Abstract
Scedosporium apiospermum and Scedosporium prolificans are fungal pathogens that can cause severe human infections, including disseminated mycosis in immunocompromised patients. Two real-time PCR (RT-PCR) assays for the diagnosis of these species were developed and validated for the classification of clinical strains and for the detection of DNA in clinical samples by use of a murine model of invasive infection. A total of 14 clinical strains and 141 samples, including blood, serum, and lung samples from infected CD1 mice, were analyzed. Each RT-PCR methodology used a species-specific molecular beacon probe targeting a highly conserved region of the fungal ribosomal DNA gene. Results showed 100% specificity and a detection limit of 10 fg of DNA for both assays. The sensitivities for the S. prolificans-specific PCR assay were 100% for cultured clinical strains, 95.5% for lung tissues, 85% for serum, and 83.3% for blood. For S. apiospermum, the sensitivities were 100% for clinical strains and 97.2%, 81.8%, and 54.5% for lung tissues, serum, and blood, respectively. Both techniques can be useful for clinical diagnosis, and further studies are warranted.
Scedosporium apiospermum (telemorph, Pseudallescheria boydii) and Scedosporium prolificans are members of the hyphomycetous genus Scedosporium (17). These molds are pathogenic in humans. They may cause asymptomatic colonization or localized or disseminated infection following trauma, surgery, and immunosuppression (17).
Infections by S. prolificans have been described more recently than those by S. apiospermum. Initially S. prolificans was found to cause mainly bone and soft tissue infections in immunocompetent individuals. However, the importance of S. prolificans has increased in the last few years, as it can cause disseminated infections, specifically in neutropenic patients (2, 7). Most reports of these infections have been from Belgium, Australia, Spain, and North America. The causes for this highly localized prevalence are not yet known, although climate conditions could be related (16).
Invasive Scedosporium infections are characterized by high mortality and poor response to antifungal agents. The majority of the available antifungal drugs show low in vitro activity against S. apiospermum, and only some new triazoles, such as voriconazole and posaconazole, seem to be active in vitro (10). On the other hand, S. prolificans is a multiresistant fungus, tolerating virtually all systemically active antifungal agents, including the new triazoles and echinocandins (5, 6, 10). Because of the usually fatal outcome of these infections, a correct and early diagnosis and identification of these fungi are necessary.
Current diagnosis methods for Scedosporium infections have some limitations. In tissue sections, these species appear as septated and branched hyphae and can be easily misidentified as Aspergillus, Fusarium, and even species of black fungi. A technique based on hybridization in situ using DNA probes has been described to improve these methods (12). In addition, culture and identification of the fungus are necessary to perform a correct diagnosis, but other morphologically similar species can make the classification of these fungi difficult (9). Serological methods like immunodiffusion tests are not commercially available yet (10, 21). An assay based on the detection of a peptidorhamnomannan antigen for S. apiospermum is available (15); however, cross-reactivity with other fungal species has been reported (13). Finally, conventional PCR assays have been developed to detect Scedosporium DNA (1, 11, 22), but to date no methods based on quantitative PCR methodologies to detect these species have been described.
The aim of this study was to develop a rapid and sensitive technique for the detection of Scedosporium spp. in cultures and from clinical samples. On that basis, two specific real-time PCR (RT-PCR)-based assays for the detection of S. apiospermum and S. prolificans DNA were developed. Each assay used a fluorescently labeled molecular beacon probe, which allows a specific and quantitative detection of Scedosporium DNA without postamplification manipulation steps. The two RT-PCR assays were validated in vitro and in a murine model of invasive infection. The sensitivities, specificities, and reproducibilities of both methodologies were evaluated.
(This work was presented in part at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 2006.)
MATERIALS AND METHODS
Design of the RT-PCR-based assay. (i) Strains.
Six clinical strains of S. prolificans and nine strains of S. apiospermum (including P. boydii and Graphium fructicola) belonging to the collection of the Spanish National Center of Microbiology (CNM-CM) were used. The specificity of the technique was assessed, as other fungal species were included in the study. Clinical strains employed were as follows: Aspergillus fumigatus (CNM-CM-AF237), Aspergillus flavus (CNM-CM-2669), Aspergillus terreus (CNM-CM-2013), Fusarium verticillioides (CNM-CM-2975), Fusarium oxysporum (CNM-CM-2914), and Candida albicans (ATCC 64551). In addition, human and murine genomic DNAs (Promega, Madrid, Spain) were included in the experiments.
(ii) DNA extraction.
DNA extraction from cultured organisms was done as described previously by Tang et al. (21).
(iii) Primer and probe design.
Primers and molecular beacon probes were designed on the basis of the nucleotide sequence of the internal transcribed spacer (ITS) ribosomal DNA (rDNA) region for 20 strains of S. prolificans and 15 strains of S. apiospermum by use of the Beacon Designer 4.0 software (Premier Biosoft, Palo Alto, CA). Primers and probes selected were subjected to a BLAST search in the GenBank sequence database (http://www.ncbi.nlm.nih.gov/GenBank/) and in the database of the Department of Mycology of the Spanish National Center for Microbiology (more than 3,000 distinct sequences) to avoid cross-homology with other microorganisms, including dematiaceous species. All primers and probes were species specific, except for primers Sp1 and Sp2, which were designed in a conserved region (rDNA 18S and 5.8S). Analysis was done with the help of Fingerprinting II Informatix software, version 3.0 (Bio-Rad, Madrid, Spain). The oligonucleotide sequences of primers and probes are shown in Table 1.
TABLE 1.
Sequences of the primers and probes used in RT-PCR
| Organism assayed | RT-PCR primer or probe
|
|
|---|---|---|
| Name | Sequence (5′-3′)a | |
| S. apiospermum | Sap1 (forward primer) | 5′-CCATTGTGAACCTTACCTATG-3′ |
| Sap2 (reverse primer) | 5′-TCAAATCAGAACTGTAATCCG-3′ | |
| Sap-MB1 (probe) | FAM-CGCGATACGCCGCCGAGGCAACAGATCGCG-BHQ1 | |
| S. prolificans | Sp1 (forward primer) | 5′-AACCCTTTGTGAACCTTACC-3′ |
| Sp2 (reverse primer) | 5′-CGATGCCAGAACCAAGAG-3′ | |
| Sp-MB1 (probe) | FAM-CGCGATCCACCAAACTCTTGCATTTATAGCGGATTAGATCGCG-BHQ1 | |
Underlining indicates the PCR target sequence.
(iv) PCR assay.
PCRs were performed in the Chromo 4 system (MJ Research, Bio-Rad). The kit 2× SensiMix DNA (Quantace, Ecogen, Madrid, Spain) was used as described by the manufacturer. The PCR mixture (20 μl) contained 0.5 μM of each of the primers, 4.5 mM MgCl2, and 0.4 μM of the probe labeled with 5′ 6-carboxyfluorescein (FAM) and 3′ Black Hole Quencher 1 (BHQ1) (Sigma Genosys, Spain). Each reaction mixture contained 18 μl of the master mix and a 2-μl aliquot of DNA from the extracted sample. The cycling conditions included a first step for preincubation (activation of the enzyme) and denaturation of the template DNA at 95°C for 10 min. The next steps included an amplification program of 40 cycles as follows: denaturation at 95°C for 15 s, annealing at 54°C for S. apiospermum or 52°C for S. prolificans at 15 s, and extension at 72°C for 5 s. Quantification standards were run in conjunction with each set of samples. The amplicons generated were 132 bp for S. apiospermum and 176 bp for S. prolificans.
PCR products were subjected to electrophoresis in 2% agarose gels (Pronadisa, Madrid, Spain) following the protocols of Sambrook et al. (18) to confirm the PCR results. Amplified fragments were sequenced (ABI Prism 377 DNA sequencer; Applied Biosystems, Madrid, Spain), and the obtained sequences were compared to an S. prolificans and S. apiospermum ITS1 sequence database available in the laboratory. Each PCR run contained both negative and positive controls consisting of water and different concentrations of genomic DNA from S. prolificans (CMN-CM-1627) and S. apiospermum (CMN-CM-3169).
(v) Standardization.
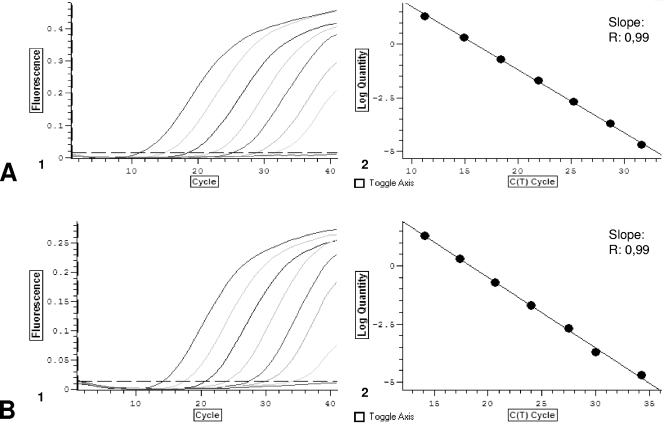
Standard curves were constructed with PCR results from five repetitions of different dilutions of S. prolificans (CMN-CM-1627) and S. apiospermum (CMN-CM-3169) genomic DNA. Dilutions ranged between 10 ng and 1 fg DNA/μl. The crossing point (threshold cycle [CT]; cycle at which fluorescence becomes detectable above background) values were plotted against the logarithmically converted DNA concentrations, and a linear regression coefficient was calculated (Fig. 1). An average value and 99% confidence interval for crossing-point values obtained for each DNA concentration were calculated. The reproducibility was defined as the percentage of values inside the confidence interval. In addition, a range of assay validation was calculated per DNA concentration, taking into account the average values and the confidence intervals.
FIG. 1.
(A) S. prolificans RT-PCR. (1) Amplification plot obtained for dilutions of standard DNA (from 20 ng to 10 fg/20 μl). (2) Standard curve obtained plotting CTs against the logarithmically converted DNA concentration. (B) S. apiospermum RT-PCR. (1) Amplification plot obtained for dilutions of standard DNA (from 20 ng to 10 fg/20 μl). (2) Standard curve obtained plotting CTs against the logarithmically converted DNA concentration.
RT-PCR assays of cultured clinical strains.
The utility of the assays of cultured clinical strains was evaluated. DNA extraction and RT-PCRs were performed as described above. Two microliters of the extracted DNA was used in each reaction.
Murine model.
A standardized murine model was performed based on the model described by Smith et al. (19). Male 6- to 7-week-old ICR (CD1 specific-pathogen-free) mice (Criffa, Barcelona, Spain) weighing about 30 g each at the time of inoculation were used in the experiment. Upon arrival, mice were separated into groups of up to eight per cage, housed in covered plastic cages with filters, and allocated in a ventilation rack with HEPA filters and a positive ventilation system (Techniplast, Barcelona, Spain). Sterilized food, bedding, and bottles were used throughout the experiment. Sterilized tap water with tetracycline at 1 mg/ml (Sigma-Aldrich, Madrid, Spain) was also used to avoid bacterial contamination. All procedures with mice were performed in accordance with the Real Decreto 223/1988 for the protection of experimental animals.
The following groups were studied: (i) healthy controls, (ii) immunosuppressed control mice, and (iii) immunosuppressed infected mice. Immunosuppression was performed as follows. Cyclophosphamide (Genoxal; Prasfarma, Barcelona, Spain) at 200 mg/kg of body weight was administered intraperitoneally in 0.2 ml of saline buffer on days −3 and every 3 days until study completion (day 14). Cortisone acetate (Sigma-Aldrich) was administered at 112.5 mg/kg subcutaneously in 0.1 ml of 0.02% Tween 80 solution on days −3 and −1. S. apiospermum (CNM-CM-3169) and S. prolificans (CMN-CM-1627) were used in the experiment. For preparation of the inoculum, the organisms were subcultured in agar potato dextrose tubes (TEC-Laim, Madrid, Spain) and incubated at 37°C for 3 to 5 days. Conidia were harvested with 5 ml of 0.85% sterile saline-0.01% Tween 80 solution. The suspension was filtered through an 11-μm filter and then adjusted in order to give each mouse 3 × 104 conidia of S. apiospermum or S. prolificans in a final volume of 30 μl. Inoculation was performed intranasally on day 0 under general anesthesia with 0.1 ml of a mixture of a 9:1 (vol/vol) mixture of 12.5 mg/ml ketamine (Ketolar, 50 mg/ml; Parke-Davis S.L., Madrid, Spain) and xylazine (Rompum, 2%; Quimica Farmaceutica Bayer S.A., Barcelona, Spain). A total of 41 mice were inoculated with S. apiospermum, and 51 were inoculated with S. prolificans. Cages were checked twice daily for dead or moribund mice. Animals were euthanized when symptoms of pulmonary or disseminated infection were detected. Any animal that had severely reduced mobility, that was unable to reach the drinker, or that was otherwise in substantial distress was sacrificed by intracardiac puncture under general anesthesia. Lung, whole-blood, and serum samples were recovered under aseptic conditions. For animals that were found dead, no blood or serum samples could be recovered.
Lung tissues were homogenized in 2 ml of 0.85% sterile saline. An aliquot of this homogenate was cultured in Sabouraud dextrose chloramphenicol gentamicin (Oxoid, Madrid, Spain) and blood agar (Oxoid) plates and incubated at 30°C. The rest of the homogenate was stored at −20°C until DNA extraction was performed.
RT-PCR assays of animal samples.
DNA extraction from mice samples (blood, serum, and lung tissue) was done using the QIAamp DNA kit (Qiagen, Izasa, Madrid, Spain) following the manufacturer's recommendations. Elution was performed in 50 μl of elution buffer. All samples were stored at −20°C, and they were allowed to thaw at room temperature before testing. PCRs were performed as described above. Positive controls with Scedosporium DNA and negative controls with murine DNA were routinely included.
RESULTS
In vitro sensitivities, reproducibilities, and specificities of the RT-PCR assays.
The designed PCR-based assays specifically detected S. prolificans and S. apiospermum DNA. The sensitivities for both assays were 10 fg of DNA per μl of sample. The determination coefficient (r2) of the linear regression between crossing-point values and different dilutions of genomic DNA was 0.99 (P < 0.01). The average coefficients of variation were 3.3% and 3.5% for S. prolificans and S. apiospermum PCR assays, respectively.
The specificities for both PCR-based assays were 100%. No positive signal was detected when 2 ng of DNA from other fungi (different species of Aspergillus, Fusarium, and Candida) and genomic DNA from human and mouse were tested. In addition, no cross-reaction was observed between Scedosporium species.
RT-PCR assays for cultured clinical strains.
RT-PCR results were positive for all cultured clinical strains tested. The PCR-based assays specifically detected DNA from eight clinical strains of S. apiospermum and six of S. prolificans. Sequencing of the amplified fragments confirmed the RT-PCR results. The crossing-point values varied according to the amount of DNA obtained for each strain.
Animal model.
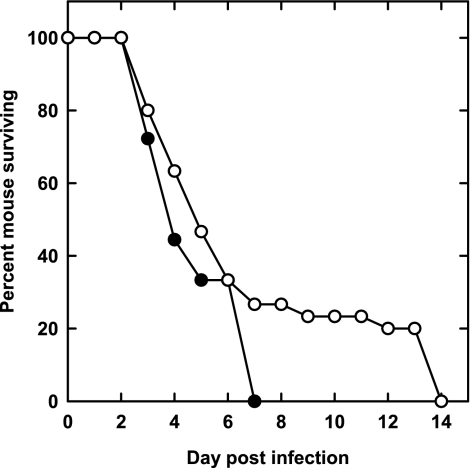
Infection in mice was confirmed when lung tissues were positive in culture. From a total of 51 mice inoculated with S. prolificans (CMN-CM-1627), 45 had proven infections. Similarly, for S. apiospermum, 36 mice from a total of 41 inoculated developed infection. Figure 2 shows the survival curves for the two murine models. Control and immunosuppression groups had no symptoms of infection and were sacrificed on the 14th day. A total of 141 samples from infected mice were tested, and the number and type per animal model are stated below.
FIG. 2.
Survival curves of mice infected with 3 × 104 CFU of S. prolificans (○) and S. apiospermum (•).
RT-PCR results in samples from the Scedosporium prolificans murine model.
RT-PCR was performed for serum (n = 18), blood (n = 20), and lungs (n = 45) from S. prolificans-infected mice. The assay was positive for 43 of 45 lung samples (95.5%), for 17 of 20 blood samples (85%), and for 15 of 18 serum samples (83.3%). The average DNA concentrations per μl of samples were 230 pg in lungs, 15 fg in blood, and 4.6 fg in sera. Samples from control and immunosuppression groups were negative in the RT-PCR assay.
RT-PCR results in samples from the Scedosporium apiospermum murine model.
Samples of serum (n = 11), blood (n = 11), and lungs (n = 36) from S. apiospermum-infected mice were collected, and RT-PCR assays were performed. Results were positive for 35 of 36 lung samples (97.2%), 6 of 11 blood samples (54.5%), and 9 of 11 serum samples (81.8%). The average DNA concentrations per μl of samples were 600 pg in lungs, 12 fg in blood, and 21 fg in sera. Samples from control and immunosuppression groups were negative in the RT-PCR assay.
DISCUSSION
There are very few reports that describe molecular diagnostic methods for S. apiospermum and S. prolificans. Wedde et al. (22) developed a conventional PCR assay that consisted in the amplification of an rDNA region and detection with hybridization probes for the clinically relevant Scedosporium species. Other authors designed PCR-based assays for the detection of Scedosporium DNA from infected tissues (11, 14). Recently, a microarray assay for the detection of fungal DNA from 14 pathogen species, including S. prolificans, has been designed (20). These assays rely on previous fungal isolation or on additional techniques such as gel electrophoresis or DNA sequencing. In addition, to date there are very few data on the sensitivity and validation of these techniques in clinical samples.
We have developed two RT-PCR-based assays using molecular beacon probes targeting the ITS1 region of rDNA for the detection of S. prolificans and S. apiospermum DNA. The two RT-PCR assays were specific (100%) and had good sensitivities and reproducibilities.
The developed techniques were tested first for six cultured clinical strains of S. prolificans and eight of S. apiospermum. All Scedosporium strains gave positive results in each specific assay, while other control species included in the assay were negative. These results show one of the clinical uses of these techniques, that is, the correct classification of cultured strains. Both species appear as filamentous fungi with unspecified septated branched hyphae and can be easily misidentified as other species of hyaline or black fungi.
In order to validate both RT-PCR assays, animal model experiments were done. Other animal models of disseminated Scedosporium infection have been previously reported (3, 4, 8). However, here we described an easy and reproducible method using intranasal inoculation. Blood, serum, and lung samples from infected mice were recovered to perform the RT-PCR assays. In the S. prolificans model, the PCR assay was positive in 95.5% of lung samples, 83.3% of serum samples, and 85% of blood samples. Similarly, in the S. apiospermum model, the PCR assay was positive in a very high percentage of samples of lung (97.2%) and serum (81.8%), but sensitivity was lower (54.5%) in blood samples, probably due to the presence of inhibitors. However, new studies including an internal control should be carried out to detect inhibition of the PCR.
In summary, the methods described here allow a quick and sensitive detection of S. apiospermum and S. prolificans. The differentiation between these two species is clinically important, as they show marked differences in their in vitro susceptibilities to the currently used antifungal agents. These assays allow a rapid confirmation of cultured strains suspected to be Scedosporium and differentiation from other fungi. The use of these techniques in clinical samples will contribute to the establishment of an early diagnosis, which is necessary for successful treatment.
Acknowledgments
This work was supported by research project grants from Fundación Ramón Areces (MPY 1397/04), from Instituto de Salud Carlos III (MPY 1279/05), and from RED Española de Investigación en Patología Infecciosa (REIPI, MPY 1022/07). M. V. Castelli has a research contract from AECI (Agencia Española de Cooperación Internacional). L Bernal-Martinez has a research contract from REIPI (Red Española de Investigación de Patología Infecciosa, project MPY 1022/07_1).
We declare that we have no potential conflicts of interest.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Bagyalakshmi, R., K. L. Therese, S. Prasanna, and H. N. Madhavan. 2008. Newer emerging pathogens of ocular nonsporulating molds (NSM) identified by polymerase chain reaction (PCR)-based DNA sequencing technique targeting internal transcribed spacer (ITS) region. Curr. Eye Res. 33139-147. [DOI] [PubMed] [Google Scholar]
- 2.Berenguer, J., J. L. Rodriguez-Tudela, C. Richard, M. Alvarez, M. A. Sanz, L. Gaztelurrutia, J. Ayats, J. V. Martinez-Suarez, et al. 1997. Deep infections caused by Scedosporium prolificans. A report on 16 cases in Spain and a review of the literature. Medicine (Baltimore) 76256-265. [DOI] [PubMed] [Google Scholar]
- 3.Cano, J., J. Guarro, E. Mayayo, and J. Fernandez-Ballart. 1992. Experimental infection with Scedosporium inflatum. J. Med. Vet. Mycol. 30413-420. [PubMed] [Google Scholar]
- 4.Capilla, J., C. Serena, F. J. Pastor, M. Ortoneda, and J. Guarro. 2003. Efficacy of voriconazole in treatment of systemic scedosporiosis in neutropenic mice. Antimicrob. Agents Chemother. 473976-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuenca-Estrella, M., A. Gomez-Lopez, E. Mellado, M. J. Buitrago, A. Monzon, and J. L. Rodriguez-Tudela. 2006. Head-to-head comparison of the activities of currently available antifungal agents against 3,378 Spanish clinical isolates of yeasts and filamentous fungi. Antimicrob. Agents Chemother. 50917-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuenca-Estrella, M., B. Ruiz-Diez, J. V. Martinez-Suarez, A. Monzon, and J. L. Rodriguez-Tudela. 1999. Comparative in-vitro activity of voriconazole (UK-109,496) and six other antifungal agents against clinical isolates of Scedosporium prolificans and Scedosporium apiospermum. J. Antimicrob. Chemother. 43149-151. [DOI] [PubMed] [Google Scholar]
- 7.de Batlle, J., M. Motje, R. Balanza, R. Guardia, and R. Ortiz. 2000. Disseminated infection caused by Scedosporium prolificans in a patient with acute multilineal leukemia. J. Clin. Microbiol. 381694-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez, G. M., R. Tijerina, L. Najvar, M. Rinaldi, I. T. Yeh, and J. R. Graybill. 2002. Experimental murine model of disseminated Pseudallescheria infection. Med. Mycol. 40243-248. [DOI] [PubMed] [Google Scholar]
- 9.Guarro, J., and J. Gene. 2002. Acrophialophora fusispora misidentified as Scedosporium prolificans. J. Clin. Microbiol. 403544. [PubMed] [Google Scholar]
- 10.Guarro, J., A. S. Kantarcioglu, R. Horre, J. L. Rodriguez-Tudela, E. M. Cuenca, J. Berenguer, and G. S. de Hoog. 2006. Scedosporium apiospermum: changing clinical spectrum of a therapy-refractory opportunist. Med. Mycol. 44295-327. [DOI] [PubMed] [Google Scholar]
- 11.Hagari, Y., S. Ishioka, F. Ohyama, and M. Mihara. 2002. Cutaneous infection showing sporotrichoid spread caused by Pseudallescheria boydii (Scedosporium apiospermum): successful detection of fungal DNA in formalin-fixed, paraffin-embedded sections by seminested PCR. Arch. Dermatol. 138271-272. [DOI] [PubMed] [Google Scholar]
- 12.Hayden, R. T., P. A. Isotalo, T. Parrett, D. M. Wolk, X. Qian, G. D. Roberts, and R. V. Lloyd. 2003. In situ hybridization for the differentiation of Aspergillus, Fusarium, and Pseudallescheria species in tissue section. Diagn. Mol. Pathol. 1221-26. [DOI] [PubMed] [Google Scholar]
- 13.Jabado, N., J. L. Casanova, E. Haddad, F. Dulieu, J. C. Fournet, B. Dupont, A. Fischer, C. Hennequin, and S. Blanche. 1998. Invasive pulmonary infection due to Scedosporium apiospermum in two children with chronic granulomatous disease. Clin. Infect. Dis. 271437-1441. [DOI] [PubMed] [Google Scholar]
- 14.Lau, A., S. Chen, T. Sorrell, D. Carter, R. Malik, P. Martin, and C. Halliday. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J. Clin. Microbiol. 45380-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinto, M. R., B. Mulloy, R. M. Haido, L. R. Travassos, and B. E. Barreto. 2001. A peptidorhamnomannan from the mycelium of Pseudallescheria boydii is a potential diagnostic antigen of this emerging human pathogen. Microbiology 1471499-1506. [DOI] [PubMed] [Google Scholar]
- 16.Ponton, J., R. Ruchel, K. V. Clemons, D. C. Coleman, R. Grillot, J. Guarro, D. Aldebert, P. Ambroise-Thomas, J. Cano, A. J. Carrillo-Munoz, J. Gene, C. Pinel, D. A. Stevens, and D. J. Sullivan. 2000. Emerging pathogens. Med. Mycol. 38(Suppl. 1)225-236. [DOI] [PubMed] [Google Scholar]
- 17.Rippon, J. W. 1988. Pseudallescheriasis, p. 651-680. In J. W. Rippon (ed.), Medical mycology: the pathogenic fungi and the pathogenic actinomycetes. W. B. Saunders Co., Philadelphia, PA.
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Smith, J. M., C. M. Tang, S. Van Noorden, and D. W. Holden. 1994. Virulence of Aspergillus fumigatus double mutants lacking restriction and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 625247-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiess, B., W. Seifarth, M. Hummel, O. Frank, A. Fabarius, C. Zheng, H. Morz, R. Hehlmann, and D. Buchheidt. 2007. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. J. Clin. Microbiol. 453743-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang, C. M., J. Cohen, and D. W. Holden. 1992. An Aspergillus-fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol. Microbiol. 61663-1671. [DOI] [PubMed] [Google Scholar]
- 22.Wedde, M., D. Muller, K. Tintelnot, G. S. de Hoog, and U. Stahl. 1998. PCR-based identification of clinically relevant Pseudallescheria/Scedosporium strains. Med. Mycol. 3661-67. [PubMed] [Google Scholar]