Abstract
p-Cresol methylhydroxylases (PCMH) from aerobic and facultatively anaerobic bacteria are soluble, periplasmic flavocytochromes that catalyze the first step in biological p-cresol degradation, the hydroxylation of the substrate with water. Recent results suggested that p-cresol degradation in the strictly anaerobic Geobacter metallireducens involves a tightly membrane-bound PCMH complex. In this work, the soluble components of this complex were purified and characterized. The data obtained suggest a molecular mass of 124 ± 15 kDa and a unique αα′β2 subunit composition, with α and α′ representing isoforms of the flavin adenine dinucleotide (FAD)-containing subunit and β representing a c-type cytochrome. Fluorescence and mass spectrometric analysis suggested that one FAD was covalently linked to Tyr394 of the α subunit. In contrast, the α′ subunit did not contain any FAD cofactor and is therefore considered to be catalytically inactive. The UV/visible spectrum was typical for a flavocytochrome with two heme c cofactors and one FAD cofactor. p-Cresol reduced the FAD but only one of the two heme cofactors. PCMH catalyzed both the hydroxylation of p-cresol to p-hydroxybenzyl alcohol and the subsequent oxidation of the latter to p-hydroxybenzaldehyde in the presence of artificial electron acceptors. The very low Km values (1.7 and 2.7 μM, respectively) suggest that the in vivo function of PCMH is to oxidize both p-cresol and p-hydroxybenzyl alcohol. The latter was a mixed inhibitor of p-cresol oxidation, with inhibition constants of a Kic (competitive inhibition) value of 18 ± 9 μM and a Kiu (uncompetitive inhibition) value of 235 ± 20 μM. A putative functional model for an unusual PCMH enzyme is presented.
Monomethylphenols (cresols) are produced in large amounts by petrochemical processes as constituents of resins, solvents, disinfectants, and wood-preserving chemicals but also by anaerobic tyrosine fermentation via p-hydroxyphenylacetate (37). Cresols represent toxic pollutants and are therefore of environmental concern. Biological degradation of p-cresol (4-methylphenol) has been described for both aerobic and anaerobic bacteria (2, 8, 15, 23, 25, 29, 32, 34).
In aerobic and denitrifying bacteria, the initial reaction of p-cresol degradation is the hydroxylation of the methyl group with water, yielding p-hydroxybenzyl alcohol; cytochrome c or azurin serves as the electron acceptor (see references above). The reaction is catalyzed by a periplasmic, soluble p-cresol methylhydroxylase (PCMH) which was initially purified from and characterized for the aerobic bacterium Pseudomonas putida (17). All PCMH enzymes characterized so far are flavocytochromes of a common α2β2 composition. The α subunit contains a flavin adenine dinucleotide (FAD) cofactor, which is covalently linked to a conserved tyrosine residue (26, 27); the β subunit represents a c-type cytochrome (21, 28). The crystal structure of PCMH has been solved, and many details about the structure/function relationship have been elucidated in the last 2 decades (some selected references include 6, 7, 12, and 21). The further reactions involved in p-cresol catabolism comprise two oxidation steps yielding 4-hydroxybenzoate; both are suggested to be catalyzed in vivo by soluble NAD(P)+-dependent alcohol and aldehyde dehydrogenases (5, 19). In aerobic organisms, 4-hydroxybenzoate is further metabolized by oxygenases (13). In contrast, facultative anaerobes activate the aromatic carboxylic acid first to 4-hydroxybenzoyl-coenzyme A by an ATP-dependent ligase (1, 14), after which the hydroxy functionality is removed by reduction catalyzed by the Mo cofactor containing 4-hydroxybenzoyl-coenzyme A reductase (3, 36).
In sulfate-reducing bacteria, the terminal electron acceptors are considered to be too negative to accept electrons from p-cresol oxidation (29, 35). For this reason, it was surprising that a sulfate-reducing bacterium was reported to metabolize p-cresol by means of methyl group hydroxylation (23). In contrast, the sulfate-reducing Desulfobacterium cetonicum initiates p-cresol degradation by an alternative reaction, the addition of the methyl group to fumarate, yielding p-hydroxybenzylsuccinate (29). This reaction is analogous to the first step in toluene degradation and is suggested to be catalyzed by a glycyl radical enzyme.
In a recent study, the related Fe(III)-reducing deltaproteobacterium Geobacter metallireducens was shown to use the p-cresol degradation pathway of aerobes and facultative anaerobes (31). A proteomic approach revealed a p-cresol-induced gene cluster in G. metallireducens (pcmA to pcmX; p-cresol metabolism) comprising genes with a high level of similarity to those for PCMH enzymes from other bacteria.
The initial analysis of p-cresol metabolism of G. metallireducens revealed some unprecedented features, which are briefly summarized here (25, 31). (i) Both the p-cresol and p-hydroxybenzyl alcohol-oxidizing activities were fully located in the membrane protein fraction. No additional NAD(P)+-dependent p-hydroxybenzyl alcohol dehydrogenase activity was present in soluble cell extracts, indicating that p-cresol and p-hydroxybenzyl alcohol oxidation are both catalyzed by membrane-bound PCMH in vivo. (ii) Analysis of the genes putatively coding for PCMH (pcmG, pcmI, pcmJ) suggested an αα′β2 rather than an α2β2 subunit composition. The pcmI (gi 78194587) and pcmJ (gi 78194588) genes were proposed to code for two isoforms of the FAD-containing subunit, referred to as the α and α′ subunits and showing 62% amino acid sequence identity to each other. Notably, the α′ subunit lacks the conserved tyrosine residue (Tyr384 in the α subunit of PCMH from P. putida and Tyr394 in the α subunit from G. metallireducens) which is involved in covalent FAD attachment (26, 27). The β subunit, a c-type cytochrome, is suggested to be encoded by pcmG (gi 78194585), whereas the function of the pcmH open reading frame (gi 78194586), which shows no significant similarities to other genes, remains unknown. (iii) The p-cresol-induced pcmCDEF genes putatively code for a cytochrome bc membrane complex. An association of PCMH with this complex was suggested, and such an association would explain why the PCMH activity was found completely within the membrane fraction. An electron transfer from p-cresol or, thermodynamically more conceivably, from p-hydroxybenzyl alcohol to menaquinone mediated by an unorthodox cytochrome bc complex was discussed (31).
In this work, the soluble components of the membrane-bound PCMH complex were separated from the cytoplasmic membrane in an active form, purified, and characterized. The results obtained shed light on the function of an unusual PCMH enzyme from an obligately anaerobic model organism.
MATERIALS AND METHODS
Growth of G. metallireducens and preparation of cell extracts.
G. metallireducens (DSMZ no. 7210; Deutsche Sammlung von Mikroorganismen und Zellkulturen) was grown anaerobically in a 200-liter fermenter in a mineral salt medium containing 3 mM p-cresol and 30 mM Fe(III)-citrate as carbon and energy sources as described previously (31). Cell extracts were prepared by resuspending 12 g of frozen cells (wet mass) in 24 ml of buffer (50 mM Tris, pH 6.8, containing 2.5 mM MgCl2, 0.1 mg DNase I, and 1 mM phenylmethylsulfonyl fluoride). After suspension, cells were disrupted by passage through a French pressure cell at 137 MPa. Membrane fractions were obtained by centrifugation at 100,000 × g (1 h, 4°C).
Enzyme assays. (i) Discontinuous HPLC assay.
p-Cresol and/or p-hydroxybenzyl alcohol consumption and product formation were monitored by high-performance liquid chromatography (HPLC). The standard assay (350 μl) contained 0.2 mM phenazine methosulfate, 0.2 mM p-cresol, and 500 mM NaCl in 50 mM Tris, pH 8.0. The reaction was started by adding 5 to 50 μl of crude extract or membrane fraction. Fifty-microliter aliquots were taken at 0, 1, 3, 6, and 12 min and added to 7 μl of 10% formic acid (vol/vol). Protein was pelleted by centrifugation at 4°C (15 min at 15,000 × g), and 40 μl of the supernatant was subjected to HPLC analysis. Separation was accomplished on a LiChrospher RP-C18e column (125 by 4.6 mm; Wicom, Heppenheim, Germany) at a flow rate of 1 ml min−1 (Agilent 1200 series HPLC system, Agilent 1200 series UV detector). p-Cresol, p-hydroxybenzaldehyde, and p-hydroxybenzyl alcohol all eluted in a linear gradient from 10 to 40% methanol in 40 mM formic acid within 20 min, at 19.2, 10.3, and 5.0 min, respectively. Detection was at 275 nm, and the amounts of substrates and products were calculated by comparing the peak area integrals to those from calibration curves of p-cresol, p-hydroxybenzaldehyde, and p-hydroxybenzyl alcohol standards.
(ii) Spectrophotometric assay.
The p-cresol oxidation activity assay described by Hopper and Taylor (17) was not applicable for determining PCMH activity for enriched protein fractions of G. metallireducens. Instead, an alternative test system using 200 μM ferrocenium hexafluorophosphate (ferrocenium) as the electron acceptor (ɛ300 = 3,600 M−1 cm−1; our own determination) in 350 μl of 50 mM Tris, 500 mM NaCl (pH 8.0) at 30°C was established. The reaction was started by the addition of 0.2 mM p-cresol. The assay allowed the determination of the initial rate of p-cresol oxidation, but it was not applicable for monitoring the oxidation of p-hydroxybenzyl alcohol to p-hydroxybenzaldehyde due to the absorbance of the latter at 300 nm. The oxidation of p-hydroxybenzyl alcohol was determined by directly monitoring p-hydroxybenzaldehyde formation (ɛ330 = 22,100 M−1 cm−1; our own determination) by use of the same assay mixture as for the p-cresol oxidation.
Purification of PCMH. (i) Membrane-washing steps.
After cell disruption and centrifugation at 100,000 × g (1 h, 4°C), a 4.9-g pellet from the first centrifugation step was homogenized in 10 ml of 50 mM Tris, pH 8.0 (referred to as basal buffer), by use of a Potter homogenizer on ice for 10 min and centrifuged again (100,000 × g, 1 h, 4°C). The following detergents were used for solubilization experiments (incubation times were from 30 min to 4 h on ice and at 26°C): dodecyl-β-d-maltoside (0.5 to 2% [wt/vol]), Triton X-100 (1% and 3%), Big-Chap (2 to 8 mM), and lauryldimethylamine N-oxide (LDAO; 0.05 to 2% [wt/vol]). As no solubilization was achieved, the resulting pellet (5.1 g) was suspended after ultracentrifugation in 10.2 ml of basal buffer containing LDAO (1% [wt/vol] final concentration), homogenized, and centrifuged as described above.
(ii) Separation of PCMH from the membrane.
An LDAO-treated pellet (3.5 g) was suspended in 7 ml of basal buffer containing 500 mM NaCl, homogenized, and centrifuged as before. The NaCl treatment was repeated, and the supernatants obtained (7 ml and 5.8 ml) were pooled and concentrated to 3 ml by use of Vivaspin concentrators with a 10-kDa molecular mass cutoff (Sartorius AG, Göttingen, Germany).
(iii) Gel filtration.
The NaCl pool was filtered and applied to a Superdex 200 prep-grade column (XK16/60, 120 ml; GE Healthcare, Munich, Germany) in three runs with a 1-ml sample volume in 20 mM Tris, 150 mM KCl, pH 8.0 (1 ml min−1). PCMH eluted between 65 and 90 ml. The activity-containing fractions were pooled and concentrated to 6 ml as described above.
(iv) Hydrophobic interaction chromatography.
Solid ammonium sulfate powder (1.75 g) was added to the pooled fractions from gel filtration (50% saturation) and incubated on ice for 30 min under soft shaking. Precipitated protein was pelleted by centrifugation (3,000 × g, 10 min, 4°C). Seven milliliters of the clear supernatant was applied to an Octyl Sepharose column (XK16, 10 ml; GE Healthcare) equilibrated in basal buffer containing 2 M ammonium sulfate at 2 ml min−1 in two independent runs. PCMH eluted in a linear gradient from 2 M ammonium sulfate to 150 mM KCl in basal buffer containing between 400 and 0 mM ammonium sulfate. The activity-containing fractions were pooled, concentrated to a final volume of 2 ml as described above, and stored at −20°C for several months without loss of activity.
Molecular mass determination.
The native molecular mass of PCMH was determined by gel filtration on a Superdex 200 prep-grade column (XK16/60, 120 ml; GE Healthcare, Munich, Germany). A calibration curve with lysozyme (14.4 kDa), carboanhydrase (29 kDa), ovalbumin (43 kDa), bovine serum albumin (67 kDa), catalase (250 kDa), and apoferritin (443 kDa) as standards was performed. The run conditions were as described for the purification protocol.
Gel electrophoresis and fluorescence detection of enzyme-bound FAD.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% and 16.5% acrylamide) was performed as described previously (22). The fluorescence of FAD was detected on an 8% polyacrylamide gel by use of an Ettan Dige imager (Cy2 filter, 0.4 s, excitation wavelength of 488 nm, detection wavelength of 520 nm; GE Healthcare). After fluorescence scanning, proteins were visualized by use of Coomassie blue staining (38).
FAD determination.
For the extraction of noncovalently bound FAD, 200 μl of PCMH (0.1 mg ml−1) was precipitated by the addition of 20 μl of 55% (wt/vol) trichloroacetic acid followed by incubation on ice for 10 min. The protein pellet obtained after centrifugation (15,000 × g, 5 min) was washed twice with 100 μl 1% trichloroacetic acid (wt/vol), and 100-μl portions of the supernatants were subjected to reversed-phase HPLC analysis (Knauer Eurospher RP-18 125- by 4.6-mm column; Waters Alliance 2695 system with Waters 2996 photodiode array detector). Elution was carried out in isocratic runs by use of 9% acetonitrile in 20 mM ammonium acetate, pH 6.0, at a flow rate of 0.75 ml min−1. FAD eluted at 9.8 min. The detection limit was 0.1 nmol FAD by UV detection at 260 nm. For the extraction of covalently linked FAD, the method of McIntire et al. (28) was slightly modified. The washed pellet was resuspended in 110 μl of 50 mM Tris-HCl, pH 8.0, anaerobized by flushing with dinitrogen for 5 min, and subsequently reduced by adding 12 μl of a freshly prepared anaerobic 50 mM sodium dithionite solution. After 30 min of incubation on ice, precipitated protein was pelleted by centrifugation (15,000 × g, 5 min), and 100 μl of the supernatant was analyzed by HPLC.
To test whether the addition of FAD to PCMH stimulated PCMH activity, two different experiments were performed as follows. (i) Purified PCMH (0.2 mg ml−1) was incubated with 0.5 mM FAD for up to 18 h at 20°C. Aliquots were tested for activity in the spectrophotometric assay. The photometric assay mixture did not contain additional FAD. (ii) The HPLC assay described above was carried out with 100 μM FAD added to the assay mixture. For this experiment, purified enzyme was not incubated with FAD prior to the activity test.
UV/vis spectroscopy.
UV/visible (vis) spectra of the purified enzyme were recorded on a Shimadzu 1650 PC double-beam photometer (Shimadzu, Duisburg, Germany) and analyzed using UVProbe and Prism 4.0 software (Shimadzu and GraphPad Software, San Diego, CA). Anaerobic samples were generated in sealed gastight cuvettes (80-μl ultramicrocuvettes, 1-cm light path; Hellma, Müllheim, Germany) by flushing with dinitrogen gas for 5 min. Additions of substrates or dithionite to the cuvette were made using gastight syringes (Hamilton, Bonaduz, Switzerland).
LC/MS.
Bands from Coomassie blue-stained SDS gels were excised and extracted twice with methanol-acetic acid (50%/5% [vol/vol]) in ultrapure water, washed, and digested with trypsin at 37°C overnight (33). The resulting peptide mixture was extracted twice with acetonitrile-85% formic acid (50%/5.8% [vol/vol]) and subsequently dried by vacuum centrifugation. Tryptic peptides were dissolved in 0.1% trifluoroacetic acid and used for mass spectrometry (MS).
Matrix-assisted laser desorption ionization MS analysis was performed on a dual time-of-flight instrument (Ultraflex III TOF/TOF MS; Bruker Daltonics, Bremen, Germany) as described previously (18). The measurements were performed in positive ion reflector mode and externally calibrated in the peptide range using peptide calibration standard II (Bruker Daltonics). All samples were prepared on MTP AnchorChip target plates with transponder technology (Bruker Daltonics, Bremen, Germany) by using a dried droplet preparation with α-cyano-4-hydroxycinnamic acid dissolved in ethanol-acetone (2:1 [vol/vol]) as the matrix. The peak selection was defined by use of masses from 800 to 4,000 Da for evaluation and processing. The peptide mass fingerprint spectra were acquired by summing up values for 560 laser shots done in 70 shot steps, and the m/z values are given as [M + 1H]+.
A database search against all NCBI entries (National Center for Biotechnology Information, Bethesda, MD) was performed using the peptide mass fingerprint and the tandem MS ion search (MASCOT; Matrix Science, London, United Kingdom). The following parameters were selected: (i) tryptic digestion, (ii) carbamidomethylated cysteines and (iii) oxidized methionines. The search was restricted to peptides containing single charges and was conducted with a peptide tolerance of ±1 Da and a tandem MS tolerance of ±0.5 Da.
Determination of kinetic constants.
The Km values for p-cresol and p-hydroxybenzyl alcohol were determined in the spectrophotometric assay using constant amounts of protein and various amounts of substrate. The resulting initial substrate concentration-dependent rates were analyzed by nonlinear regression to the Michaelis-Menten equation by use of Prism 4.0 (GraphPad Software, San Diego, CA).
Inhibition by p-hydroxybenzyl alcohol was tested by incubating the enzyme with various concentrations of p-hydroxybenzyl alcohol in the assay cuvette for 1 min followed by the addition of p-cresol. From the regression curves, specific activities at 1.7, 5.2, 10.4, 20.9, 40.0, and 120.0 μM p-cresol were extrapolated. The initial rates were analyzed by Dixon and Cornish-Bowden plots (4, 10). A pattern for a mixed inhibition was observed, with Kic representing the competitive inhibition constant and Kiu representing the uncompetitive inhibition constant; they were determined by transformations of the equations V(app) = V/(1 + i/Kiu) and Km(app) = Km·[1 + i/Kic]/[(1 + i)/Kiu], with V(app) representing apparent maximum velocity and i representing the inhibitor concentration.
RESULTS
Enzyme activity measurements.
Due to high background reaction levels, the spectrophotometric assay for PCMH described by McIntire et al. (28) was not applicable for membrane fractions from G. metallireducens cells. Instead, phenazine methosulfate was used as the electron acceptor in a discontinuous assay, which monitored p-cresol consumption and product formation by HPLC analysis (31). As PCMH activity was unstable at low ionic strength, the assay mixture always contained 500 mM NaCl to prevent protein precipitation. The specific activities of PCMH in cell extracts varied between 6 and 12 nmol min−1 mg−1 in different cell batches. At later purification stages with soluble PCMH preparations, ferrocenium served as the electron acceptor for PCMH in a continuous spectrophotometric assay.
Solubilization assays and purification of the soluble PCMH complex.
After the ultracentrifugation of cell extracts, all PCMH activity was found in the membrane fraction, as reported earlier (31). Even though no activity was found in the soluble supernatant, only one-third of the initial activity was recovered by this procedure. The PCMH activity could not be solubilized by the use of a number of detergents under different conditions (see Materials and Methods). However, as incubation with the strong zwitterionic detergent LDAO resulted in a 30% recovery of total PCMH activity in the membrane fraction, a 1% LDAO washing step was included in the purification protocol for the removal of other membrane proteins. The total recovery after this step with respect to crude extracts dropped to 9% (Table 1); obviously, the PCMH membrane complex was partially but reversibly (see below) inactivated by LDAO.
TABLE 1.
Purification of PCMH from G. metallireducens
| Purification step | Total protein (mg) | Enzymatic activityb
|
Yield of recovery (%) | Purification (fold) | |
|---|---|---|---|---|---|
| Total (nmol min−1) | Specific (nmol min−1 mg−1) | ||||
| Crude extracta | 819 | 4,750 | 5.8 | 100 | 1 |
| Washed membrane | 88 | 1,473 | 16.7 | 31 | 2.9 |
| LDAO-treated membrane | 66.2 | 417 | 6.3 | 9 | 1.1 |
| NaCl supernatant | 46.9 | 1,186 | 25.3 | 25 | 4.4 |
| Gel filtration pool | 9.6 | 988 | 103 | 21 | 17.8 |
| Octyl Sepharose pool | 0.4 | 880 | 2,200 | 18 | 380 |
Crude extract was from 12 g of G. metallireducens cells grown on p-cresol with Fe(III) as the electron acceptor.
Enzymatic activities were determined with the HPLC assay in crude extract and membrane fractions and with the spectrophotometric assay in soluble fractions.
As attempts to solubilize the whole PCMH membrane complex failed, the active-site-containing components of the complex were released from the membrane by the addition of 500 mM NaCl, which resulted in an increase of specific activity, most probably by the removal of LDAO. PCMH was then further purified by gel filtration and hydrophobic interaction chromatography as described in Materials and Methods.
The purity of the enzyme fraction obtained was estimated to be higher than 95% by SDS-PAGE analysis (Fig. 1A, lane 8). The specific activity was 2.2 μmol min−1 mg−1, corresponding to a 380-fold enrichment with a recovery of 18% (Table 1).
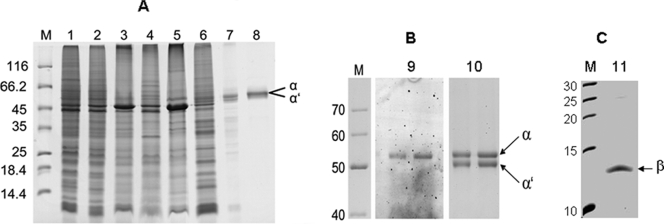
FIG. 1.
SDS-PAGE of the samples obtained during purification of PCMH and content of covalently bound FAD. (A) Shown is a 12.5% polyacrylamide gel. Lanes: M, molecular mass standard; 1, membrane fraction after ultracentrifugation; 2, washed membrane fraction; 3, LDAO-treated membrane; 4, supernatant after LDAO treatment; 5, NaCl-treated membrane; 6, supernatant after NaCl treatment; 7, gel filtration pool; 8, pool after hydrophobic interaction chromatography. (B) Shown is an 8% polyacrylamide gel of purified PCMH. Lanes: M, molecular mass standard; 9, fluorescence of 1 and 1.5 μg purified PCMH (excitation at 488 nm, detection at 520 nm); 10, Coomassie blue staining of the same gel. (C) Shown is a 16.5% polyacrylamide gel of purified PCMH. Lanes: M, molecular mass standard; 11, purified PCMH. For lanes M, masses in kDa are indicated to the left.
Molecular architecture and FAD content.
Analysis of p-cresol-induced genes putatively coding for PCMH suggested that the enzyme is composed of two isoformic α subunits and a heme c-containing β subunit (31). The molecular masses of the individual subunits deduced from the amino acid sequences were as follows: for the α subunit encoded by pcmI, 59.4 kDa; for the α′ subunit encoded by pcmJ, 57.0 kDa; and for the β subunit encoded by pcmG, 11.96 kDa (including one covalently bound heme c and the cleavage of an N-terminal 31-amino-acid signal peptide). The role of the pcmH open reading frame was unknown; the deduced amino acid sequence did not show similarities to that of any other protein.
On standard 12.5% polyacrylamide gels of PCMH, a major band of around 58 kDa was observed (Fig. 1A). On an 8% polyacrylamide gel, this band could be resolved into two bands of equal intensities (Fig. 1B, lane 10). The β subunit of around 12.5 kDa was identified using a 16.5% SDS-polyacrylamide gel (Fig. 1C). The α, α′, and β subunits were excised from the gel, digested by trypsin, and analyzed by MS. The results obtained unambiguously identified them as the pcmI, pcmJ, and pcmG gene products, respectively, with sequence coverages of more than 49%. No evidence for the presence of any other protein component, such as the putative pcmH gene product, was obtained. The native molecular mass of PCMH was determined by gel filtration on a 120-ml Superdex 200 prep-grade column. The elution volume of 72 ± 2 ml correlated to a native molecular mass of 124 ± 15 kDa, which was interpreted as representing a tetrameric αα′β2 composition.
PCMH from P. putida contains an FAD cofactor covalently linked to Tyr384 in both identical α subunits (26, 27). This tyrosine residue is conserved only in the larger α subunit and not in the smaller α′ subunit of PCMH from G. metallireducens (31). Thus, the FAD cofactor content of PCMH was at issue.
PCMH was analyzed for the presence of a noncovalently bound FAD cofactor by HPLC and UV/vis spectroscopic analysis of enzyme extracts, but no evidence for a flavin cofactor was obtained from the supernatants of protein precipitates. To check the presence of a covalently linked FAD cofactor, purified PCMH was subjected to SDS-PAGE and analyzed for fluorescence at 520 nm (the excitation wavelength was 488 nm). Coomassie blue staining of the gel revealed that only the band corresponding to the α subunit exhibited fluorescence, indicating the presence of a covalently linked FAD cofactor in the α but not in the α′ subunit (Fig. 1B, lane 9). To verify the covalent linkage, FAD was extracted from the enzyme by reductive cleavage of the assumed 8α-O-tyrosyl bond as described previously (28). HPLC analysis of such extracts determined a level of 0.93 ± 0.18 mol FAD per mol αα′β2 tetramer, which was in accordance with the presence of only one fluorescent protein band on SDS-polyacrylamide gels.
For further analysis, the peptides obtained from the α subunit after trypsin digestion were analyzed for the presence of a covalently bound flavin cofactor by MS. A peptide mass of 2,602.32 Da was detected, which was derived from the tryptic peptide QLMMGGSTLQEFGLYNWR. This peptide was assigned to a peptide from the α subunit (PcmI), with the tyrosine residue representing Tyr394. The calculated mass of this peptide is 2,131.01 Da. Assuming the oxidation of a methionine residue (+16 Da) and the covalent attachment of FMN (456.4 Da minus 1 Da) to the Tyr394 of the peptide, a mass of 2,602.4 Da is obtained, which fits well to the detected mass. During MS analysis, the 8α-O-tyrosyl bond obviously remained intact, while the AMP moiety of FAD was cleaved, as reported earlier (11).
We further checked whether the α′ subunit might have lost a noncovalently bound FAD cofactor during the purification procedure. No increase of activity was observed by incubating PCMH with 0.5 mM FAD for up to 8 h, irrespective of whether the FAD added was removed by gel filtration or whether it was still present in the activity assay (not shown). The results obtained suggest that a single flavin cofactor is covalently attached to the Tyr394 of the α subunit (corresponding to Tyr384 in P. putida), whereas the α′ subunit does not contain any flavin cofactor.
UV/vis spectroscopic properties.
To identify the assumed heme cofactor-containing β subunit, a UV/vis spectroscopic analysis of PCMH was carried out. The FAD and heme c cofactors of PCMHs from aerobic and facultatively anaerobic bacteria are responsible for characteristic UV/vis spectra, with major absorption maxima at 405 to 420 nm (Soret band), 520 to 530 nm (β band), and 540 to 560 nm (α band) caused by heme c and a shoulder around 450 nm caused by FAD. The UV/vis spectrum of oxidized PCMH unambiguously confirmed the presence of a heme c cofactor-containing subunit (Fig. 2A). By use of a molecular mass of 130 kDa, the absorption coefficients were determined as 20,800 M−1 cm−1 for the α band (552 nm), 26,700 M−1 cm−1 for the β band (522 nm), and 182,100 M−1 cm−1 for the Soret band (410 nm) of the heme c moiety. By use of a ɛ552 value of 19,000 M−1 cm−1 for PCMH from P. putida as reported previously (28), a content of 2.2 mol heme c per mol of enzyme was estimated for PCMH from G. metallireducens. This result is in accordance with the presence of two c-type cytochrome subunits per PCMH, supporting the proposed αα′β2 composition.
FIG. 2.
UV/vis spectra of PCMH (0.2 mg ml−1, pH 8.0). (A) Oxidized (straight line) and dithionite reduced (dotted line). (B) Oxidized (straight line) and p-cresol reduced (1 mM, 15 min, dotted line). (C) Oxidized (straight line) and p-hydroxybenzaldehyde reduced (1 mM, 15 min, dotted line). Insets in panels A to C show difference spectra (reduced value minus oxidized value).
Upon reduction of PCMH by dithionite, the Soret band shifted to 416 nm, with a molar extinction coefficient of 206,800 M−1 cm−1; the molar extinctions of the α and β bands increased to 42,200 M−1 cm−1 and 30,600 M−1 cm−1, respectively. The shoulder at 450 nm was bleached upon reduction by dithionite. The detected spectral difference of 23,400 M−1 cm−1 at 450 nm appeared to be too high for the reduction of one FAD cofactor (Δɛ450[FADox-FADred] = 11,300 M−1 cm−1 [9]) but can be explained by the broad maximum of the Soret band, which overlays the absorption of FAD (Fig. 2A). Upon the reduction of PCMH by p-cresol or p-hydroxybenzyl alcohol, the shoulder at 450 nm disappeared in a manner similar to that seen upon reduction by dithionite. In contrast, the heme absorptions at 552 nm increased only to 60% and 55% of the value obtained with dithionite as reductant, respectively (Fig. 2B and C). The addition of excess substrate (1 mM) or prolonged incubation for up to 1 h did not result in a further change of the UV/vis spectrum. Obviously, both substrates reduced the FAD cofactor completely but did this for only one of the two heme cofactors of PCMH. The spectral properties of PCMH are summarized in Table 2.
TABLE 2.
UV/vis spectral properties of PCMH
| Spectral band | Absorption coefficient (M−1 cm−1)
|
||
|---|---|---|---|
| Oxidized | Reduced | Difference | |
| α (552 nm) | 20,800 | 42,200 | 21,400 |
| β (522 nm) | 26,700 | 30,600 | 3,900 |
| Soret (410/416 nm) | 182,100 (410 nm) | 206,800 (416 nm) | 140,400a |
| FAD (450 nm) | 48,100 | 24,700 | 23,400 |
The molar absorption coefficient for the Soret is calculated from the maximum (418 nm) and the minimum (406 nm) of the difference spectrum.
Kinetic studies and stoichiometry of reaction.
Purified PCMH from G. metallireducens catalyzed both the hydroxylation of p-cresol to p-hydroxybenzyl alcohol and the subsequent oxidation to p-hydroxybenzaldehyde. The stoichiometry of ferrocenium reduction and p-cresol or p-hydroxybenzyl alcohol oxidation was tested by sampling from a running spectrophotometric assay and subsequently analyzing substrate consumption by HPLC. The amounts of substrate consumed and electron acceptor reduced clearly pointed to there being four electrons transferred per p-cresol oxidized to p-hydroxybenzaldehyde (not shown).
The Km values of PCMH from aerobic and facultatively anaerobic bacteria vary from 3.6 to 21 μM for p-cresol and from 15 to 47 μM for p-hydroxybenzyl alcohol (16, 17, 20). The Km values for the substrates of PCMH from G. metallireducens were 1.7 ± 0.3 μM at a Vmax of 2.18 ± 0.05 μmol min−1 mg−1 for p-cresol and 2.7 ± 0.7 μM at a Vmax of 0.23 ± 0.02 μmol min−1 mg−1 for p-hydroxybenzyl alcohol.
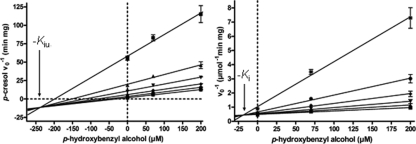
Surprisingly, p-hydroxybenzyl alcohol was a potent inhibitor of p-cresol oxidation. To test whether this inhibitory effect was caused by competitive binding of the two substrates to the active site, the Km values for p-cresol were determined in the presence of different p-hydroxybenzyl alcohol concentrations. The results were analyzed by both Dixon and Cornish-Bowden plots (Fig. 3) (4, 10). The analysis of the data indicated that p-hydroxybenzyl alcohol was a mixed inhibitor of p-cresol oxidation, with inhibition constants of a Kic value of 18 ± 9 μM for the competitive inhibition and a Kiu value of 235 ± 20 μM for the uncompetitive inhibition.
FIG. 3.
Inhibition of p-cresol oxidation by p-hydroxybenzyl alcohol. The Km values for p-cresol were determined in the presence of 0, 70, and 200 μM p-hydroxybenzyl alcohol by varying the p-cresol concentration in the presence of constant concentrations of p-hydroxybenzyl alcohol; the initial rates were was extrapolated from the regression curves for 1.7 (✸), 5.2 (•), 10.4 (⧫), 20.9 (▾), 40.0 (▴), and 120 (▪) μM p-cresol. (Left) Cornish-Bowden plot (10); (right) Dixon plot (4). Values for Kic (18 ± 9 μM) and Kiu (235 ± 20 μM) were estimated from the intersection points of the regression lines.
DISCUSSION
In a previous study, we demonstrated that methyl group hydroxylation is the initial step of p-cresol degradation in G. metallireducens (31). In contrast to what has been seen for all other PCMHs known so far, the activity was found completely in the membrane fraction. Though attempts to solubilize the whole membrane complex failed, we now provide insights into the active-site-containing soluble components of a proposed PCMH membrane complex. The properties of PCMH from G. metallireducens are summarized in Table 3 and are discussed below.
TABLE 3.
Molecular and kinetic properties of PCMH
| Property | Value |
|---|---|
| Substrates (Km values [μM]) | p-Cresol (1.7 ± 0.3); p-hydroxybenzyl alcohol (2.7 ± 0.7) |
| Products | p-Hydroxybenzyl alcohol; p-hydroxybenzaldehyde |
| Subunit composition | αα'β2; α, 57 kDa; α′, 59 kDa; β, 8.5 kDa |
| Native mol mass | 124 ± 15 kDa |
| Cofactors | One covalently linked FAD, two heme c's |
| Absorption maxima | Reduced, 416, 523, and 552 nm; oxidized, 410 and 522 nm, shoulder at 450 nm |
| Specific activities (μmol min−1 mg−1)/turnover nos. (s−1) | 2.2/4.8 (p-cresol); 0.2/0.48 (p-hydroxybenzyl alcohol) |
| kcat/Km (mol−1 s−1) | 2.98 × 106 (p-cresol); 0.20 × 106 (p-hydroxybenzyl alcohol) |
| Inhibition constants for p-hydroxybenzyl alcohol | Kic = 18 ± 9 μM; Kiu = 235 ± 20 μM |
Molecular properties.
In contrast to all PCMHs described so far, the enzyme from G. metallireducens has a unique αα′β2 subunit composition, with the α subunit carrying an FAD cofactor which is lacking in the α′ subunit. This composition is in accordance with a prior in silico analysis of the p-cresol-induced pcm gene cluster in which two isoforms of the FAD-containing subunit were predicted (31). As FAD serves as ultimate hydride ion acceptor for both p-cresol and p-hydroxybenzyl alcohol oxidation, the FAD-free α′ subunit is considered to be catalytically inactive, and its role remains elusive.
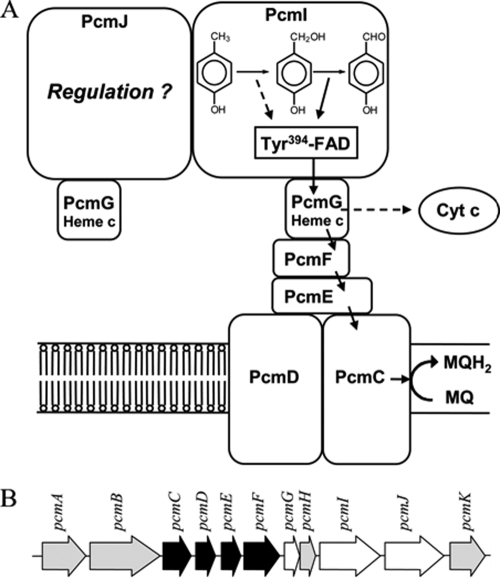
Both substrates of PCMH, p-cresol and p-hydroxybenzyl alcohol, were able to completely reduce the FAD cofactor but reduced the heme cofactor only by about 50%. This observation cannot be explained by a thermodynamic equilibrium, as the redox potentials of p-cresol oxidation (E0′ of +80 mV [29]) and p-hydroxybenzyl alcohol oxidation (E0′ of between −150 mV and −200 mV) are much more negative than that of the electron-accepting heme cofactor (E0′ of +234 mV [12, 35]). An explanation for this phenomenon might be given by the asymmetric architecture of the enzyme: only the α and not the α′ subunit contains the catalytically essential FAD cofactor and should therefore mediate electron transfer only from the substrate to the heme moiety of the attached β subunit (Fig. 4). In PCMH from P. putida, the edge-to-edge distance from the FAD to the heme of one functional unit is about 7.8 Å, while the distance from the FAD of one functional unit to the heme cofactor of the other functional unit is 47.3 Å (calculated from crystallographic data presented in reference 7), which is much too far for an efficient electron transfer (30). Assuming a similar overall structure of the soluble components of PCMH from G. metallireducens and PCMH from P. putida (7), the heme cofactor of the β subunit attached to the noncatalytic α′ subunit appears not to be involved in an electron transfer reaction catalyzed by the α subunit (Fig. 4).
FIG. 4.
Model for a putative PCMH/cytochrome bc complex. (A) Proposed alternative electron transfer routes during p-cresol oxidation (dashed line) and p-hydroxybenzyl alcohol oxidation (solid line) are shown. Electrons derived from alcohol oxidation can be transferred to menaquinone, whereas those derived from p-cresol oxidation may be transferred to a periplasmic cytochrome c. Alternatively, the latter could also be transferred to menaquinone by means of energy-driven reversed electron transfer. The areas of the symbolized subunits do not represent the exact relative molecular masses. MQ, menaquinone; MQH2, menaquinol. Cyt c, cytochrome c. (B) Part of the pcm gene cluster. PCMH is encoded by pcmG (β subunit), pcmI (α subunit), and pcmJ (α′ subunit), and the putative cytochrome bc complex is encoded by pcmC (product similar to the N-terminal part of transmembrane cytochrome b), pcmD (product similar to the C-terminal part of transmembrane cytochrome b), pcmE (Rieske protein), and pcmF (cytochrome c). The function of the other putative gene products is unknown.
Catalytic properties.
In contrast to other p-cresol-degrading organisms, G. metallireducens appears to use PCMH for both p-cresol and p-hydroxybenzyl alcohol oxidation in vivo, which is supported by the following observations. First, no p-hydroxybenzyl alcohol dehydrogenase activity was determined for soluble extracts of G. metallireducens by use of various artificial electron acceptors (31). In contrast, for P. putida and other aerobic or facultatively anaerobic p-cresol-degrading bacteria, soluble NAD+-dependent p-hydroxybenzyl alcohol dehydrogenases were supposed to catalyze the second step in p-cresol metabolism in vivo (19, 28). For this reason, the p-hydroxybenzyl alcohol-oxidizing activity of PCMH was considered to be physiologically irrelevant in these organisms. Second, the Km values of PCMH from G. metallireducens for p-cresol and p-hydroxybenzyl alcohol were both extremely low (Table 3). In contrast, the Km values for p-hydroxybenzyl alcohol are 1 order of magnitude higher for PCMHs from other organisms. Third, the inhibition of the p-cresol-oxidizing activity of PCMH by p-hydroxybenzyl alcohol can be explained by the need for keeping both partial activities of the enzyme in balance: as p-cresol is oxidized at a rate 10-fold higher than that for p-hydroxybenzyl alcohol, an accumulation of the latter has to be avoided.
Surprisingly, the inhibition by p-hydroxybenzyl alcohol was not purely competitive, but at high concentrations an additional uncompetitive effect was determined. The obvious explanation for the latter effect might be that p-hydroxybenzyl alcohol forms a tight complex with the active site of PCMH in which the affinity to p-cresol is decreased. However, the detection of p-hydroxybenzyl alcohol as a free intermediate during turnover suggests that its binding to PCMH is reversible. An alternative explanation for the uncompetitive effect is that the FAD-free α′ subunit plays a regulatory role for PCMH activity. The α and α′ subunits are highly similar to each other, and therefore a binding of accumulating substrates to the α′ subunit is conceivable. Such a binding could modulate the α subunit with regard to its preference for p-cresol or p-hydroxybenzyl alcohol oxidation. In this case, the unusual αα′β2 composition in PCMH from G. metallireducens could be explained by the unique function of PCMH to catalyze both p-cresol and p-hydroxybenzaldehyde oxidation in vivo.
Role of membrane association.
Even though the amino acid sequences deduced from the genes coding for the α, α′, and β subunits clearly identified these subunits as soluble proteins, the PCMH activity was tightly bound to the membrane and could be released to the soluble fraction only after incubation with 500 mM NaCl. This finding suggested a strong binding to integral membrane components, which are supposed to be components of a cytochrome bc-like membrane complex encoded by the p-cresol-induced pcmCDEF genes. Cytochrome bc complexes usually mediate electron transfer between quinols (in the case of G. metallireducens menaquinol [24]) and cytochrome c. The putative PCMH/cytochrome bc complex in G. metallireducens (PcmCDEFG2IJ) would be involved in electron transfer from the substrate to menaquinone. However, the redox potential of the enzyme-bound redox pair p-hydroxybenzyl alcohol/p-cresol (+80 mV) is too positive for electron transfer to menaquinone (E0′ = −80 mV [35]). In contrast, electrons derived from further oxidation of the alcohol to the aldehyde (E0′ < −150 mV) could indeed be transferred to the menaquinone pool via the PcmCDEF complex. The reduction of the terminal acceptor Fe(III) (E0′ of ∼100 mV under in vivo conditions) by menaquinol is clearly exergonic and may be coupled to energy conservation. In contrast, this is not the case for the reduction of Fe(III) by cytochrome c. The noncatalytic α′ subunit may be involved in mediating both alternative electron transfer routes either to the periplasmic cytochrome c pool (in the case of p-cresol oxidation) or to the menaquinone pool (in the case of p-hydroxybenzyl alcohol oxidation [Fig. 4]). Initial attempts to demonstrate the latter electron transfer route with crude membrane preparations failed due to high background reaction levels. For example, membrane preparations from G. metallireducens grown on p-cresol always contained considerable amounts of p-cresol and numerous electron-scavenging cytochromes.
In summary, the unusual membrane association and molecular architecture of PCMH from G. metallireducens may be explained by its in vivo role in oxidizing both p-cresol and p-hydroxybenzyl alcohol. Furthermore, an electron transfer from p-hydroxybenzyl alcohol to menaquinone instead to cytochrome c would significantly increase the energy yield, which appears to be particularly important for an obligately anaerobic organism with a low energy yield from the overall metabolism.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft.
We thank Nasser Gad'on (Freiburg) and Simon Wischgoll (Leipzig) for help with the cultivation of G. metallireducens.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Biegert, T., U. Altenschmidt, C. Eckerskorn, and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur. J. Biochem. 213555-561. [DOI] [PubMed] [Google Scholar]
- 2.Bossert, I. D., and L. Y. Young. 1986. Anaerobic oxidation of p-cresol by a denitrifying bacterium. Appl. Environ. Microbiol. 521117-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brackmann, R., and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from a denitrifying Pseudomonas species. Eur. J. Biochem. 213563-571. [DOI] [PubMed] [Google Scholar]
- 4.Cornish-Bowden, A. 1974. A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 137143-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronin, C. N., J. Kim, J. H. Fuller, X. Zhang, and W. S. McIntire. 1999. Organization and sequences of p-hydroxybenzaldehyde dehydrogenase and other plasmid-encoded genes for early enzymes of the p-cresol degradative pathway in Pseudomonas putida NCIMB 9866 and 9869. DNA Seq. 107-17. [DOI] [PubMed] [Google Scholar]
- 6.Cunane, L. M., Z. W. Chen, W. S. McIntire, and F. S. Mathews. 2005. p-Cresol methylhydroxylase: alteration of the structure of the flavoprotein subunit upon its binding to the cytochrome subunit. Biochemistry 442963-2973. [DOI] [PubMed] [Google Scholar]
- 7.Cunane, L. M., Z. W. Chen, N. Shamala, F. S. Mathews, C. N. Cronin, and W. S. McIntire. 2000. Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism. J. Mol. Biol. 295357-374. [DOI] [PubMed] [Google Scholar]
- 8.Dagley, S., and D. S. Patel. 1957. Oxidation of p-cresol and related compounds by a Pseudomonas. Biochem. J. 66227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and M. J. Jones. 1986. Data for biochemical research, 3rd ed. Clarendon Press, Oxford, United Kingdom.
- 10.Dixon, M. 1953. The determination of enzyme inhibitor constants. Biochem. J. 55170-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efimov, I., C. N. Cronin, D. J. Bergmann, V. Kuusk, and W. S. McIntire. 2004. Insight into covalent flavinylation and catalysis from redox, spectral, and kinetic analyses of the R474K mutant of the flavoprotein subunit of p-cresol methylhydroxylase. Biochemistry 436138-6148. [DOI] [PubMed] [Google Scholar]
- 12.Efimov, I., C. N. Cronin, and W. S. McIntire. 2001. Effects of noncovalent and covalent FAD binding on the redox and catalytic properties of p-cresol methylhydroxylase. Biochemistry 402155-2166. [DOI] [PubMed] [Google Scholar]
- 13.Entsch, B., and W. J. van Berkel. 1995. Structure and mechanism of para-hydroxybenzoate hydroxylase. FASEB J. 9476-483. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, J., M. Dispensa, G. C. Fogg, D. T. Evans, and C. S. Harwood. 1994. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J. Bacteriol. 176634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopper, D. J. 1976. The hydroxylation of p-cresol and its conversion to p-hydroxybenzaldehyde in Pseudomonas putida. Biochem. Biophys. Res. Commun. 69462-468. [DOI] [PubMed] [Google Scholar]
- 16.Hopper, D. J., I. D. Bossert, and M. E. Rhodes-Roberts. 1991. p-Cresol methylhydroxylase from a denitrifying bacterium involved in anaerobic degradation of p-cresol. J. Bacteriol. 1731298-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopper, D. J., and D. G. Taylor. 1977. The purification and properties of p-cresol-(acceptor) oxidoreductase (hydroxylating), a flavocytochrome from Pseudomonas putida. Biochem. J. 167155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jehmlich, N., F. Schmidt, M. von Bergen, H.-H. Richnow, and C. Vogt. Protein based stable isotope probing (protein-SIP) reveals active species within anoxic mixed cultures. ISME J., in press. [DOI] [PubMed]
- 19.Keat, M. J., and D. J. Hopper. 1978. The aromatic alcohol dehydrogenases in Pseudomonas putida N.C.I.B. 9869 grown on 3,5-xylenol and p-cresol. Biochem. J. 175659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keat, M. J., and D. J. Hopper. 1978. p-Cresol and 3,5-xylenol methylhydroxylases in Pseudomonas putida N.C.I.B. 9896. Biochem. J. 175649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J., J. H. Fuller, V. Kuusk, L. Cunane, Z. W. Chen, F. S. Mathews, and W. S. McIntire. 1995. The cytochrome subunit is necessary for covalent FAD attachment to the flavoprotein subunit of p-cresol methylhydroxylase. J. Biol. Chem. 27031202-31209. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 23.Londry, K. L., J. M. Suflita, and R. S. Tanner. 1999. Cresol metabolism by the sulfate-reducing bacterium Desulfotomaculum sp. strain Groll. Can. J. Microbiol. 45458-463. [PubMed] [Google Scholar]
- 24.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. Phillips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159336-344. [DOI] [PubMed] [Google Scholar]
- 25.Lovley, D. R., and D. J. Lonergan. 1990. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism, GS-15. Appl. Environ. Microbiol. 561858-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McIntire, W., D. E. Edmondson, D. J. Hopper, and T. P. Singer. 1981. 8 Alpha-(O-tyrosyl)flavin adenine dinucleotide, the prosthetic group of bacterial p-cresol methylhydroxylase. Biochemistry 203068-3075. [DOI] [PubMed] [Google Scholar]
- 27.McIntire, W., D. E. Edmondson, T. P. Singer, and D. J. Hopper. 1980. 8 Alpha-O-tyrosyl-FAD: a new form of covalently bound flavin from p-cresol methylhydroxylase. J. Biol. Chem. 2556553-6555. [PubMed] [Google Scholar]
- 28.McIntire, W., D. J. Hopper, and T. P. Singer. 1985. p-Cresol methylhydroxylase. Assay and general properties. Biochem. J. 228325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller, J. A., A. S. Galushko, A. Kappler, and B. Schink. 2001. Initiation of anaerobic degradation of p-cresol by formation of 4-hydroxybenzylsuccinate in Desulfobacterium cetonicum. J. Bacteriol. 183752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page, C. C., C. C. Moser, X. Chen, and P. L. Dutton. 1999. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 40247-52. [DOI] [PubMed] [Google Scholar]
- 31.Peters, F., D. Heintz, J. Johannes, A. van Dorsselaer, and M. Boll. 2007. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens. J. Bacteriol. 1894729-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudolphi, A., A. Tschech, and G. Fuchs. 1991. Anaerobic degradation of cresols by denitrifying bacteria. Arch. Microbiol. 155238-248. [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 34.Smolenski, W. J., and J. M. Suflita. 1987. Biodegradation of cresol isomers in anoxic aquifers. Appl. Environ. Microbiol. 53710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unciuleac, M., E. Warkentin, C. C. Page, M. Boll, and U. Ermler. 2004. Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow. Structure 122249-2256. [DOI] [PubMed] [Google Scholar]
- 37.Yu, L., M. Blaser, P. I. Andrei, A. J. Pierik, and T. Selmer. 2006. 4-Hydroxyphenylacetate decarboxylases: properties of a novel subclass of glycyl radical enzyme systems. Biochemistry 459584-9592. [DOI] [PubMed] [Google Scholar]
- 38.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182157-159. [DOI] [PubMed] [Google Scholar]