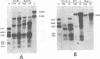Abstract
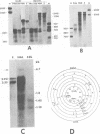
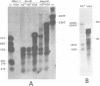
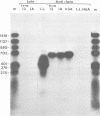
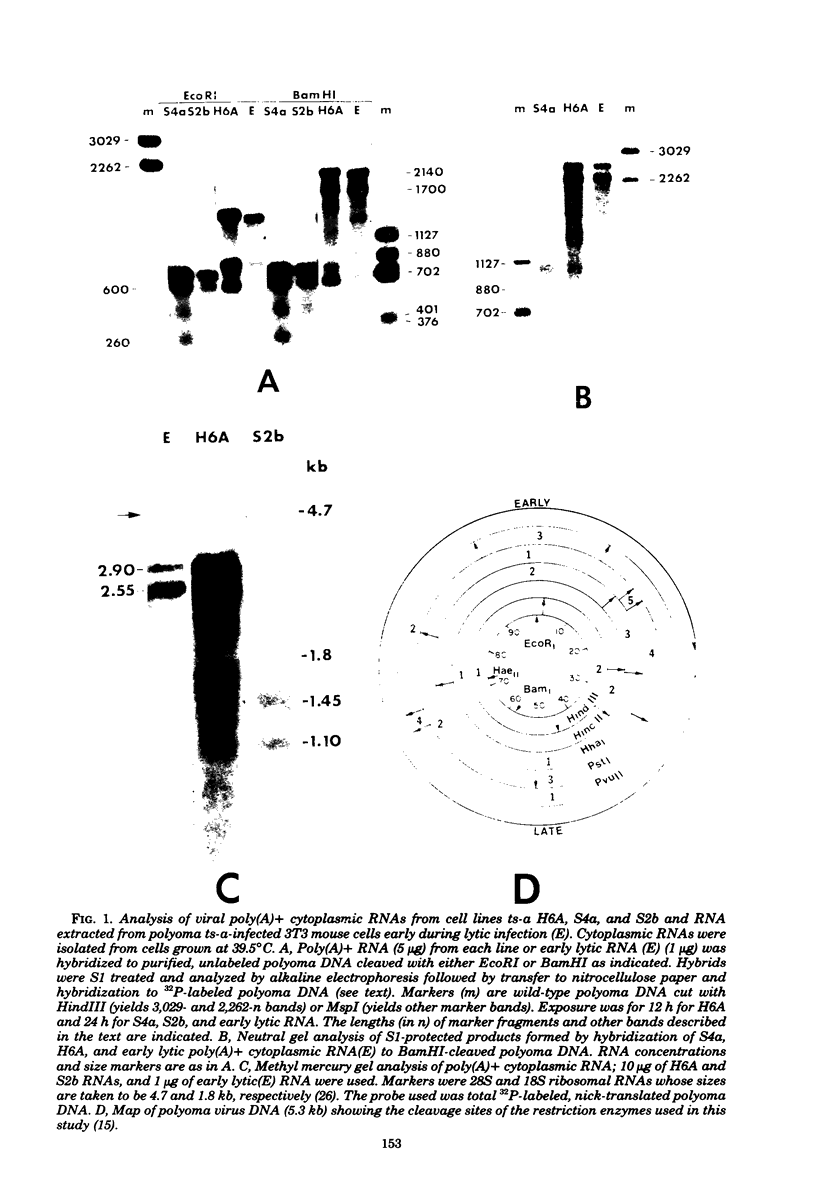
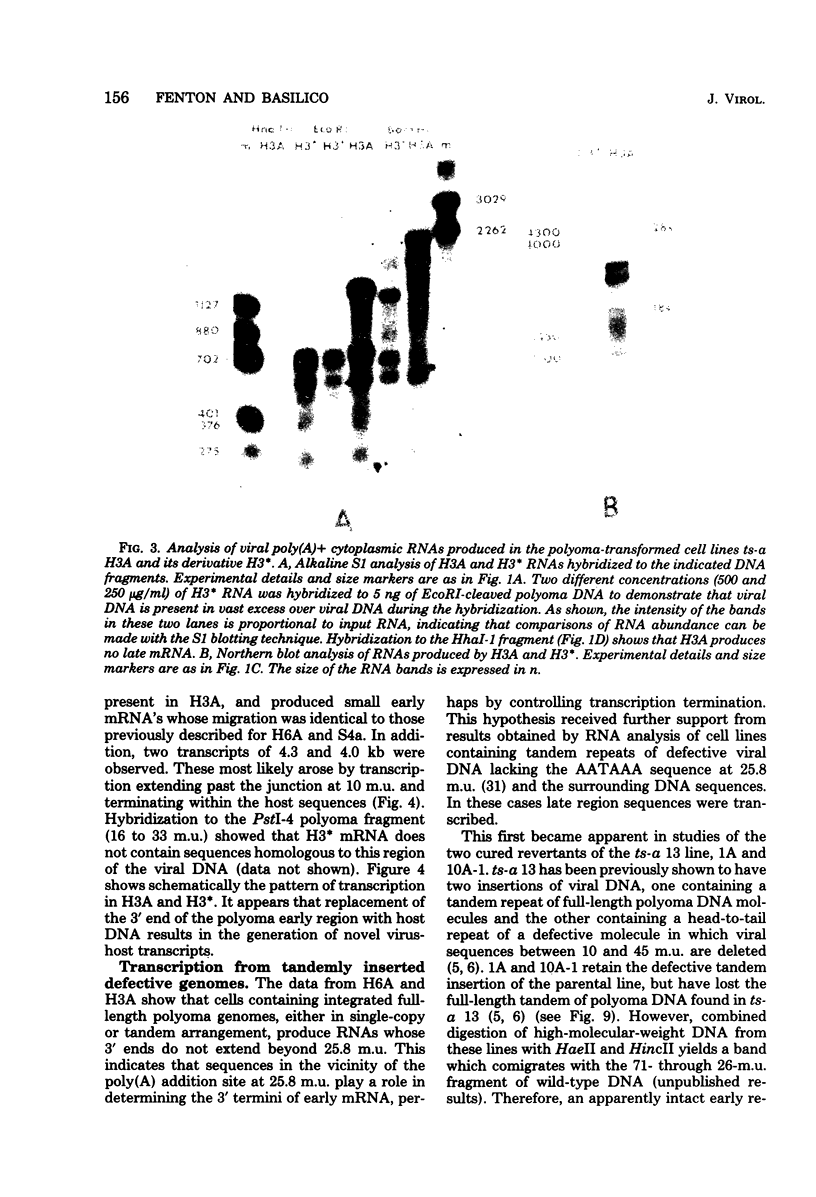
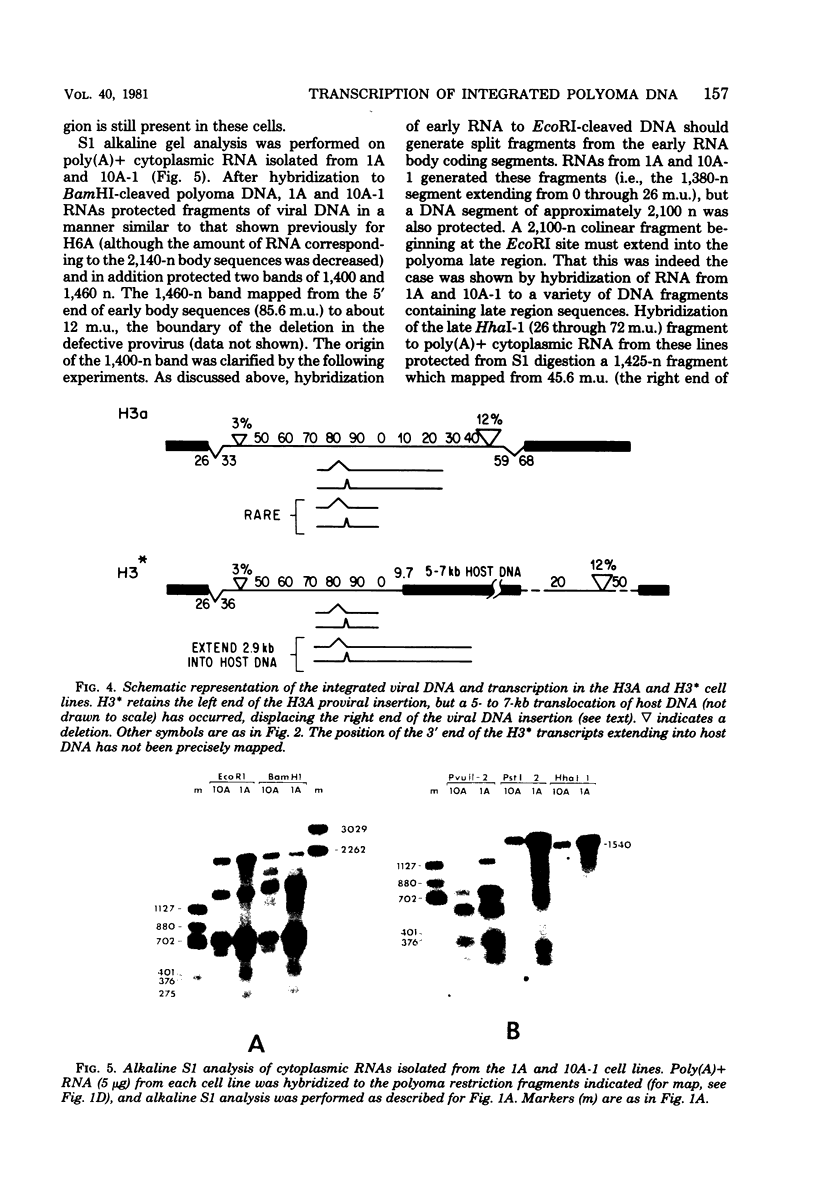
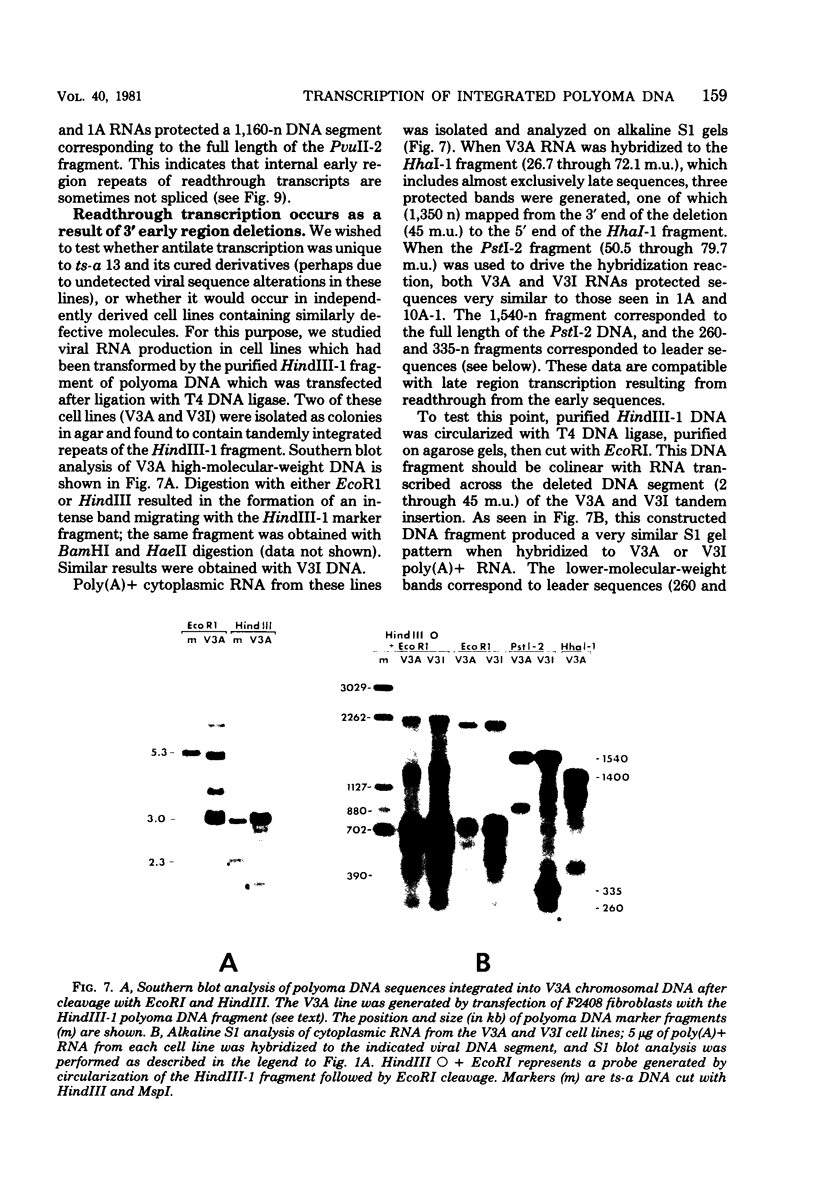
We have studied transcription of integrated viral DNA sequences in a variety of ts-a polyoma virus-transformed rat cells and cured revertants (which had undergone excision of variables amounts of integrated viral DNA) to characterize the structure of viral mRNA's produced in these lines under conditions in which integrated DNA is stable. Our results indicate that cells containing intact early region sequences, either in single-copy or tandem insertions, produce mRNA's indistinguishable from those observed early in lytic infections; sequences complementary to the polyoma late region were not transcribed from integrated viral DNA. Cured revertants no longer encoded full-length early mRNA's , but produced viral transcripts whose 3' ends mapped at an alternative early region polyadenylic acid attachment site at 99 map units or extended in to flanking host sequences. The phenotype of these revertant cells correlated with the abundance of these transcripts, suggesting that the transforming function(s) of polyoma virus controls the cellular phenotype in a dose-dependent manner. Unexpected results were obtained from studies of cells containing tandem repeats of defective viral DNA in which the polyadenylic acid attachment signal at 25.8 map units and surrounding sequences were deleted. In these cases, polyadenylated mRNA's were observed that contained sequences complementary to the early strand of the polyoma late region. These mRNA's (some larger than 8 kilobases) originated at the viral early promoter, extended into the late region, and continued into the early region of the contiguous repeat in the tandem. The multimeric mRNA's produced contained defective early regions in tandem with late region sequences. S1 analysis indicated that whereas the 5' early region sequences of readthrough transcripts were spliced in the usual manner, internal early region repeats were either unspliced or used only one of the small early region splices. When deletions in the viral readthrough transcripts were observed. This suggests that sequences nearby the AAUAAA sequence at 26 map units may control transcription termination of the polyoma early region.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler L. T. Virus-secific transcription in 3T3 cells transformed by the ts-a mutant of polyoma virus. J Virol. 1977 Apr;22(1):54–64. doi: 10.1128/jvi.22.1.54-64.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Basilico C., Zouzias D., Della-Valle G., Gattoni S., Colantuoni V., Fenton R., Dailey L. Integration and excision of polyoma virus genomes. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):611–620. doi: 10.1101/sqb.1980.044.01.064. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen B. Virus-specific early RNA in 3T6 cells infected by a tsA mutant of polyoma virus. Virology. 1978 Mar;85(1):222–230. doi: 10.1016/0042-6822(78)90426-9. [DOI] [PubMed] [Google Scholar]
- Colantuoni V., Dailey L., Basilico C. Amplification of integrated viral DNA sequences in polyoma virus-transformed cells. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3850–3854. doi: 10.1073/pnas.77.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Valle G., Fenton R. G., Basilico C. Polyoma large T antigen regulates the integration of viral DNA sequences into the genome of transformed cells. Cell. 1981 Feb;23(2):347–355. doi: 10.1016/0092-8674(81)90130-6. [DOI] [PubMed] [Google Scholar]
- Eckhart W., Hutchinson M. A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979 Dec;18(4):925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffin B. E. Organization of the genomes of polyoma virus and SV40. Adv Cancer Res. 1977;24:67–113. doi: 10.1016/s0065-230x(08)61013-1. [DOI] [PubMed] [Google Scholar]
- Gattoni S., Colantuoni V., Basilico C. Relationship between integrated and nonintegrated viral DNA in rat cells transformed by polyoma virus. J Virol. 1980 Jun;34(3):615–626. doi: 10.1128/jvi.34.3.615-626.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J. Topography of the three late mRNA's of polyoma virus which encode the virion proteins. J Virol. 1980 Feb;33(2):637–651. doi: 10.1128/jvi.33.2.637-651.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Kamen R., Lindstrom D. M., Shure H., Old R. W. Virus-specific RNA in cells productively infected or transformed by polyoma virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):187–198. doi: 10.1101/sqb.1974.039.01.025. [DOI] [PubMed] [Google Scholar]
- Kamen R., Shure H. Topography of polyoma virus messenger RNA molecules. Cell. 1976 Mar;7(3):361–371. doi: 10.1016/0092-8674(76)90165-3. [DOI] [PubMed] [Google Scholar]
- Khoury G., Alwine J. C., Dhar R., Gruss P., Lai C. J., Segal S., Seif I. Regulation of SV40 gene expression through RNA splicing. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):41–54. doi: 10.1101/sqb.1980.044.01.007. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moore J. L., Chowdhury K., Martin M. A., Israel M. A. Polyoma large tumor antigen is not required for tumorigenesis mediated by viral DNA. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1336–1340. doi: 10.1073/pnas.77.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Dilworth S. M., Griffin B. E. Coding capacity of a 35 percent fragment of the polyoma virus genome is sufficient to initiate and maintain cellular transformation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3278–3282. doi: 10.1073/pnas.77.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E., Coleclough C., Seidman J. G., Leder P., Tonegawa S., Matthyssens G., Weigert M. Transcription of mouse kappa chain genes: implications for allelic exclusion. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1937–1941. doi: 10.1073/pnas.77.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad I., Zouzias D., Basilico C. State of the viral DNA in rat cells transformed by polyoma virus. I. Virus rescue and the presence of nonintergrated viral DNA molecules. J Virol. 1976 May;18(2):436–444. doi: 10.1128/jvi.18.2.436-444.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Phosphorylation of polyoma T antigens. Cell. 1979 Dec;18(4):935–946. doi: 10.1016/0092-8674(79)90206-x. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Griffin B., Fried M. Protein kinase activity associated with polyoma virus middle T antigen in vitro. Cell. 1979 Dec;18(4):915–924. doi: 10.1016/0092-8674(79)90204-6. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouzias D., Prasad I., Basilico C. State of the viral DNA in rat cells transformed by polyma virus. II. Identification of the cells containing nonintegrated viral DNA and the effect of viral mutations. J Virol. 1977 Oct;24(1):142–150. doi: 10.1128/jvi.24.1.142-150.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]