Abstract
It was found that S. meliloti strain SmA818, which is cured of pSymA, could not grow on defined medium containing only formate and bicarbonate as carbon sources. Growth experiments showed that Rm1021 was capable of formate/bicarbonate-dependent growth, suggesting that it was capable of autotrophic-type growth. The annotated genome of S. meliloti Rm1021 contains three formate dehydrogenase genes. A systematic disruption of each of the three formate dehydrogenase genes, as well as the genes encoding determinants of the Calvin-Benson-Bassham, cycle was carried out to determine which of these determinants played a role in growth on this defined medium. The results showed that S. meliloti is capable of formate-dependent autotrophic growth. Formate-dependent autotrophic growth is dependent on the presence of the chromosomally located fdsABCDG operon, as well as the cbb operon carried by pSymB. Growth was also dependent on the presence of either of the two triose-phosphate isomerase genes (tpiA or tpiB) that are found in the genome. In addition, it was found that fdoGHI carried by pSymA encodes a formate dehydrogenase that allows Rm1021 to carry out formate-dependent respiration. Taken together, the data allow us to present a model of how S. meliloti can grow on defined medium containing only formate and bicarbonate as carbon sources.
Sinorhizobium meliloti is a gram-negative rhizobium that is capable of forming nodules on alfalfa, sweet clover, and trigonella. Bacterial infection occurs at the root hairs of the host plant, involving a signal exchange between the rhizobia and the plant (6, 55). Root hair curling encapsulates the bacteria, which subsequently travel through an infection thread and are finally released into plant cells, where they differentiate into bacteroids (24). In this environment, the bacteria reduce atmospheric nitrogen in exchange for nutrients and carbon compounds (37).
From the sequencing of its genome, it has become apparent that S. meliloti retains an array of genes that are capable of encoding enzymes for many metabolic pathways that allow the organism to scavenge energy from many different substrates (7, 23, 25). As a result of the whole-genome sequence data, greater effort has been made to address the functionality of many of the unknown open reading frames. Systems that have utilized the construction of either genetic deletions or genetic fusions of reporter genes have been developed for this purpose (15, 28, 40, 52).
Earlier efforts to uncover functionality associated with the megaplasmids in S. meliloti relied on the creation of precise deletions or plasmid-curing experiments (12, 45). These works showed that a number of carbon-catabolic loci, as well as transport systems, were associated with these plasmids (5, 12, 13, 45). It was also observed that an S. meliloti strain cured of pSymA had a reduced ability to respire formate (I. J. Oresnik, unpublished observation).
Formate is a ubiquitous compound in the environment. Many plants and bacterial species produce and excrete formate into their environments. The high negative reduction potential (−420 mV) of formate allows it to be a ready source of energy. For example, Escherichia coli is able to respire formate when grown under anaerobic conditions in the presence of nitrate as the terminal electron acceptor (18, 30, 31). Its oxidation results in the generation of a proton-motive force and the production of CO2.
It has long been known that members of the Rhizobiaceae capable of forming nodules on legumes have an obligate need for CO2 (27, 38). A variety of carboxylating enzymes needed for anapleurotic pathways have been shown to be present in these organisms (16), yet only S. meliloti and Bradyrhizobium japonicum have been shown to have ribulose bisphosphate carboxylase activity (39). To date, only B. japonicum has been shown to grow chemoautotrophically using H2 gas as a sole source of electrons (27, 36). In contrast, S. meliloti has been shown to be unable to grow chemolithoautotrophically unless it is provided with a cosmid containing the hydrogen uptake genes from B. japonicum (34). S. meliloti, however, is capable of formate-dependent CO2 fixation (39), but the molecular basis for this growth has never been elucidated. More recently, it has been shown that S. meliloti contains the necessary genetic components for the Calvin-Benson-Bassham pathway, as well as three annotated formate dehydrogenases (23, 32).
In this work, we characterize the ability of S. meliloti to grow on a medium containing only formate and bicarbonate as carbon sources. We show that two of the three annotated formate dehydrogenase gene clusters, fdo (consisting of sma0002, sma0005, and sma0007) and fds (consisting of smc02524, smc03086, smc03085, smc04444, smc02525, and smc02524), in S. meliloti affect formate-dependent growth. Moreover, we show that, in addition to the formate dehydrogenase genes, this growth is also dependent upon the cbb genes carried on pSymB.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains, plasmids, and primers are listed in Tables 1 and 2. S. meliloti strains were grown at 30°C in either Luria-Bertani (LB) medium (42), Vincent's minimal medium (VMM) (59), or Rhizobium minimal medium (RMM) (9). Generally, carbon sources were filter sterilized prior to use and were added to defined media to a final concentration of 15 mM. To grow strains in defined medium containing only formate or bicarbonate as carbon sources, the strains were first grown in LB medium supplemented with 15 mM sodium formate and subsequently subcultured into VMM or RMM containing 60 mM sodium formate and 5 mM sodium bicarbonate. This medium was routinely supplemented with biotin and methionine (5 μg ml−1 each), as previously suggested (60); in addition, sodium selenate and sodium molybdate were added to a final concentration of 20 μM. All cultures grown in this manner were streaked for purity. When required, the final concentrations of antibiotics were as follows: tetracycline (Tc), 5 μg ml−1; gentamicin (Gm), 60 μg ml−1; neomycin (Nm), 100 μg ml−1; streptomycin (Sm), 200 μg ml−1; kanamycin (Kan), 40 μg ml−1; ampicillin (Ap), 200 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| Rm1021 | SU47 str-21 Smr | 41 |
| Rm2011 | Smr wild type of RCR2011 (SU47) | 10 |
| SmA818 | Rm2011; pSymA cured | 45 |
| SRmA330 | Rm1021; fdoG::pKnock-Gmr | This work |
| SRmA402 | Rm1021; sma0478::pKanr | This work |
| SRmA411 | Rm1021; fdsA::pKnock-Tcr | This work |
| SRmA442 | Rm1021; cah::pKnock-Gmr | This work |
| SRmA493 | fdsA fdoGHI ΦM12(SRmA411) → SRmA330a Gmr Tcr | This work |
| SRmA495 | sma0478 fdsA ΦM12(SRmA402) → SRmA411 Nmr Tcr | This work |
| SRmA497 | fdoG sma0478 ΦM12(SRmA330) → SRmA402 Gmr Nmr | This work |
| SRmA571 | fdoG sma0478 fdsA ΦM12(SRmA411) → SRmA497 Gmr Nmr Tcr | This work |
| SRmA580 | Rm1021; cbbF1::Kanr | This work |
| E. coli | ||
| MM294A | pro-82 thi-1 hsdR17 supE44 | 22 |
| S17-1 | recA derivative of MM294A with integrated RP4-2 (Tc::Mu::Km::Tn7) | 54 |
| MT607 | MM294A; recA56 | 22 |
| MT616 | MT607(pRK600) | 22 |
| DH5α | λ− φ80dlacZ°M15 °(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 26 |
| DH5αλpir | λpir lysogen of DH5α | 28 |
| Plasmids | ||
| pBluescriptII SK | Cloning vector; ColE1 oriV Apr | Stratagene |
| pRK600 | pRK2013 npt::Tn9 Cmr | 22 |
| pRK415 | Tcr IncP broad-host-range cloning vector | 33 |
| pPH1JI | IncP plasmid; Gmr | 8 |
| pKnock-Tc | Suicide vector for insertional mutagenesis; R6K ori RP4 oriT Tcr | 2 |
| pKnock-Gm | Suicide vector for insertional mutagenesis; R6K ori RP4 oriT Gmr | 2 |
| pKan | pKNOCK-Gmr replaced with Kmr | This work |
| pBP1 | fdoG internal fragment in pKnock-Gm | This work |
| pBP3 | fdsA internal fragment in pBlueScript | This work |
| pBP6 | cbbF fragment in pBlueScript | This work |
| pBP9 | sma0478 internal fragment in pKnock-Gm | This work |
| pBP13 | sma0478 internal fragment in pKan | This work |
| pBP15 | fdsA internal fragment in pKnock-Tc | This work |
| pBP19 | cah internal fragment in pKnock-Gm | This work |
| pBP20 | Kanr cassette cloned into cbbF fragment in pBP6 | This work |
| pBP32 | cbbF::Kanr from pBP20 cloned into pRK415 | This work |
| pMM22 | Kanr fragment cloned as a SmaI fragment into pBlueScript | This work |
SRmA493 was constructed by ΦM12 transduction by transducing Tcr from SRmA411 into SRmA330. The transductants were subsequently screened for Gmr.
TABLE 2.
Primers used in this work
| No. | Primer name | Sequence (5′→3′) |
|---|---|---|
| 1 | fdsAFwd | TGAAGCAGGCGCGTTTCG |
| 2 | fdsARvs | GCTGTGATGGGGCTCGG |
| 3 | fdoGFwd | ATATGAATTCATCATTCACATCGAAGGCG |
| 4 | fdoGRvs | ATATGAATTCAGCAGATTCCAGACACAAC |
| 5 | sma0478Fwd | ATATGAATTCAGGTCGAAGGTTCG |
| 6 | sma0478Rvs | ATATGAATTCATGCAGCGCTGCGGCC |
| 7 | cbbFwd | ATATGGATCCCGGCCTCGTCGTGCAC |
| 8 | cbbRvs | ATATGATATCTGTCAGGCGCGACTCTC |
Genetic manipulations.
Conjugations of S. meliloti and E. coli were carried out as previously described (49). Mutants that were constructed and recombined into the genome of S. meliloti were retransduced into Rm1021 prior to experimental use, as previously described (21).
DNA manipulations, mutant construction, and Southern analysis.
Standard techniques were used for plasmid isolation, restriction enzyme digestion, and agarose gel electrophoresis (51). Mutations were routinely verified by Southern blot analysis, as previously described (14).
The vector pKan was constructed by modification of pKnock-Gm. Briefly, pKnock-Gm, containing an internal EcoRV site in the gentamicin cassette, was restricted with EcoRV and blunt-end cloned with a SmaI kanamycin cassette obtained from pMM22. The final construct was verified by restriction and designated pKan.
To construct a strain carrying a mutation in fdsA (smc04444), a 2,020-bp internal fragment of fdsA was PCR amplified using primers 1 and 2 (Table 2) from Rm1021 genomic DNA, isolated, digested with EcoRI, and cloned into pBluescript II SK, yielding pBP3. pBP3 was digested with KpnI and PstI to yield a 1,023-bp fragment, subsequently cloned into pKnock-Tc (2), and transformed into DH5αλpir, yielding pBP15 (Table 1). pB15 was mobilized into Rm1021 with MT616, and single-crossover recombinants were selected as previously described (49). The resultant strain was verified by Southern blot analysis and named SRmA411 (Table 1).
Similarly, an fdoG (sma0002) mutation was constructed by utilizing primers 3 and 4 (Table 2). The 869-bp fragment generated from these primers was restricted with EcoRI, gel isolated, and cloned into pKnock-Gm, yielding pBP1 (Table 1), which was subsequently conjugated and recombined into Rm1021. The resultant strain was verified by Southern blot analysis and named SRmA330 (Table 1).
A fragment internal to sma0478 was PCR amplified from genomic DNA using primers 5 and 6 (Table 2) to yield a 1,050-bp fragment. This fragment was digested with BamHI/PstI, and the resulting 750-bp product was gel isolated and cloned into pKan to give pBP14. pBP14 was transformed into DH5αλpir, verified by restriction, and conjugated into Rm1021. This construct was conjugated into the wild type, and Smr Nmr single-crossover recombinants were selected. The resultant strain was verified by Southern blot analysis and designated SRmA411.
To construct a cbb mutation, primers 7 and 8 were used to amplify a portion of cbbF. The fragment isolated was restricted with BamHI/EcoRV and cloned into pBlueScript, yielding pBP6. A Kanr cassette was cut as a SmaI fragment from pMM22, gel isolated, and ligated into pBP6, which was digested with StuI, yielding pBP20. The Kanr cassette and the flanking DNA (from cbbF) were removed as a BamHI/EcoRV fragment from pBP20 and recloned into pRK415, yielding pBP32 (Table 1). pBP32 was conjugated into Rm1021, and allelic exchange was carried out using pPH1JI as previously described (46). The resultant strain, SRmA580, was verified by amplifying the insertion junctions and sequencing the PCR products.
Whole-cell CO2 incorporation and formate uptake.
To assay carbon fixation, cells were grown in RMM containing 60 mM sodium formate and 5 mM sodium bicarbonate to an optical density at 600 nm of approximately 0.3. The cells were pelleted, washed twice with phosphate buffer, and finally resuspended in 10 ml of buffer. Incorporation was initiated by the addition of Na214CO3. Aliquots of 1 ml were taken at 0, 2, 5, 10, 15, 30, 45, and 60 min; quenched with perchloric acid (10% [vol/vol] final concentration); and allowed to degas overnight. Acid-stable counts were determined using a Beckman LS 6500 liquid scintillation spectrophotometer and were 3 orders of magnitude above those of dead-cell controls. Linear portions of the data were used to determine fixation rates, and incorporation was normalized to the total cell protein, measured using an enhanced Lowry protein assay (56).
Uptake of formate was carried out essentially as previously described (47). Cells were grown as described above, and assays were initiated with the addition of Na14COOH. Some assays were allowed to continue for over 15 min. Incorporation and normalization of data were as described above.
Native gel formate dehydrogenase assay.
Cells were grown in either complex or defined medium, depending upon the experiment. The preparation of cell extracts was carried out as previously described (44, 47). Native polyacrylamide gel electrophoresis (PAGE) gels were run as previously described (45). The PAGE gels were developed using a p-nitroblue tetrazolium (p-NBT)-based formate dehydrogenase stain (35). Samples were stained with and without both formate and NAD+ to determine the specificities of the activity stains.
Determination of oxygen consumption.
Cells were grown to an optical density at 600 nm of at least 0.20. The cultures were washed twice and resuspended in a phosphate buffer (containing K2HPO4, KH2PO4, and KNO3, pH 7). Oxygen consumption was measured using a Gilson-Clark-type oxygen electrode. Formate-dependent oxygen consumption rates were determined by subtracting the endogenous rates measured prior to the addition of sodium formate from the measured rates following the addition of formate. To ensure that cells were coupled, each assay was concluded by the addition of 2 μM carbonyl cyanide m-chlorophenylhydrazone. Oxygen consumption was normalized to the total cell protein, measured using an enhanced Lowry protein assay (56).
Phylogenetic analysis.
Phylogenetic analysis was carried out essentially as previously described (50). Briefly, sequences were selected on the basis of BLASTP scores (3), aligned using CLUSTAL-X (57), and analyzed using PHYLIP version 3.6a (19, 20). The resulting distance matrix was used to construct a phylogenetic tree and was evaluated using a bootstrap procedure (SEQBOOT; 1,000 replicates).
RESULTS
Formate-dependent chemoautotrophic growth.
The generation of SmA818, a strain cured of the megaplasmid pSymA, allowed the initial identification of a number of pSymA-associated catabolic phenotypes prior to the release of the completed sequence of Rm1021 (45). Many of the putative phenotypes that were observed using BioLog plates, however, were difficult to confirm because of the difficulty in growing the wild-type strain well enough using the identified carbon or nitrogen source. One such observed phenotype was the inability of SmA818 to reduce the indicator dye found within the BioLog plates in the presence of formate as a sole carbon source.
Formate is a one-carbon carboxylic acid. Formate can be oxidized to CO2, and the energy can be used to generate a proton-motive force that in some bacteria can also be used to generate reductant. The resultant CO2 is either reduced or released as a waste product. In other bacteria, formate can be reduced directly and used as a carbon source. The availability of the genomic sequence of Rm1021 provided the opportunity to systematically pursue the observed phenotype of SmA818 grown in the presence of formate. As an initial experiment, we set out to grow Rm1021 in a defined medium with formate as a carbon source.
Wild-type Rm1021 (also Rm2011) and SmA818 were grown on VMM supplemented with 60 mM formate. Since Rm1021 has known deficiencies with respect to biotin and cobalt (60), the medium was supplemented with these, as well as a mixture of trace metals, because many of the enzymes that utilize formate require trace metals (18). It was also possible that formate might be utilized only as an electron source and that the carbon source would be derived from the product of formate oxidation, CO2. To alleviate the possibility that growth would not be observed because of low dissolved CO2 concentrations, bicarbonate was added to increase the amount of dissolved CO2, thus allowing the possibility of autotrophic growth.
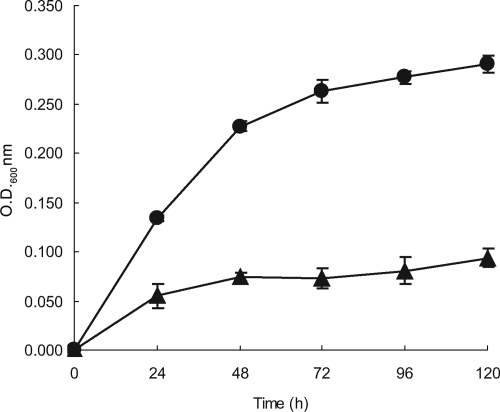
The growth curves clearly showed that Rm1021 does have the capability of growing on a defined medium with formate and bicarbonate as the only sources of carbon (Fig. 1). Growth was dependent upon the presence of bicarbonate, as well as formate. Omission of formate from the medium resulted in Rm1021 growth curves that resembled those of SmA818, characterized by a single doubling following subculture into formate/bicarbonate-containing medium. To determine the effect of reduced levels of bicarbonate, Rm1021 was grown without the addition of bicarbonate to the medium. Growth was greatly decreased and increased only slightly over time compared to the control (data not shown). These results are consistent with the premise that the CO2 concentration within the growth medium slowly increased as formate was oxidized to CO2.
FIG. 1.
Formate-dependent autotrophic growth. Growth curves of S. meliloti strains grown in VMM containing 60 mM formate and 5 mM bicarbonate. Rm1021, •; SmA818, ▴. The data were pooled from four independent experiments; standard deviations are shown. OD600, optical density at 600 nm.
To control for growth that might have been due to carryover of nutrients from the starter culture, Rm1021 was also tested for growth on the base medium without formate or bicarbonate and only the amount of complex medium that would be introduced during subculture. The growth of Rm1021 that occurred under these conditions resembled that of SmA818 (data not shown). Interestingly, if an amount of complex medium equivalent to the amount that was used for the initial subculture was added when wild-type cells reached stationary phase, growth would resume. This suggests that the complex medium must contain some vitamin or nutrient that is not found in our defined medium. The elucidation of this compound(s) was not pursued.
pSymA contains an inducible NAD+ formate dehydrogenase activity.
Nondenaturing protein gels incubated with substrates that lead to the visualization of specific enzyme activities has long been used as a tool in population genetics studies to quantitatively score loci in Rhizobium (17, 53). This technique has also been used to characterize mutations (4, 11, 13, 45). Since the genome of S. meliloti is predicted to contain multiple formate dehydrogenase genes, it was reasoned that this technique would allow us to distinguish the different formate dehydrogenase activities.
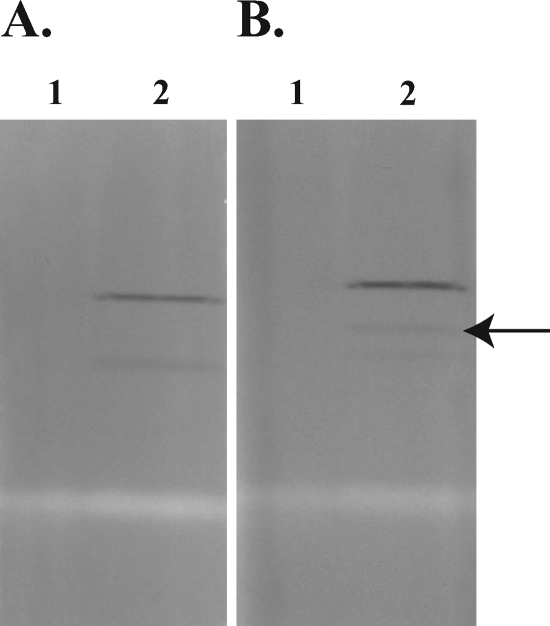
The annotated sequence of fdoGHI suggests that these genes encode a periplasmic formate dehydrogenase complex that is NAD+ independent (7). To test the hypothesis that SmA818 lacked NAD+-independent formate dehydrogenase activity, SmA818 was cultured on defined medium containing formate/bicarbonate. Since SmA818 showed some limited growth in this medium, cells from approximately 2 liters of culture were pooled to provide enough bacterial cells to make cell extracts. Equal amounts of SmA818 and Rm1021 proteins were loaded onto nondenaturing polyacrylamide gels, run, and stained in the presence or absence of NAD+ and formate in an effort to show functionality.
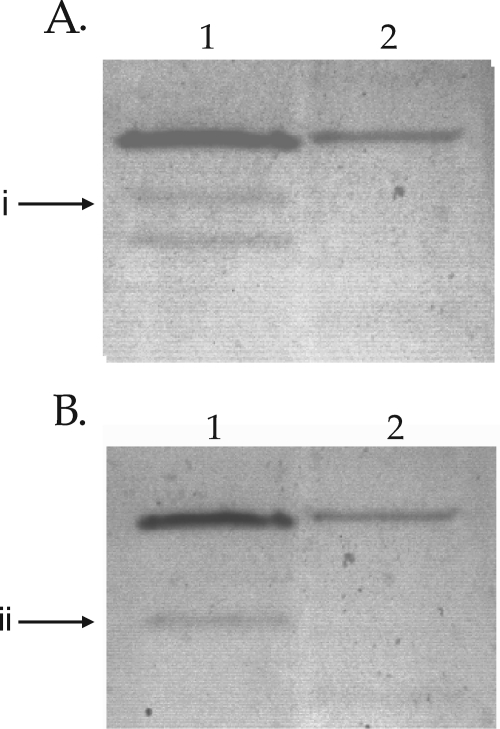
Consistent with the gene annotation, the results showed that Rm1021 contained a formate-dependent, NAD+-independent activity band that was absent in SmA818 (Fig. 2). To determine if this formate-dependent, NAD+-independent activity was constitutive or inducible, extracts from Rm1021 that were grown with either glucose or formate were compared. The presence of this activity band was dependent upon being grown in the presence of formate and was not seen in cell extracts from glucose-grown cells (Fig. 3). Moreover, this activity band was absent in cell extracts from cells grown on complex media, such as LB or tryptone-yeast extract medium, but was present if formate was added to the medium (data not shown). Together, these data suggest that the band of formate dehydrogenase activity being visualized is inducible by the presence of formate. Interestingly, we noted that a second activity band was detectable (Fig. 3). This activity band was not dependent on the presence of either formate or NAD+ and was detectable when the cells were grown in the presence of formate (Fig. 3).
FIG. 2.
Native PAGE formate dehydrogenase activity. S. meliloti strains were grown in defined medium containing 5 mM bicarbonate and 60 mM formate. Gels were stained for dehydrogenase activity with either the absence of formate and the presence of NAD+ (A) or the absence of NAD and the presence of formate (B). Lanes 1, SmA818; lanes 2, Rm1021. The arrow indicates an activity band that is formate dependent and NAD+ independent. Equivalent amounts of protein were loaded in all lanes.
FIG. 3.
Native PAGE gels depicting formate-inducible enzyme activity. Shown are extracts from S. meliloti Rm1021 grown in defined medium containing either 5 mM bicarbonate and 60 mM formate (lanes 1) or 5 mM bicarbonate and 15 mM glucose (lanes 2). (A) Stained in the presence of 30 mM formate and in the absence of NAD+. (B) Stained in the presence of NAD+ and the absence of formate. i, a formate-dependent activity band that was present when the bacteria were grown with formate; ii, an activity band that was dependent upon growth in formate but was not dependent on the presence of either formate or NAD+. Note that extracts from glucose-grown cells contained two times the amount of protein in formate grown cells.
The S. meliloti Rm1021 genome contains genes encoding three probable formate dehydrogenases.
A text search of the descriptions of the annotated open reading frames in Rm1021 revealed that genes for three putative formate dehydrogenases might exist in the S. meliloti genome. Two sets of genes, fdoGHI and sma0478, are carried on the megaplasmid pSymA. The third, fdsABCDG, is located on the chromosome.
FdoGHI consists of three subunits. The alpha subunit (FdoG) contains the active site, and the beta subunit (FdoH) contains Fe-S clusters and is involved in electron transfer from the active-site subunit to the gamma subunit (FdoI), which is an integral membrane protein that can donate electrons to quinones (31). The alpha subunit is also predicted to contain a molybdopterin cofactor, as well as an in-frame opal codon that is used for selenocysteine incorporation into the active site (7). Many of these types of formate dehydrogenase enzymes are targeted to the periplasm (29). We noted that fdoG also contains a putative twin-arginine leader and the only codon in the entire S. meliloti genome for selenocysteine (7). Interestingly, the genes annotated as encoding selenocysteine biosynthesis lie immediately adjacent to the fdoGHI operon (23).
The other formate dehydrogenase gene found on pSymA, sma0478, is annotated as encoding a probable formate dehydrogenase, and a simple BLASTP search revealed that it also appeared moderately similar to hydroxy-acid dehydrogenases. This gene is not found in any other sequenced member of the Rhizobiaceae.
The chromosomal formate dehydrogenase, encoded by fdsABCDG, is conserved among other Rhizobiaceae. It is predicted to contain a molybdopterin cofactor and to be NAD+ dependent and may reside on the cytosolic face of the inner membrane.
Since formate dehydrogenases have not been previously characterized in S. meliloti, insertional mutants were constructed in each of the putative formate dehydrogenases to assess what roles they may have in growing in defined medium containing only formate and bicarbonate as sole carbon sources (Table 1). From these, three double formate dehydrogenase mutants, SRmA493, SRmA495, and SRmA497, containing fdsA/fdoG, sma0478/fdsA, and fdoG/sma0478, respectively, were constructed by transduction. Similarly, SRmA571 was constructed to be mutated for each of the predicted formate dehydrogenase alleles. Because it appeared that Rm1021 was growing autotrophically (Fig. 1), the cbb genes present on pSymB were also mutated so that it could be unambiguously determined whether S. meliloti was growing using the Calvin-Benson cycle.
Growth using formate and bicarbonate is dependent upon the Calvin-Benson cycle.
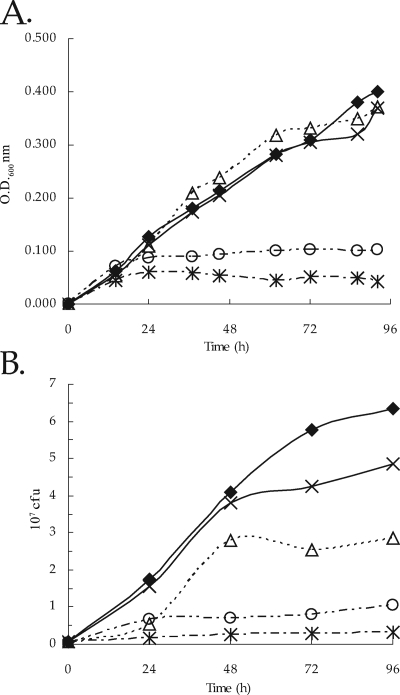
Growth experiments clearly showed that SRmA330 (containing an fdoGHI mutation) and SRmA402 (containing an sma0478 mutation) appeared to grow very similarly to Rm1021 (Fig. 4A). SRmA411 (containing an fdsA mutation), however, did not show any growth on this medium (Fig. 4A). Similarly, SRmA580 (containing a cbbF mutation) did not grow. These data suggest that growth on defined medium containing formate and bicarbonate is dependent upon the Calvin-Benson cycle genes and the chromosomally located fds locus. Surprisingly, a mutation in either the fdoGHI locus or the sma0478 locus did not appear to have the same phenotype as SmA818 (Fig. 4A). Also, strain SRmA497 (containing fdoGHI and sma0478 mutations) did not have the same growth phenotype as SmA818 (data not shown).
FIG. 4.
Growth and viability of S. meliloti in defined medium. (A) Growth curve for S. meliloti strains grown in RMM containing 60 mM formate and 5 mM bicarbonate. (B) Viability of cultures at each time point. The data represent three independent experiments; standard deviations are shown. ⧫, Rm1021; ▵, SRmA330; ×, SRmA402; *, SRmA411; ○, SRmA580. OD600, optical density at 600 nm.
Measurement of growth using optical density assumes a direct correlation between the optical density and cell viability. Since there was a discrepancy between the growth of SmA818 and our working hypothesis that fdoGHI and/or sma0478 would be necessary for growth on this medium, we wished to test whether the culture optical density was correlated with cell viability. To carry this out, aliquots were taken at each time point and plated for viability. The results showed that the viability results appeared to be mostly congruent with those in which optical density was used as a measure of growth (Fig. 4). In most experiments, a strain carrying a mutation in sma0478 was either very close to or indistinguishable from the wild type (Fig. 4B and data not shown). SRmA330 (containing an fdoGHI mutation) consistently showed viable counts that were half of those from Rm1021. Moreover, the colonies of SRmA330 formed more slowly when taken from the defined broth medium and plated onto LB medium. Taken together, these data suggest that fdoGHI does play a role in growth on defined medium containing only formate and bicarbonate as carbon sources but that its role is dispensable (Fig. 4B).
Organisms using the Calvin-Benson cycle reduce CO2 utilizing ribulose 1,5-bisphosphate carboxylase/oxygenase. The cell removes carbon from this cycle as glyceraldehyde 3-phosphate. Glyceraldehyde 3-phosphate is subsequently isomerized by triose phosphate isomerase to dihydroxyacetone phosphate, and a hexose is synthesized via an aldolase reaction, yielding fructose 1,6-bisphosphate. It has recently been shown that S. meliloti contains two triose phosphate isomerases (48). It has been shown that tpiA plays a role in central metabolic metabolism (48), whereas tpiB appears to be necessary for erythritol catabolism in S. meliloti and Rhizobium leguminosarum (48, 61). Although TpiB activity appears to be specifically induced by erythritol, there appears to be enough residual activity from TpiB so that only a strain carrying both a tpiA and a tpiB mutation was unable to grow on carbon sources that necessitated the use of gluconeogenesis (48). It was reasoned that if S. meliloti growth was dependent upon CO2 fixation, strains carrying either a tpiA mutation or a tpiB mutation would be capable of growing on defined medium with formate and bicarbonate as sole carbon sources but a strain carrying both mutations would be unable to grow. This hypothesis was tested, and it was found that strains carrying either tpiA or tpiB grew in a manner similar to the wild type, whereas a double tpiA-tpiB mutant had the same growth characteristics as SRmA580 (cbbF), strongly suggesting that S. meliloti does utilize the Calvin-Benson cycle for growth on defined medium that contains only formate and bicarbonate as carbon sources (data not shown).
fdoGHI and fdsABCDG mutants lack formate dehydrogenase activity.
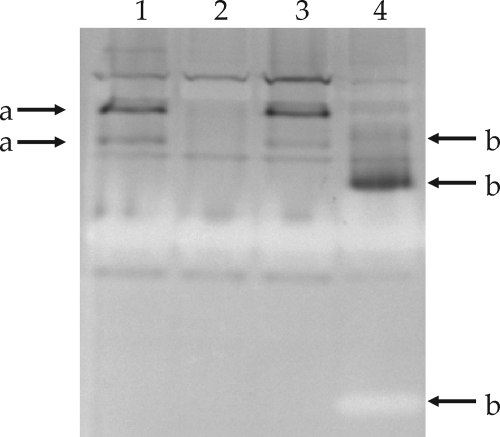
Since it was not possible to grow all the formate dehydrogenase insertional mutants on defined medium with formate and bicarbonate (Fig. 4) but we had evidence that formate dehydrogenase activity was inducible (Fig. 3), and there is evidence for multiple-enzyme complexes having similar enzymatic activities, it was reasoned that the most straightforward method to separate these activities would be to carry out activity stains on induced extracts of the mutants and compare these to the wild type (Fig. 5). The results clearly showed that SRmA330 (fdoG) and SRmA411 (fdsA) zymograms differed from that of Rm1021. In SRmA330, two bands of activity (Fig. 5, bands a) that were NAD+ independent were missing, whereas the pattern yielded by SRmA411 (Fig. 5, bands b) was different and complex, suggesting that the fdsABCDG locus is correlated with formate dehydrogenase activity. The pattern yielded by an extract of SRmA402 (sma0478) did not show any discernible differences from that of the wild type.
FIG. 5.
Native PAGE analysis of formate dehydrogenase mutants. Extracts from S. meliloti strains were grown in LB medium supplemented with 60 mM sodium formate. Gels were stained for dehydrogenase activity in the presence of formate and NAD+. Lane 1, Rm1021; lane 2, SRmA330 (fdoG::pKnock-Gm); lane 3, SRmA402 (sma0478::pKan); lane 4, SRmA411 (fdsA::pKnock-Tc). a, fdoG-dependent activity bands; b, fdsA-dependent activity bands. Equal amounts of protein were loaded in all lanes.
We also noted the appearance of achromatic bands associated with the fdsABCDG mutant (Fig. 5). Since the activity stain works on the principle that electrons from dehydrogenases reduce NBT, forming a blue precipitate at the point of activity, an achromatic band is often associated with enzymes that use reduced NBT as a provider of electrons. It is not clear from our data whether these achromatic bands could be due to the formation of incomplete formate dehydrogenase complexes or whether they might be due to other regulatory changes that might occur in the absence of fdsABCDG. These may lead to the expression of other genes that encode enzymes, such as superoxide dismutases, that are noted for this type of staining pattern (45).
CO2 incorporation activity is reduced in an fdoGHI mutant.
Two carbon sources are available in our defined growth medium, formate and bicarbonate. To try to resolve whether S. meliloti grew by using formate or CO2, Rm1021 was assayed for incorporation of either formate or bicarbonate into an acid-stable product. Incubation of Rm1021 with labeled formate during a 15-min assay resulted in amounts of incorporation that were barely over the level that was found with the dead-cell controls (data not shown). Incubation of Rm1021 with Na14CO3 yielded radiolabel incorporation that showed saturation (Table 3). In contrast, SRmA580 (cbbF) had rates that were less than one-fifth that of the wild type (Table 3). These data show that the majority of the carbon incorporated from our defined medium is dependent upon the presence of the Calvin-Benson cycle.
TABLE 3.
Acid-stable CO2 fixationa
| Strain | Relevant genotype | Rate (nmol/min/mg protein)b | % of wild type |
|---|---|---|---|
| Rm1021 | Wild type | 54.2 | 100 |
| SRmA330 | fdoGHI | 21.3 | 39.4 |
| SRmA402 | sma0487 | 52.2 | 96.3 |
| SRmA411 | fdsABC | ND | ND |
| SRmA580 | cbbF | 9.4 | 17.4 |
The table represents the data from a single experiment; the numbers represent the averages of two independent cultures for each strain. The experiment was repeated three times, showing consistent results.
The rates are directly compared to that of Rm1021. ND, not determined.
To determine if the products encoded by either fdoGHI or sma0478 play roles in carbon fixation, whole cells carrying these mutations were grown with formate and bicarbonate and were also assayed for 14CO2 incorporation. Whereas SRmA402 incorporation rates were identical to those of the wild type, the rates of CO2 incorporation in SRmA330 were consistently less than half those of the wild type. This is consistent with the hypothesis that fdoGHI does indeed play a role in formate-dependent autotrophic growth but that its presence is not essential (Table 3).
Formate-dependent oxygen consumption is dependent on fdoGHI.
Based on the annotation of fdoGHI, as well as simple bioinformatic searches, it was predicted that FdoGHI should form a complex that is capable of oxidizing formate and donating the electrons directly to the ubiquinol and subsequently to a terminal oxidase, leading to formate-dependent oxygen consumption. To directly test the functionality of FdoGHI, it was hypothesized that Rm1021 should be capable of formate-dependent oxygen reduction, whereas an fdoGHI mutant should be impaired in this ability.
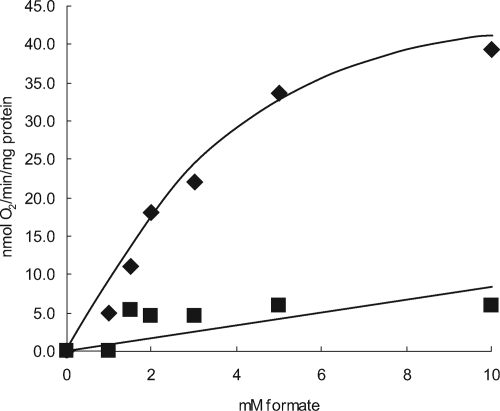
To verify this, cells were grown in minimal media containing formate and bicarbonate as their sole carbon sources and tested for formate-dependent oxygen consumption. Experiments carried out with the wild type showed that S. meliloti did display formate-dependent respiration, and the whole-cell kanamycin level for formate was approximately 2.4 mM (Fig. 6). It was found that oxygen consumption was completely abolished in a strain carrying an fdoGHI mutation (Fig. 6). Immediately downstream of fdoGHI, and presumably within the same operon, are the genes fdhE, selA, and selB. Whereas SelA and SelB are likely involved in the biosynthesis of the selenocysteine necessary for FdoG activity (7), FdhE has been identified as a maturation protein likely involved in FdoGHI maturation (58). Interestingly, unlike E. coli, in which an fdhE mutation completely blocks formate dehydrogenase activity (1), a strain carrying a deletion of the fdhE open reading frame so that transcription of selAB was not affected still showed formate-dependent oxygen consumption, albeit at reduced rates (B. S. Pickering and I. J. Oresnik, unpublished data). These results suggest that the FdoGHI complex does play a direct role in oxidizing formate and that the electrons are donated to the electron transport chain and play a role in generating a proton-motive force across the cell membrane.
FIG. 6.
Oxygen consumption is FdoG dependent. Cells were grown in RMM containing 60 mM formate and 5 mM bicarbonate. ⧫, Rm1021; ▪, SRmA330 (fdoG). Note that endogenous respiration rates were subtracted from both the wild type and SRmA330.
fdoGHI is not found in other Rhizobium species.
A BLASTP search revealed that the presence of fdoGHI is not common within the alphaproteobacteria, whereas the fdsABCDG genes are found in every member of the Rhizobiaceae that has been sequenced. To better assess fdoGHI, a phylogeny of FdoG was carried out with the top BLAST hits. The results clearly showed that FdoG is found in only four other alphaproteobacteria: Xanthobacter autotrophicus, Azorhizobium caulinodans, Ocrhobactrum anthropi, and Paracoccus denitrificans. The FdoG found in S. meliloti is most closely related to that of the alphaproteobacterial species P. denitrificans. Interestingly, the genes fdoGHI are usually associated with either beta- or gammaproteobacterial species that are usually facultative anaerobes. The phylogenetic data strongly supports the idea that fdoGHI were acquired by a lateral gene transfer event from either a beta- or gammaproteobacterium. Strikingly, the genes for fdoGHI, as well as the associated genes for selenocysteine biosynthesis, are not found in the closely related species Sinorhizobium medicae. It may be noteworthy that S. medicae strain WSM419 is capable of formate-dependent autotrophic growth (data not shown).
DISCUSSION
The basic premise for carrying out this work was to investigate which of the formate dehydrogenase genes on pSymA were responsible for a delayed formate respiration phenotype that was observed with a strain that was devoid of pSymA. It was reasoned that this information would provide functional evidence that would increase our physiological understanding of the sequenced S. meliloti strain Rm1021. The data presented show that Rm1021 can grow on a formate bicarbonate defined medium (Fig. 1). Moreover, we have shown that this growth is not solely dependent upon the presence of pSymA but on genetic determinants that are found on both the chromosome and pSymB (Table 3 and Fig. 4A).
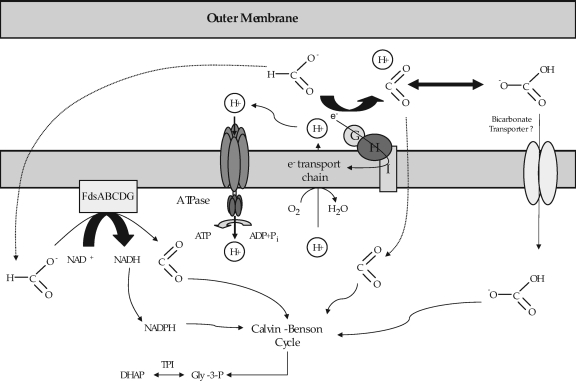
Taken together, the data provide sufficient evidence to allow us to construct a model of how S. meliloti can grow by utilizing only formate and bicarbonate (Fig. 7). Uptake assays clearly showed that the rates of formate uptake are consistent with this compound entering into the cell by diffusion and that very little carbon is assimilated into cell material (data not shown). In contrast, labeling experiments with bicarbonate showed that carbon incorporation occurs rapidly and is dependent upon the cbb operon found on pSymB (Table 3). In addition to the genes found on pSymB, growth was also dependent upon the presence of tpiA and tpiB. This observation is consistent with our understanding of how carbon is taken from the Calvin-Benson-Bassham cycle and utilized for gluconeogenesis. These data are all consistent with the hypothesis that the carbon source for growth on our defined formate-bicarbonate medium is CO2. In some organisms, the uptake of bicarbonate has been shown to be an active process (43). It may be a noteworthy observation that an ABC transporter is found in close proximity to the cbb operon on pSymB.
FIG. 7.
Model for formate-dependent autotrophic growth in S. meliloti Rm1021. See the text for details.
Whereas SmA818 was unable to grow on our defined medium, growth experiments with strains carrying defined mutations in each of the annotated formate dehydrogenase genes clearly showed that fdsABCDG is essential for growth on our formate-bicarbonate defined medium whereas fdoGHI plays a nonessential role (Fig. 4 and Table 3). It was shown, however, that FdoGHI is responsible for formate-dependent oxygen consumption (Fig. 6). Although it is theoretically possible to produce reductant directly from formate using FdoGHI, our data suggest that the primary source for reductant using our growth conditions must be provided by FdsABCDG, since any strains that carried a mutation in fdsA did not grow on our defined medium (Fig. 4). FdoGHI must therefore contribute to the overall energetics of the cell by contributing to the proton-motive force (Fig. 6). These data are also consistent with the decreased rates of CO2 fixation and lower viability observed in SRmA330 (Fig. 4B and Table 3). We noted that S. medicae strain WSM419 was also capable of formate-dependent autotrophic growth, although it does not carry fdoGHI and the associated genes for the biosynthesis of selenocysteine (data not shown).
Clearly there are determinants on pSymA that affect growth in a defined medium containing only formate and bicarbonate. We noted that a carbonic anhydrase (cah; sma0045) gene is also found in close proximity to fdoGHI. Defined mutations of this gene did not affect rates of carbon fixation (data not shown). Also, we were unable to ascribe any function that was related to formate utilization to sma0478. It is currently unclear whether the inability of SmA818 to grow in this defined medium is a direct affect of a single locus on pSymA or if it is an effect of a number of independent loci that cumulatively give rise to the inability to grow. These questions are currently unresolved.
The presence of fdoGHI in S. meliloti is curious. To date, this complex has not been found in any other sequenced member of the genus Rhizobium and very few other alphaproteobacteria. As might be expected, all of the FdoG proteins within the alphaproteobacteria appear to be from a distinct branch on our phylogenetic tree (data not shown). With a greater number of the Rhizobiaceae being sequenced, it will be of interest to determine what other members have this complex as well as the associated selenocysteine biosynthesis genes. The fact that these genes give rise to functional proteins that can affect the physiology of S. meliloti suggests that there must be some selective advantage to maintaining the genes. It may be noteworthy that all of our assays and growth were carried out aerobically, suggesting that FdoGHI also functions aerobically, in contrast to what is often found in many enteric bacteria.
It was previously thought that the presence of two formate dehydrogenase genes on pSymA might indicate that formate respiration was important within the nodule environment (7). Testing our formate dehydrogenase mutants carrying either a single, double, or triple mutation in any of the dehydrogenase genes did not show a visual symbiotic defect (data not shown). It may be that this region plays a role in allowing the bacteria to stay competitive within the rhizosphere, for example, by allowing them to withstand varying environmental conditions, such as water-saturated soils, which might lead to brief periods of hypoxia that would favor the accumulation of formate due to metabolic products from other organisms. Hypotheses such as these have not yet been addressed.
Our work on formate utilization and the demonstration of formate-dependent autotrophic growth has revealed an area of metabolism and physiology that had not been previously recognized in S. meliloti at the molecular level. The questions that this work raises concerning the regulation of these determinants, as well the physiological roles they play in the rhizosphere, are unanswered. Ongoing work in these areas should provide answers that will allow us to better understand the physiology of the agronomically important soil bacterium S. meliloti.
Acknowledgments
We thank R. Sparling for advice concerning medium formulation and for the kind gifts of labeled bicarbonate and formate. We also thank Michelle Sille for the construction of pMM22.
This work was funded by an NSERC Discovery grant to I.J.O.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Abaibou, H., J. Pommier, S. Benoit, G. Giordano, and M. A. Mandrand-Berthelot. 1995. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for the aerobic formate dehydrogenase. J. Bacteriol. 1777141-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques. 26824-828. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldani, J. I., R. W. Weaver, M. F. Hynes, and B. D. Eardly. 1992. Utilization of carbon substrates, electrophoretic enzyme patterns, and symbiotic performance of plasmid-cured rhizobia. Appl. Environ. Microbiol. 582308-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardin, S., S. Dan, M. Osteras, and T. M. Finan. 1996. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 1734540-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, M. J., and R. F. Fisher. 2006. Global gene expression in the rhizobial-legume symbiosis. Symbiosis 421-24. [Google Scholar]
- 7.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 989883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beringer, J. E., J. L. Beynon, A. V. Buchanan-Wollason, and A. W. B. Johnston. 1978. Transfer of the drug resistance transposon Tn5 to Rhizobium. Nature 276633-634. [Google Scholar]
- 9.Broughton, W. J., C. H. Wong, U. Lewin, U. Samrey, H. Myint, H. Meyer, D. N. Dowling, and R. Simon. 1986. Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J. Cell Biol. 1021173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casse, F., C. Boucher, J. S. Huillot, M. Michel, and J. Dénarié. 1979. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J. Bacteriol. 113229-242. [Google Scholar]
- 11.Charles, T. C., G. Q. Cai, and P. Aneja. 1997. Megaplasmid and chromosomal loci for the PHB degradation pathway in Rhizobium (Sinorhizobium) meliloti. Genetics 1461211-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles, T. C., and T. M. Finan. 1991. Analysis of a 1600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics 1275-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles, T. C., R. S. Singh, and T. M. Finan. 1990. Lactose utilization and enzymes encoded in Rhizobium meliloti: implications for population studies. J. Gen. Microbiol. 1362497-2502. [Google Scholar]
- 14.Clark, S. R. D., I. J. Oresnik, and M. F. Hynes. 2001. RpoN of Rhizobium leguminosarum bv. viciae strain VF39SM plays a central role in FnrN-dependent microaerobic regulation of genes involved in nitrogen fixation. Mol. Gen. Genet. 264623-633. [DOI] [PubMed] [Google Scholar]
- 15.Cowie, A., J. Cheng, C. D. Sibley, Y. Fong, R. Zaheer, C. L. Patten, R. M. Morton, B. Golding, and T. M. Finan. 2006. An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti. Appl. Environ. Microbiol. 727156-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, M. F. 1998. Tricarboxylic acid cycle and anapleurotic enzymes in rhizobia. Fems Microbiol. Rev. 22105-123. [DOI] [PubMed] [Google Scholar]
- 17.Eardly, B. D., L. A. Materon, N. H. Smith, D. A. Johnson, M. D. Rumbaugh, and R. K. Selander. 1990. Genetic structure of natural population of the nitrogen-fixing bacterium Rhizobium meliloti. Appl. Environ. Microbiol. 56187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enoch, H. G., and R. L. Lester. 1975. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J. Biol. Chem. 2506693-6705. [PubMed] [Google Scholar]
- 19.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-789. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 2002. PHYLIP (Phylogeny Inference Package) version 3.6a. Department of Genetics, University of Washington, Seattle.
- 21.Finan, T. M., E. Hartwieg, K. Lemieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finan, T. M., B. Kunkel, G. F. de Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 16766-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernández-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Pühler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont S. meliloti. Proc. Natl. Acad. Sci. USA 989889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dréano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernández-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F. J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293668-672. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-570. [DOI] [PubMed] [Google Scholar]
- 27.Hanus, F. J., R. J. Maier, and H. J. Evans. 1979. Autotrophic growth of H2-uptake-positive strains of Rhizobium japonicum in an atmosphere supplied with hydrogen gas. Proc. Natl. Acad. Sci. USA 761788-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.House, B. L., M. W. Mortimer, and M. L. Kahn. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 702806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jormakka, M., B. Byrne, and S. Iwata. 2003. Formate dehydrogenase—a versatile enzyme in changing environments. Curr. Opin. Struct. Biol. 13418-423. [DOI] [PubMed] [Google Scholar]
- 30.Jormakka, M., B. Byrne, and S. Iwata. 2003. Protonmotive force generation by a redox loop mechanism. FEBS Lett. 54525-30. [DOI] [PubMed] [Google Scholar]
- 31.Jormakka, M., S. Toronoth, B. Byrne, and S. Iwata. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 2951863-1868. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9189-197. [DOI] [PubMed] [Google Scholar]
- 33.Keen, N. T., S. Tamaski, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host range plasmids for DNA cloning in gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 34.Lambert, G. R., M. A. Cantrell, F. J. Hanus, S. A. Russell, K. R. Hadad, and H. J. Evans. 1985. Intra- and interspecies transfer and expression of Rhizobium japonicum hydrogen uptake genes and autotrophic growth capability. Proc. Natl. Acad. Sci. USA 823232-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latner, A. L., and A. W. Skillen. 1968. Isoenzymes in biology and medicine. Academic Press, New York, NY.
- 36.Lepo, J. E., F. J. Hanus, and H. J. Evans. 1980. Chemoautotrophic growth of hydrogen-uptake-positive strains of Rhizobium japonicum. J. Bacteriol. 141664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodwig, E., and P. Poole. 2003. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 2237-78. [Google Scholar]
- 38.Lowe, R. H., and H. J. Evans. 1962. Carbon dioxide requirement for growth of legume nodule bacteria. Soil Sci. 94351-356. [Google Scholar]
- 39.Manian, S. S., and F. O'Gara. 1982. Derepression of ribulose bisphosphate carboxylase activity in Rhizobium meliloti. FEMS Microbiol. Lett. 1495-99. [Google Scholar]
- 40.Mauchline, T. H., J. E. Fowler, A. K. East, A. L. Sartor, A. H. F. Hosie, P. S. Poole, and T. M. Finan. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. USA 10317933-17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meade, H. M., S. R. Long, G. B. Ruvkin, S. E. Brown, and F. M. R. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Omata, T., G. D. Price, M. R. Badger, M. Okamura, S. Gohta, and T. Ogawa. 1999. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechoccus sp. strain PCC 7942. Proc. Natl. Acad. Sci. USA 9613571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oresnik, I. J., and D. B. Layzell. 1994. Composition and distribution of adenylates in soybean (Glycine max L.) nodule tissue. Plant Physiol. 104217-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oresnik, I. J., S. L. Liu, C. K. Yost, and M. F. Hynes. 2000. Megaplasmid pRme2011a of Sinorhizobium meliloti is not required for viability. J. Bacteriol. 1823582-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oresnik, I. J., L. A. Pacarynuk, S. A. P. O'Brien, C. K. Yost, and M. F. Hynes. 1998. Plasmid encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant-Microbe Interact. 111175-1185. [Google Scholar]
- 47.Poysti, N. J., E. D. Loewen, Z. Wang, and I. J. Oresnik. 2007. Sinorhizobium meliloti pSymB carries genes necessary for arabinose transport and catabolism. Microbiology 153727-736. [DOI] [PubMed] [Google Scholar]
- 48.Poysti, N. J., and I. J. Oresnik. 2007. Characterization of Sinorhizobium meliloti triose phosphate isomerase genes. J. Bacteriol. 1893445-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson, J. S., M. F. Hynes, and I. J. Oresnik. 2004. A genetic locus necessary for rhamnose uptake and catabolism in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 1868433-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson, J. S., and I. J. Oresnik. 2007. l-Rhamnose transport in Rhizobium leguminosarum is dependent upon RhaK, a sugar kinase. J. Bacteriol. 1898437-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Schroeder, B. K., B. L. House, M. W. Mortimer, S. N. Yurgel, S. C. Maloney, K. L. Ward, and M. L. Kahn. 2005. Development of a functional genomics platform for Sinorhizobium meliloti: construction of an ORFeome. Appl. Environ. Microbiol. 715858-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo engineering: transposon mutagenesis in gram-negative bacteria. BioTechniques 1784-791. [Google Scholar]
- 55.Spaink, H. P. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54257-288. [DOI] [PubMed] [Google Scholar]
- 56.Stoschek, C. M. 1990. Quantitation of protein. Methods Enzymol. 18250-68. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner, R. J., A. L. Papish, and F. Sargent. 2004. Sequence analysis of bacterial redox enzyme maturation proteins (REMPs). Can. J. Microbiol. 50225-238. [DOI] [PubMed] [Google Scholar]
- 59.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. Blackwell Scientific, Oxford, United Kingdom.
- 60.Watson, R. J., R. Heys, T. Martin, and M. Savard. 2001. Sinorhizobium meliloti cells require biotin and either cobalt or methionine for growth. Appl. Environ. Microbiol. 673767-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yost, C. K., A. M. Rath, T. C. Noel, and M. F. Hynes. 2006. Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 1522061-2074. [DOI] [PubMed] [Google Scholar]