icaADBC transcription but that only SarA was relevant for biofilm formation (27), contradicting previous information that attributed to σB a relevant role in S. aureus biofilm formation (22). Later, Handke et al. showed in an experiment using the icaADBC operon controlled by a cadmium-inducible promoter that at least in S. epidermidis, both SarA and σB were important for biofilm formation (11), although they suggested that while SarA has a direct effect on ica transcription, the σB effect is indirect. Indeed, microarray-based follow-up studies revealed that σB strongly affected SarA expression in S. aureus, but the same strong effect was not found for any of the ica genes (3, 21). Furthermore, another study showed that sarA transcription shifts from the σA-dependent promoter during the exponential growth phase to the σB-dependent promoter during the late exponential and stationary phases (28), clarifying even more the dependence of SarA on σB and suggesting an explanation for the variability of results regarding its role in icaADBC expression. Our results showed that for S. aureus, deletion of σB influences icaR expression, icaADBC expression, and biofilm formation in two distinct strains.
Although most members of the TetR family of regulatory proteins that are divergently transcribed from the structural gene that they regulate appear to self-regulate their own gene (12), it was described that in S. epidermidis, icaR had no effect on its own expression (9). Similarly, in S. aureus deletion of icaR did not have a significant effect on icaR promoter activity (results for SA113 are shown in Fig. 2; results for Newman were similar and are not shown).
The reporter constructs were translational fusions, so β-galactosidase activities were a function of both transcription and translation. To assess the regulation of transcription alone, we used real-time PCR. Total cellular RNA was prepared using the FastRNA Pro Blue kit (MP Biomedicals, Solon, OH) accordingly to manufacturer's instructions, and contaminating DNA was removed by two treatments with Turbo DNase (Ambion, Austin, TX). Reverse transcription of 2 μg RNA was performed using AccuScript (Stratagene, La Jolla, CA) and 10 pmol bgaB-RTREV (5′-ATCGATCGGCAAAGAATCTG-3′) or 16S-RTREV (5′-CCACTTTCCTCTTCTGCACTCA-3′). SensiMixPlus (Quantace, Norwood, MA) was used for real-time reverse transcription-PCR with the primers bgaB-RTFW (5′-GGGATTTTCAGTTGGAGCAA-3′) and bgaB-RTREV or 16S-RTFW (5′-TCCGGAATTATTGGGCGTAA-3′) and 16S-RTREV. Control reactions lacked reverse transcriptase enzyme to evaluate the level of DNA contamination. Real-time PCR results were similar to ONPG assay results (Fig. 2), suggesting that the observed effects are largely due to transcriptional rather than posttranscriptional control.
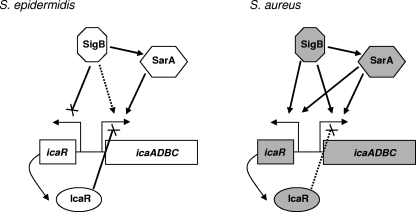
Altogether, the results indicate that in contrast to the case in S. epidermidis, SarA and σB were positive regulators of icaR expression in S. aureus whereas IcaR had little effect on its own expression and a relatively weak repressive effect on expression of the icaADBC operon (Fig. 3). Other regulators of biofilm formation have been described. In a recent study, Pamp et al. demonstrated by Northern blot analysis that in the absence of Spx, icaR transcript levels were reduced, while icaADBC expression was augmented (20). This illustrates the careful and complex orchestration of this process; biofilm formation is important under certain conditions, but the unregulated expression of PNAG would be metabolically wasteful. This study also revealed distinctions in regulation in different strains. Cafiso et al. found a relationship between Agr type and the relative roles of IcaR and σB in biofilm formation, suggesting one potential explanation for strain variability (5).
FIG. 3.
Diagram of icaR and icaADBC regulation in S. epidermidis (9, 11, 16, 26) versus S. aureus (based on work presented here). Solid lines ending in arrowheads represent activation, solid lines ending in “×” represent repression, and dotted lines represent weak activation or repression.
Acknowledgments
This work was supported by NIH grant R01 AI068892.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 5558-63. [DOI] [PubMed] [Google Scholar]
- 2.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 714206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 1864085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brückner, R., E. Wagner, and F. Götz. 1993. Characterization of a sucrase gene from Staphylococcus xylosus. J. Bacteriol. 175851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cafiso, V., T. Bertuccio, M. Santagati, V. Demelio, D. Spina, G. Nicoletti, and S. Stefani. 2007. agr-genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 51220-227. [DOI] [PubMed] [Google Scholar]
- 6.Cerca, N., and K. K. Jefferson. 2008. Effect of growth conditions on poly-N-acetylglucosamine expression and biofilm formation in Escherichia coli. FEMS. Microbiol. Lett. 28336-41. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. L., and A. C. Manna. 2005. Role of the distal sarA promoters in SarA expression in Staphylococcus aureus. Infect. Immun. 734391-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon, K., H. Humphreys, and J. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS. Microbiol. Lett. 216171-177. [DOI] [PubMed] [Google Scholar]
- 9.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 1844400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 675427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handke, L. D., S. R. Slater, K. M. Conlon, S. T. O'Donnell, M. E. Olson, K. A. Bryant, M. E. Rupp, J. P. O'Gara, and P. D. Fey. 2007. Sigma B and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can. J. Microbiol. 5382-91. [DOI] [PubMed] [Google Scholar]
- 12.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48345-369. [DOI] [PubMed] [Google Scholar]
- 13.Jäger, S., D. Mack, H. Rohde, M. A. Horstkotte, and J. K. Knobloch. 2005. Disintegration of Staphylococcus epidermidis biofilms under glucose-limiting conditions depends on the activity of the alternative sigma factor σB. Appl. Environ. Microbiol. 715577-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jefferson, K., D. Pier, D. Goldmann, and G. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 1862449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasatiya, S. S., and J. N. Baldwin. 1967. Nature of the determinant of tetracycline resistance in Staphylococcus aureus. Can. J. Microbiol. 131079-1086. [DOI] [PubMed] [Google Scholar]
- 16.Knobloch, J. K., S. Jäger, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 723838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J. C. 1995. Electrotransformation of staphylococci. Methods Mol. Biol. 47209-216. [DOI] [PubMed] [Google Scholar]
- 18.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339520-532. [DOI] [PubMed] [Google Scholar]
- 19.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270179-188. [DOI] [PubMed] [Google Scholar]
- 20.Pamp, S. J., D. Frees, S. Engelmann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 1884861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pane-Farre, J., B. Jonas, K. Forstner, S. Engelmann, and M. Hecker. 2006. The sigma B regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296237-258. [DOI] [PubMed] [Google Scholar]
- 22.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 1826824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos, J. L., M. Martinez-Bueno, A. J. Molina-Henares, W. Teran, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice, K. C., T. Patton, S. J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus Cid and Lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 1863029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stepanovic, S., D. Vukovic, I. Dakic, B. Savic, and M. Svabic-Vlahovic. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40175-179. [DOI] [PubMed] [Google Scholar]
- 26.Tormo, M. A., M. Marti, J. Valle, A. C. Manna, A. L. Cheung, I. Lasa, and J. R. Penadés. 2005. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 1872348-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penadés, and I. Lasa. 2003. SarA and not sigma B is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 481075-1087. [DOI] [PubMed] [Google Scholar]
- 28.Ziebandt, A. K., D. Becher, K. Ohlsen, J. Hacker, M. Hecker, and S. Engelmann. 2004. The influence of agr and sigma B in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 43034-3047. [DOI] [PubMed] [Google Scholar]