Abstract
CorA is the primary Mg2+ channel in Salmonella enterica serovar Typhimurium. A corA mutant is attenuated in mice and defective for invasion of and replication within epithelial cells. Microarray studies show that several virulence effectors are repressed in a corA mutant strain, which ultimately manifests itself as a decrease in virulence.
Salmonella enterica serovar Typhimurium causes infections that usually appear as a gastrointestinal ailment in more than 1.4 million Americans per year (50). Infection begins with oral consumption of live bacteria in contaminated food. In the small intestine, the bacteria are actively taken up by M cells of the Peyer's patches and/or mononuclear phagocytes. A type three secretion system (TTSS) carried on Salmonella pathogenicity island 1 (SPI-1) delivers effector proteins to the M cell to aid in invasion (8, 14). Survival within the macrophage phagosome requires another TTSS encoded on SPI-2 (16, 28, 37). Thus, S. enterica serovar Typhimurium's environment fluctuates markedly during its pathogenic life cycle and requires the coordination of expression of a multitude of virulence effectors and housekeeping genes. These gene products form dynamic signaling networks that interact to control the infection process.
We asked if intracellular Mg2+ of the bacterium is involved in virulence. To obtain Mg2+, S. enterica serovar Typhimurium has three Mg2+ transport systems. CorA, the primary or “housekeeping” Mg2+ channel, is constitutively expressed, while the MgtA and MgtB transporters are only expressed in response to activation by the PhoP/PhoQ two-component system (2, 6, 10, 15, 17, 24, 27, 39, 42, 46). CorA lacks homology to any other known protein and is widespread throughout the bacteria and archaea (18, 22, 24, 38). MgtA and MgtB are similar to eukaryotic P-type ATPases and are less widely distributed (23, 24, 42, 47). Both mgtA and mgtB are induced during infection, implicating Mg2+ transport in virulence (11, 39).
MgtA, MgtB, and CorA have different maximal velocities of Mg2+ uptake, with CorA > MgtA > MgtB as measured in single-transporter strains. Moreover, strains carrying a single transport system exhibit no growth defect in any medium. However, a triple-transporter mutant (MM281) requires additional Mg2+ for growth, at least 10 mM Mg2+ in Luria-Bertani (LB) broth (17). A corA mutant is resistant to Co2+ toxicity and has diminished transport of Mg2+, Co2+, and Ni2+ relative to the wild type even though there is no growth deficit (41). A corA mutant rescues the Fe2+ hypersensitivity phenotype of the phoP and mgtA mgtCB mutants, although CorA does not transport Fe2+ (4, 29). A corA mutant is sensitive to killing by the host lactoperoxidase system (35, 36), and expression of corA is induced upon exposure to the host lactoperoxidase system. Thus, corA appears linked to Salmonella survival in the host. We show here that mutation of corA results in a decrease in virulence.
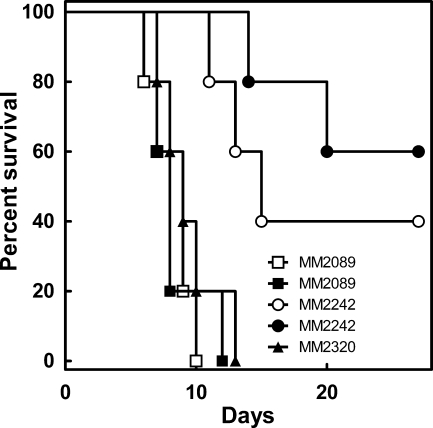
We wished to determine the virulence of a corA mutant strain compared to that of wild-type S. enterica serovar Typhimurium SL1344, a standard pathogenic strain. Groups of five BALB/c mice (8 weeks old) were infected by oral gavage with 7 × 108 or 7 × 109 cells of wild-type S. enterica serovar Typhimurium SL1344, 5 × 108 or 5 × 109 cells of a corA mutant strain (29), or 7 × 108 cells of a corA mutant strain carrying pCorA (also known as pJL10). The S. enterica serovar Typhimurium corA gene, with its native promoter from pRS170 (45), was subcloned between the XbaI and EcoRI restriction sites of low-copy-number vector pWKS30 to generate pCorA (51). The orientation of the corA allele within pCorA is the opposite of that of the lac promoter so that expression is driven by the corA promoter. Complementation of the corA mutant strain by pCorA was verified as previously described (data not shown) (41). Expression of CorA from pCorA was compared to that of wild-type S. enterica serovar Typhimurium through anti-CorA Western blot assays and found to be similar (data not shown). The bacterial strains used in this study are listed in Table 1. Additionally, C3H/HeN female mice (6 to 8 weeks old) were each infected intraperitoneally with these same strains (data not shown). Testing another mouse strain and inoculation route minimizes the possibility that the observed phenotype is either strain or inoculation route dependent. Mortality was monitored twice a day over a 25- to 28-day period. Moribund mice were immediately euthanized. Wild-type bacteria killed all of the mice by 10 to 11 days after oral administration (Fig. 1) and by 15 days after intraperitoneal administration (data not shown). When a corA mutant strain was administered to mice by the oral route, 50% of the mice survived (Fig. 1), and when it was administered by the intraperitoneal route, 30% survived (data not shown). The onset of illness and death was also delayed compared to the wild type with either route of injection. A low-copy plasmid carrying a functional S. enterica serovar Typhimurium corA allele (pCorA) was able to completely rescue the virulence defect.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Genotype | Source(s) |
|---|---|---|
| MM1364 (RM4456) | SL1344 invA::Tn10::phoA | 39 |
| MM2089 | S. enterica serovar Typhimurium SL1344 hisG rpsL xyl (wild type) | 19, B. B. Finlaya |
| MM2242 | SL1344 corA52::Tn10Δ16Δ17 | 29 |
| MM2320 | SL1344 corA52::Tn10Δ16Δ17 pCorA (pJL10) | J. Linb |
| MM3220 (WN152/CS015) | 14028s phoP102::Tn10d-Cam | 26, J. S. Gunnc |
Michael Smith Laboratories, University of British Columbia, Vancouver, British Columbia, Canada.
Case Western Reserve University, Cleveland, OH.
Center for Microbial Interface Biology, The Ohio State University, Columbus.
FIG. 1.
Attenuation of a corA mutant strain after oral administration to BALB/c mice. Mice were infected by oral gavage with either the wild type (MM2089) at doses of 7 × 109 (□) and 7 × 108 (▪) cells, a corA mutant strain (MM2242) at doses of 5 × 109 (○) and 5 ×108 (•) cells, or a corA mutant strain with pCorA (MM2320) at a dose of 7 × 108 cells (▴) as described in the text.
To obtain a more global understanding of alterations in a corA mutant strain, a microarray study comparing the corA mutant and wild-type strains was conducted with cells grown to log phase in LB broth. The microarray protocols used were adapted from those at the Pathogen Functional Genomic Resource Center at The Institute for Genomic Research (http://pfgrc.tigr.org/resources.shtml). A single microarray was run for six independent RNA samples, three for the wild type and three for the corA mutant strain. Total RNA was obtained via hot phenol-chloroform extraction. Genomic DNA was extracted with a GenElute kit (Sigma) and used as a control. Total RNA (30 μg) and 2 μg of genomic DNA were labeled with Cy3 and Cy5 dyes (Invitrogen), respectively. Prior to hybridization, slides were incubated in 25% (vol/vol) formamide-5× SSC (0.75 M sodium chloride, 75 mM sodium citrate)-0.1% sodium dodecyl sulfate (SDS)-0.1 mg/ml bovine serum albumin for 1 to 2 h in a 42°C water bath, washed, and immediately air dried by centrifugation. Probes were resuspended in hybridization buffer (25% formamide, 5× SSC, 0.1% SDS) and placed on a heated slide within a slide chamber (Corning). The slide chambers were incubated in a 42°C water bath overnight. Slides were washed twice sequentially with each of the following three buffers: hot (55°C) low-stringency buffer (2× SSC, 0.1% SDS), medium-stringency buffer (0.1× SSC, 0.1% SDS), and high-stringency buffer (0.1× SSC). Slides were immediately air dried by centrifugation. Slides were scanned on a GenePix 4000B microarray scanner to obtain a Cy3/Cy5 fluorescence ratio of 0.8 to 1.2.
The TM4 Microarray Suite produced by The Institute for Genomic Research was used for data analysis (31). Spotfinder was used to assign gene identifiers to spots and to flag spots. MIDAS was used to normalize the data (Lowess [Locfit] and Standard Deviation Regularization). The normalized data were imported into Microsoft Excel. Normalized intensity values for RNA were divided by those for the genomic DNA control for each slide to obtain a ratio. Three sets of ratios were obtained for the wild type, and three sets of ratios were obtained for the corA mutant strain. P values were calculated by t test. (See Table S1 in the supplemental material, where only data with P values of less than 0.05 are represented and the average of the ratios for the wild type was divided by the average of the ratios for the corA mutant strain and vice versa to obtain the fold changes indicated.) Microarray descriptions and data have been deposited at www.ncbi.nlm.nih.gov/geo (platform GPL4805). The trends in the array results were confirmed for 10 selected genes by reverse transcription of isolated RNA followed by quantitative PCR (data not shown).
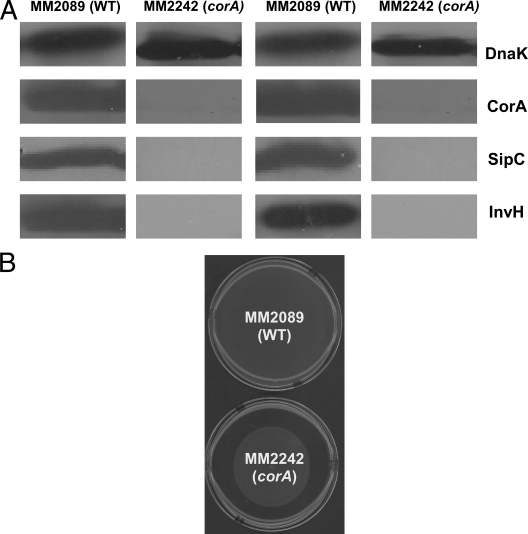
For a list of the genes that were upregulated (87 genes) or repressed (45 genes) in the corA mutant strain, sorted by function or putative function, see Table S1 in the supplemental material. SPI-1 encodes a TTSS required for entry into epithelial M cells (8, 14). Components of SPI-1 (e.g., invA, invB, and invH) are transcriptionally repressed in a corA mutant strain (see Table S1 in the supplemental material). Western blot assays against InvH and SipC, SPI-1 proteins, confirm that protein expression is also decreased in a corA mutant strain (Fig. 2A). The observation that the SipC protein but not its mRNA was decreased in the corA mutant strain suggests that additional SPI-1 proteins are affected by loss of CorA whether or not the corresponding mRNA was affected. Cells for SPI-1 Western blot assays were grown in LB broth to log phase; cells at an optical density at 600 nm of 0.2 were loaded directly onto 12% SDS polyacrylamide gels. Gels were electrophoresed and transferred onto nitrocellulose (Schleicher & Schuell, Keene, NH). The CorA antibody (40) was used at a dilution of 1:10,000, and a DnaK antibody (Bio-Rad) was used at a dilution of 1:5,000. All SPI-1 antibodies were made in rabbits and were used at a dilution of 1:5,000. Horseradish peroxidase-linked anti-rabbit and anti-mouse secondary antibodies (Amersham) were used at a dilution of 1:10,000. Proteins were visualized by enhanced chemiluminescence as recommended by the manufacturer (Amersham). Western blot assays were scanned and quantitated by densitometry with DnaK as a loading control (data not shown). Qualitatively, the fold changes represented by the microarray and Western blot assays are directly correlated. Since microarrays are a measure of global gene transcription affected by mRNA stability and degradation and Western blot assays are representative of protein levels dependent on translation, protein stability, and protein degradation, we expected a qualitative but not necessarily a quantitative correlation.
FIG. 2.
(A) Western blot assays of SPI-1 proteins were performed with total protein from the wild type (MM2089) and a corA mutant strain (MM2242) after growth in LB medium to log phase. The Western blot assays are representative of four independent experiments. Two independent experiments are shown. (B) The wild-type (MM2089) (top) and corA mutant (MM2242) (bottom) strains were plated at the centers of swim agar plates and allowed to grow at room temperature for 20 h. The data are representative of three independent experiments. WT, wild type.
Additional changes in mRNA expression are apparent in a corA mutant strain that also have relevance to virulence. Several SPI-2 genes (e.g., sseE, sseC, and sscB) required for survival within macrophages (16, 28, 37) are repressed in a corA mutant strain (see Table S1 in the supplemental material). Some flagellar genes are required for S. enterica serovar Typhimurium virulence (3). Some, although not all, flagellar genes (e.g., fljB, fliN, and fliM) are repressed in a corA mutant strain (see Table S1 in the supplemental material). This decreased expression is functionally relevant since a swimming assay with cells grown on LB broth plates with a low-percentage agar (Fig. 2B) shows that a corA mutant strain cannot swim as well as the wild type. The situation with the many flagellar genes may be similar to that with the SPI-1 genes, where both mRNA and protein levels for several are decreased (e.g., InvH) whereas only protein appears decreased for others (e.g., SipC). The swimming assay was performed by plating 10 μl of log-phase LB broth cultures of the wild type and a corA mutant strain, equivalent to an optical density at 600 nm of 0.005, at the centers of agar swim plates (10 g tryptone, 5 g yeast extract, 2.5 g agar, 0.5% glucose, 1.0 liter double-distilled H2O). Plates were incubated at room temperature for 20 h and scanned. Flagella may have some virulence properties that could aid in adherence, in bacterial movement to evade immune cells, or with movement (chemotaxis) toward beneficial surroundings. However, not all flagellar genes have been shown to be essential for Salmonella virulence; thus, it is not clear if the swimming defect would impact virulence (3, 20, 21, 32-34, 44).
Of the 87 genes upregulated in a corA mutant strain, about 20% are membrane or putative membrane proteins (see Table S1 in the supplemental material). The two other Mg2+ transporters, mgtA and mgt(C)B, are slightly upregulated in a corA mutant strain. This presumably explains the lack of an Mg2+ growth defect compared to the wild type. However, mgtA and mgtCB expression can be markedly induced via the PhoP/PhoQ two-component system upon response to certain external stimuli such as acid, a low Mg2+ concentration, and antimicrobial peptides (2, 26, 43). In the absence of Mg2+, this induction may reach 1,000-fold. Under the microarray growth conditions used, some Mg2+ is present and the expression of these genes should be generally repressed. It is noteworthy that no other PhoP/PhoQ-regulated genes seem to be affected. Thus, it is unclear whether induction of the expression of mgtA and mgtCB in a corA mutant strain is mediated by PhoP/PhoQ or another mechanism.
Two classes of membrane proteins that stand out from the set of repressed genes are those required for enterochelin-dependent iron uptake (entE, entC, fepA, etc.) and curli production. We confirmed upregulation of siderophores by a liquid chrome azurol S assay (1) and curli by a Congo red binding assay (12) (data not shown). Although iron is essential for virulence (7, 48, 49), it is unclear why a single iron uptake system should be upregulated and whether its upregulation is relevant to the attenuation of a corA mutant strain. With regard to the curli and the many other outer membrane proteins that are upregulated, most of unknown function, a significant remodeling of both membranes seems to result from the loss of corA. Despite these substantial changes in Salmonella membrane proteins, Caco-2 epithelial cells produce similar amounts of the cytokines tumor necrosis factor α, transforming growth factor β, and interleukin-1β in response to the corA mutant and wild-type strains over a similar time course (data not shown), suggesting that host cell recognition is not altered. Upregulation of iron uptake and/or modification of the bacterial cell membrane could play a role in a corA mutant's ability to rescue Fe2+ hypersensitivity of phoP and mgtA mgtCB mutants (4, 29). However, without more information on the function of these membrane proteins, no conclusions can be made as to their role in the corA virulence phenotype.
Cells for microarray analysis were grown to log phase in LB broth. This does not ideally represent bacterial gene expression within a mouse. However, the results obtained from the microarray support the data obtained from the mouse. Moreover, the results promoted additional experiments described below, which further support a role for corA in Salmonella virulence. Regardless of the growth conditions used in this study, the general trend is the same; corA is required for full virulence of Salmonella.
The microarray and Western blot assay results indicated that SPI-1-encoded effectors were downregulated in a corA mutant strain, suggesting that epithelial cell invasion might be compromised. Therefore, a corA mutant strain was tested for the ability to interact with cultured Caco-2 intestinal epithelial cells. Caco-2 epithelial cells (ATCC HTB-37) were maintained as recommended by the ATCC. Bacteria were grown in LB broth overnight at 37°C without shaking. Bacteria were washed once with phosphate-buffered saline (PBS) and suspended in cell culture growth medium without fetal bovine serum and antibiotics. Medium was removed from the cultured cells, and the bacterial suspension was added at a multiplicity of infection of 10:1. Plates were centrifuged at 1,000 rpm for 10 min at room temperature and then incubated at 37°C with 5.0% CO2. After 1 h, cultured cells were washed three times with PBS and treated with gentamicin (100 μg/ml) in cell culture growth medium without fetal bovine serum and antibiotics for at least 2 h to kill extracellular bacteria. After 6 h, the cultured cells were washed three times with PBS, lysed with 0.1% (vol/vol) Triton X-100 detergent for 5 min, collected, diluted, and plated onto LB broth plates, and grown overnight at 37°C for CFU determination. The number of bacteria obtained from within the cultured cells was divided by the total number of bacteria added to the cultured cells to obtain the percentage of gentamicin-protected bacteria.
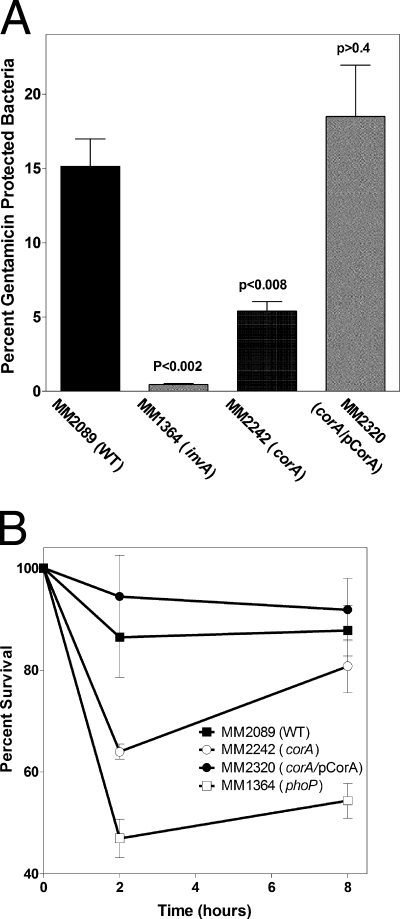
A corA mutant strain (1.1% invasion) has a statistically significantly (P < 0.03) decreased ability to invade Caco-2 epithelial cells, as measured 2 h postinvasion compared to the wild type (1.7% invasion) (data not shown). Thus, invasion by a corA mutant strain is ∼40% decreased compared to that by the wild type. In addition, by 4 to 6 h postinvasion, a corA mutant strain is defective for replication inside Caco-2 epithelial cells compared to the replication of wild-type cells (Fig. 3A). Bacterial counts of a corA mutant strain within Caco-2 epithelial cells are decreased by ∼60% compared to the wild type by 4 to 6 h after invasion, which is an ∼20% further decrease compared to invasion, suggesting an impairment of replication by a corA mutant strain within the epithelial cells. Thus, CorA is required for optimal invasion and replication within Caco-2 epithelial cells. The positive control for defective invasion, an invA strain, almost completely fails to invade and replicate within Caco-2 epithelial cells (data not shown and Fig. 3A). The corA defect can be rescued by complementation with a functional S. enterica serovar Typhimurium corA allele (pCorA), restoring replication within epithelial cells to a level similar to that of the wild type (Fig. 3A). The role of corA in epithelial cell invasion has been investigated further and is reported elsewhere (30).
FIG. 3.
(A) Epithelial cell invasion and survival by the wild-type strain (MM2089), a corA mutant strain (MM2242), a corA mutant strain with pCorA (MM2320), and a invA mutant strain (MM1364). Shown are data obtained 6 h after initiation of invasion. Similar results were obtained at other time points. The data are averages of three independent experiments. P values indicate t tests comparing the wild type to the other strains. (B) Time course of survival within J774.1 macrophages by the wild type (MM2089), a corA mutant strain (MM2242), a corA mutant strain with pCorA (MM2320), and a phoP mutant strain (MM3220). The data are from a single experiment representative of three independent experiments. WT, wild type.
Since the microarray data indicated a corA mutant strain has SPI-2 defects as well, we tested the strain's capacity to enter and survive within cultured J774A.1 macrophage-like cells. J774A.1 macrophage-like cells (ATCC TIB-67) were maintained as recommended by the ATCC. Bacteria were grown overnight in LB broth at 37°C without shaking. Bacteria were washed with PBS, pelleted, resuspended in 100% mouse serum (Innovative Research IMS-COMPL), and incubated for 20 min at 37°C prior to the experiment. The serum-treated bacteria were added to cell culture growth medium without antibiotics at a multiplicity of infection of 5:1. Plates were centrifuged at 1,000 rpm for 10 min at room temperature and then incubated at 37°C with 5.0% CO2. After 15 min, cultured cells were washed three times with cell culture growth medium without antibiotics and treated with gentamicin (12 μg/ml) in cell culture medium to kill extracellular bacteria. At each time point, the cultured cells were washed three times with complete cell culture medium without antibiotics and lysed with 1.0% Triton X-100 detergent for 5 min. The medium was collected, diluted, and plated onto LB broth plates, and the culture was grown overnight at 37°C for CFU determination.
Macrophage-like cells phagocytose S. enterica serovar Typhimurium strains with similar efficiencies: wild type, 2.7%; a corA mutant strain, 3.3%; a corA mutant strain with a low-copy plasmid carrying a functional S. enterica serovar Typhimurium corA gene (pCorA), 2.5%; a phoP mutant strain, 3.0% (data not shown). By 8 to 10 h postinfection, a corA mutant strain is weakly altered for survival inside J774A.1 macrophage-like cells compared to wild-type cells. A phoP mutant strain, a positive control, survives twofold less in J774A.1 macrophage-like cells compared to the wild type (Fig. 3B). The phoP result is comparable to those of other studies conducted with this cell line for the experimental conditions used (S. J. Libby, personal communication). A corA mutant strain carrying a low-copy plasmid with a functional S. enterica serovar Typhimurium corA gene (pCorA) survived similarly to the wild type.
Thus, downregulated expression of SPI-2 in a corA mutant strain minimally affects interactions with macrophages. The decreased SPI-2 expression is consistent with the attenuation of a corA mutant strain when mice are infected via the intraperitoneal route, bypassing the transepithelial route. However, there is only a weak effect on survival within the J774A.1 macrophage-like cell line. SPI-2 has been shown to be expressed inside epithelial cells alongside SPI-1 (25). Moreover, SPI-2 has been shown to be important for the development of enterocolitis, not just systemic disease (5). Thus, downregulation of SPI-2 in a corA mutant may contribute to the epithelial cell phenotype, as well as the weak macrophage phenotype. Alternatively, corA may be needed at some later stage of infection, a conclusion consistent with the attenuation of the corA mutant strain after intraperitoneal administration.
We have found that a corA S. enterica serovar Typhimurium strain is attenuated for virulence in the mouse. We conducted microarray experiments comparing wild-type and corA mutant strains to further examine this phenotype. The results suggest that CorA is required at multiple points during the infection process. Given the variety of pathways affected by loss of CorA, we hypothesize that CorA may be part of a broader signaling network within the cell. In addition to corA, mgtA and mgtB are also involved in virulence. After infection by oral gavage, 80% of the mice infected with an mgtA mgtB double mutant survived despite the continued presence of a functional corA allele, compared to 0% of the mice infected with the wild type (D. G. Kehres and M. E. Maguire, unpublished observations). Extracellular Mg2+ is important in terms of directly regulating mgtA and mgtCB expression (10, 46, 47). Extracellular Mg2+ also interacts with the PhoP/PhoQ two-component system, which is essential for virulence (9, 13, 26). Thus, both extracellular Mg2+ and all three Mg2+ transporters of S. enterica serovar Typhimurium are important for virulence, adding additional complexity to the network(s) responsible for Mg2+ homeostasis. The accompanying report focuses on understanding the possible mechanism(s) through which CorA is involved in virulence, with the conclusion that regulation of CorA is important for S. enterica serovar Typhimurium virulence (30). Current experiments are focused on understanding this apparent regulation of CorA expression and function and on delineating its connection to S. enterica serovar Typhimurium virulence.
Supplementary Material
Acknowledgments
K.M.P. was supported by National Institutes of Health T32 training grants AI-07024 and GM08803. These studies were supported by National Institutes of Health grants GM39447 (M.E.M.); AI44486 (F.C.F); AI48622 (S.J.L.); and AI34829, AI057733, and AI052237 (M.M.).
We thank Jun Lin for his intellectual contributions to this research focus. We thank Simon Daefler for the SPI-1 antibodies and Lakshmi Ramachandra for reagents. We thank Kerrie Lodowski and Vasily Sharov for their help analyzing the microarray data.
Footnotes
Published ahead of print on 1 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexander, D. B., and D. A. Zuberer. 1991. Use of chrome azurol S reagents to evaluate siderophore production in rhizosphere bacteria. Biol. Fertil. Soils. 1239-45. [Google Scholar]
- 2.Alpuche Aranda, C. M., J. A. Swanson, W. P. Loomis, and S. I. Miller. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. USA 8910079-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carsiotis, M., D. L. Weinstein, H. Karch, I. A. Holder, and A. D. O'Brien. 1984. Flagella of Salmonella typhimurium are a virulence factor in infected C57BL/6J mice. Infect. Immun. 46814-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamnongpol, S., and E. A. Groisman. 2002. Mg2+ homeostasis and avoidance of metal toxicity. Mol. Microbiol. 44561-571. [DOI] [PubMed] [Google Scholar]
- 5.Coburn, B., Y. Li, D. Owen, B. A. Vallance, and B. B. Finlay. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 733219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster, J. W., and H. K. Hall. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furman, M., A. Fica, M. Saxena, J. L. Di Fabio, and F. C. Cabello. 1994. Salmonella typhi iron uptake mutants are attenuated in mice. Infect. Immun. 624091-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 866383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galán, J. E., and R. Curtiss III. 1989. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb. Pathog. 6433-443. [DOI] [PubMed] [Google Scholar]
- 10.García Véscovi E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84165-174. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-del Portillo, F., J. W. Foster, M. E. Maguire, and B. B. Finlay. 1992. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol. Microbiol. 63289-3297. [DOI] [PubMed] [Google Scholar]
- 12.Gophna, U., M. Barlev, R. Seijffers, T. A. Oelschlager, J. Hacker, and E. Z. Ron. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 692659-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 867077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman, E. A., and H. Ochman. 1993. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 123779-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heithoff, D. M., C. P. Conner, U. Hentschel, F. Govantes, P. C. Hanna, and M. J. Mahan. 1999. Coordinate intracellular expression of Salmonella genes induced during infection. J. Bacteriol. 181799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269400-403. [DOI] [PubMed] [Google Scholar]
- 17.Hmiel, S. P., M. D. Snavely, J. B. Florer, M. E. Maguire, and C. G. Miller. 1989. Magnesium transport in Salmonella typhimurium: genetic characterization and cloning of three magnesium transport loci. J. Bacteriol. 1714742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hmiel, S. P., M. D. Snavely, C. G. Miller, and M. E. Maguire. 1986. Magnesium transport in Salmonella typhimurium: characterization of magnesium influx and cloning of a transport gene. J. Bacteriol. 1681444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291238-239. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, J. S., C. K. Schmitt, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, P. Adams, C. D. O'Connor, and A. D. O'Brien. 2001. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 693021-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 3081-90. [DOI] [PubMed] [Google Scholar]
- 22.Lunin, V. V., E. Dobrovetsky, G. Khutoreskaya, R. Zhang, A. Joachimiak, D. A. Doyle, A. Bochkarev, M. E. Maguire, A. M. Edwards, and C. M. Koth. 2006. Crystal structure of the CorA Mg2+ transporter. Nature 440833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguire, M. E. 1992. MgtA and MgtB: prokaryotic P-type ATPases that mediate Mg2+ influx. J. Bioenerg. Biomembr. 24319-328. [DOI] [PubMed] [Google Scholar]
- 24.Maguire, M. E. 2006. Magnesium transporters: properties, regulation and structure. Front. Biosci. 113149-3163. [DOI] [PubMed] [Google Scholar]
- 25.Månsson, L. E., K. Melican, J. Boekel, R. M. Sandoval, I. Hautefort, G. A. Tanner, B. A. Molitoris, and A. Richter-Dahlfors. 2007. Real-time studies of the progression of bacterial infections and immediate tissue responses in live animals. Cell. Microbiol. 9413-424. [DOI] [PubMed] [Google Scholar]
- 26.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 865054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, S. I., W. S. Pulkkinen, M. E. Selsted, and J. J. Mekalanos. 1990. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium. Infect. Immun. 583706-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 937800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papp, K. M., and M. E. Maguire. 2004. The CorA Mg2+ transporter does not transport Fe2+. J. Bacteriol. 1867653-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papp-Wallace, K. M., and M. E. Maguire. 2008. Regulation of CorA Mg2+ channel function affects the virulence of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1906509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeed, A. I., N. K. Bhagabati, J. C. Braisted, W. Liang, V. Sharov, E. A. Howe, J. Li, M. Thiagarajan, J. A. White, and J. Quackenbush. 2006. TM4 microarray software suite. Methods Enzymol. 411134-193. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt, C. K., S. C. Darnell, and A. D. O'Brien. 1996. The attenuated phenotype of a Salmonella typhimurium flgM mutant is related to expression of FliC flagellin. J. Bacteriol. 1782911-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt, C. K., S. C. Darnell, and A. D. O'Brien. 1996. The Salmonella typhimurium flgM gene, which encodes a negative regulator of flagella synthesis and is involved in virulence, is present and functional in other Salmonella species. FEMS Microbiol. Lett. 135281-285. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 695619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sermon, J., P. De Spiegeleer, K. Vanoirbeek, A. Aertsen, and C. W. Michiels. 2004. Characterization of lactoperoxidase stress response in Escherichia coli and involvement of corA in lactoperoxidase tolerance. Commun. Agric. Appl. Biol. Sci. 6939-42. [PubMed] [Google Scholar]
- 36.Sermon, J., E. M. Wevers, L. Jansen, P. De Spiegeleer, K. Vanoirbeek, A. Aertsen, and C. W. Michiels. 2005. CorA affects tolerance of Escherichia coli and Salmonella enterica serovar Typhimurium to the lactoperoxidase enzyme system but not to other forms of oxidative stress. Appl. Environ. Microbiol. 716515-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 932593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, R. L., J. L. Banks, M. D. Snavely, and M. E. Maguire. 1993. Sequence and topology of the CorA magnesium transport systems of Salmonella typhimurium and Escherichia coli. Identification of a new class of transport protein. J. Biol. Chem. 26814071-14080. [PubMed] [Google Scholar]
- 39.Smith, R. L., M. L. Kaczmarek, L. M. Kucharski, and M. E. Maguire. 1998. Magnesium transport in Salmonella typhimurium: induction of MgtA and MgtCB expression during invasion of epithelial and macrophage cells. Microbiology 1441835-1843. [DOI] [PubMed] [Google Scholar]
- 40.Smith, R. L., M. A. Szegedy, C. Walker, R. M. Wiet, A. Redpath, M. L. Kaczmarek, L. M. Kucharski, and M. E. Maguire. 1998. The CorA magnesium transport protein of Salmonella typhimurium: mutagenesis of conserved residues in the third transmembrane segment identifies part of a Mg2+ pore. J. Biol. Chem. 27328663-28669. [DOI] [PubMed] [Google Scholar]
- 41.Snavely, M. D., J. B. Florer, C. G. Miller, and M. E. Maguire. 1989. Magnesium transport in Salmonella typhimurium: expression of cloned genes for three distinct Mg2+ transport systems. J. Bacteriol. 1714752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snavely, M. D., S. A. Gravina, T. T. Cheung, C. G. Miller, and M. E. Maguire. 1991. Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtB expression. J. Biol. Chem. 266824-829. [PubMed] [Google Scholar]
- 43.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 1786796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stecher, B., S. Hapfelmeier, C. Muller, M. Kremer, T. Stallmach, and W. D. Hardt. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 724138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szegedy, M. A., and M. E. Maguire. 1999. The CorA Mg2+ transport protein of Salmonella typhimurium. Mutagenesis of conserved residues in the second membrane domain. J. Biol. Chem. 27436973-36979. [DOI] [PubMed] [Google Scholar]
- 46.Tao, T., P. F. Grulich, L. M. Kucharski, R. L. Smith, and M. E. Maguire. 1998. Magnesium transport in Salmonella typhimurium: biphasic time and magnesium dependence of the transcription of the mgtA and mgtCB loci. Microbiology 144655-664. [DOI] [PubMed] [Google Scholar]
- 47.Tao, T., M. D. Snavely, S. G. Farr, and M. E. Maguire. 1995. Magnesium transport in Salmonella typhimurium: mgtA encodes a P-type ATPase and is regulated by Mg2+ in a manner similar to that of the mgtB P-type ATPase. J. Bacteriol. 1772654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsolis, R. M., A. J. Baumler, F. Heffron, and I. Stojiljkovic. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 644549-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tükel, C., M. Raffatellu, A. D. Humphries, R. P. Wilson, H. L. Andrews-Polymenis, T. Gull, J. F. Figueiredo, M. H. Wong, K. S. Michelsen, M. Akcelik, L. G. Adams, and A. J. Baumler. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58289-304. [DOI] [PubMed] [Google Scholar]
- 50.Voetsch, A. C., T. J. Van Gilder, F. J. Angulo, M. M. Farley, S. Shallow, R. Marcus, P. R. Cieslak, V. C. Deneen, and R. V. Tauxe. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3)S127-S134. [DOI] [PubMed] [Google Scholar]
- 51.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.