Abstract
A portable molecular diagnostic laboratory was used to provide molecular confirmation of suspected melioidosis cases seen at Peradeniya Hospital, central Sri Lanka. Soil supernatants from rice field and rubber plantation samples also produced PCR-positive results. These procedures could be used for melioidosis field work in other remote locations.
Melioidosis was first reported in Sri Lanka in the 1920s (2). Sporadic cases have been reported since then, mainly for travelers returning to European countries (14). The Indian Ocean tsunami on Boxing Day, 2004, caused an increase in melioidosis cases and may have extended the distribution of the disease in the region (1). Two years ago an indigenous case in the Colombo area was reported (8), and several potential cases in Colombo and Peradeniya were noted. At that time, the local laboratory was unable to confirm an etiologic diagnosis. We had previously assisted the public health authorities in northeastern Brazil with an investigation into a fatal outbreak of melioidosis (15) through a series of visits in which we introduced field investigation skills, a standard case definition, and molecular diagnostic methods (6, 11). In the Peradeniya area of Sri Lanka, diagnostic microbiology services rely on classical bacteriologic methods, which are prone to a high degree of error (7). We drew on our recent experience with a deployable molecular microbiology laboratory in the remote Kimberley region of Western Australia. The methods used were developed using laboratory equipment that was sufficiently robust to operate in a severe environment, namely, a real-time TaqMan PCR assay developed in our central laboratory (11) but with amplification using a conventional thermal cycler and resolution by capillary electrophoresis on a laboratory chip device (Bioanalyzer 2100; Agilent Technologies, Little Falls, DE).
Suspected Burkholderia pseudomallei isolates were obtained from patients with gram-negative septicemia seen in the University Hospital in Peradeniya. One patient was from Central Province, one was from Uva Province, and one was from Colombo. A total of four isolates collected from the three patients were maintained on agar slants after the initial isolation. These isolates were subcultured onto blood agar and incubated at 37°C for 48 h to obtain pure growth and distinct colonies. Environmental isolates were obtained by adding 100 g of soil (collected from a depth of at least 30 cm) to 100 ml of sterile, distilled water. A 1.0-ml aliquot of soil supernatant was transferred after 18 to 24 h into Trypticase soy broth, vortex mixed, and incubated at 37°C for 24 h. A 100-μl aliquot of supernatant was subcultured to freshly prepared Burkholderia pseudomallei selective agar (3) and incubated for 48 h at 37°C, after which the plates were examined. Colonies were picked and subcultured onto blood agar. Bacterial DNA was extracted by hot alkaline lysis in 100 μl of lysis buffer-NaOH-sodium dodecyl sulfate for 15 min at 95°C and diluted in ultrapure water. A previously described B. pseudomallei-B. mallei-specific real-time PCR assay was used to produce amplicons without fluorescence acquisition (11). Briefly, 12 μl of lpxO TaqMan master mix was dispensed into thin-walled PCR tubes, and 8 μl of bacterial or soil supernatant extract was added to each tube. Samples were cycled on a thermal cycler (model 2720; Applied Biosystems, Singapore) with negative and positive controls. On completion, the amplified product was analyzed by capillary electrophoresis. Control, molecular ladder, and test samples were processed using a microfluidic laboratory chip (DNA 1000 kit; BioAnalyzer 2100, Agilent Technologies, Waldbronn, Germany). Analysis was completed by comparing products observed in the gel-like image with those seen with the positive control. Two environmental sample locations were used, one in Peradeniya close to historic plantings of Pará rubber trees (Hevea brasiliensis) for a preliminary sample run and a second location where a more detailed environmental survey of the rhizosphere of plantation rubber trees, banana palms, and rice terraces was conducted.
The deployable molecular diagnostic laboratory was successfully set up in Peradeniya in January 2008 and ran for 1 week until all molecular reagents were exhausted. One of the clinical B. pseudomallei isolates was run at a series of 10-fold dilutions to determine the sensitivity of the assay and to detect any PCR inhibition. A positive result was obtained at a dilution of 1 in 106 and used to provide a positive control for all subsequent studies (Fig. 1). Results from the series of bacterial DNA extracts provided preliminary molecular confirmation of the presence of B. pseudomallei in two of the three patient isolates (Table 1). None of the isolates from environmental samples were lpxO positive, but direct testing of soil suspension supernatants from the rice terrace and rubber plantation samples did produce several positive results.
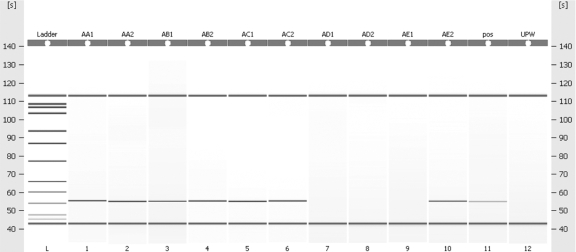
FIG. 1.
Lane L contains the DNA ladder. Lanes 1 to 10 contain DNA extracts from bacteria isolated from patient samples at undiluted and 10−1 dilution. Lane 11 contains a positive control at a dilution of 10−6, and lane 12 contains the nontemplate control (ultrapure water [UPW]). The product demonstrated is an lpxO PCR amplicon of 110 bp in length. pos, positive.
TABLE 1.
Summary of deployable laboratory results
| Samplea | Typeb | Location in Australia, source | Culture resultc | lpxO PCR result |
|---|---|---|---|---|
| A | Clinical, blood | Central Province | B. pseudomallei | + |
| B | Clinical, synovial fluid | Central Province | B. pseudomallei | + |
| C | Clinical, blood | Uva Province | B. pseudomallei | + |
| D | Clinical, blood | Colombo | Unidentified GNB | − |
| E | Clinical, blood | Colombo | Unidentified GNB | − |
| AA | Env, SS | Kandy area, rubber tree | NA | + |
| AB | Env, SS | Kandy area, rubber tree | NA | − |
| AC | Env, SS | Kandy area, banana palm | NA | + |
| AD | Env, SS | Kandy area, rice terrace | NA | + |
| AE | Env, SS | Kandy area, rice terrace | NA | − |
| BA | Env, BPSA isolate | Kandy area | Unidentified GNB | − |
| BB | Env, BPSA isolate | Kandy area | Unidentified GNB | − |
| BC | Env, BPSA isolate | Kandy area | Unidentified GNB | − |
| BD | Env, BPSA isolate | Kandy area | Unidentified GNB | − |
| BE | Env, BPSA isolate | Kandy area | Unidentified GNB | − |
Serial samples A to E, clinical isolates; AA to AE, soil supernatants; BA to BE, preliminary isolates from soil supernatants on B. pseudomallei selective agar. All serial samplings were repeated at least once.
Env, environmental; SS, soil supernatant; BPSA, B. pseudomallei selective agar.
The duration of laboratory deployment did not permit preliminary identification of soil bacteria by conventional phenotypic laboratory methods. The PCR protocol was therefore used as a rapid screening method for identification of oxidase-positive, gentamicin-resistant, gram-negative bacilli. GNB, gram-negative bacillus; NA, not applicable.
This is the first time that molecular confirmation of a diagnosis of melioidosis in Sri Lanka has been obtained. We set up and ran a portable molecular diagnostic laboratory in Peradeniya, Sri Lanka, processing bacterial DNA extracts over 4 days by the use of six consecutive laboratory chips (72 extracts analyzed). This provided preliminary molecular confirmation of two cases of melioidosis for patients admitted to the Peradeniya Hospital. Supernatants from soil samples from rice field and rubber plantation locations produced PCR-positive results, though no PCR-positive bacterial isolates were obtained from primary environmental cultures. Local staff had direct, hands-on experience in these methods and demonstrated proficiency in running the standard procedures.
Recognition of B. pseudomallei in samples from sterile cultures such as venous blood is straightforward in regions where laboratory staff are familiar with the species, but inconsistently prepared agar or a lack of familiarity with its appearance on nonselective media can cause problems at the identification stage (13). In recent years, an increasing number of centers have turned to molecular methods to confirm the identity of suspected B. pseudomallei isolates or to detect DNA of the species directly in investigations of clinical samples (4, 10-12). The use of the lpxO PCR protocol as a single-round method gave confirmation of an etiological diagnosis in one case of successfully treated septicemia and one case of fatal septicemia of previously unknown etiology. In Perth, Western Australia, we are required to provide an additional level of diagnostic certainty by obtaining a positive result with multiple PCR probes, a homologous recA sequence, or detection of 2-hydroxymyristic acid by gas liquid chromatography of bacterial fatty acid methyl esters (4). As these advanced methods are not currently available in Sri Lanka, our lpxO results are provisional and subject to confirmation by additional reference methods. The short time allowed for the laboratory deployment did not allow culture-based analysis of soil to run to conclusion—a period of around 3 to 4 weeks. It was therefore not possible to determine whether or not the lpxO-positive results obtained with soil supernatants from rice terrace and rubber plantation samples represented culturable B. pseudomallei. This potentially more sensitive method may have detected bacterial DNA in the absence of culturable bacteria, in similarity to results reported previously from the Northern Territory of Australia (9). Though we are cautious in our interpretation in the absence of culture-based data, these results indicate that rubber plantations in Sri Lanka are worth further investigation as a possible environmental source of B. pseudomallei. We proposed previously that rubber may have acted as a vehicle for the transfer of B. pseudomallei from the Americas (5). In conclusion, we have demonstrated the feasibility of assembling, preparing, and deploying a molecular diagnostic laboratory overseas.
Acknowledgments
The project is funded by the World Health Organization via a laboratory capacity-building program.
We are grateful to Agilent Technologies Australia Pty. for providing the Expert 2100 BioAnalyzer and laboratory chip reagents used in this project. We thank Shalinie Perera for assistance with movements and other logistic support in Sri Lanka, our colleagues at PathWest and the University of Peradeniya for their assistance in preparing for the laboratory deployment, and Avram Levy for his advice during manuscript preparation.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Chastel, C. 2007. Assessing epidemiological consequences two years after the tsunami of 26 December 2004? Bull. Soc. Pathol. Exot. 100139-142. (In French.) [PubMed] [Google Scholar]
- 2.Denny, C. R., and L. Nicholls. 1927. Melioidosis in a European. Ceylon J. Sci. 237-40. [Google Scholar]
- 3.Howard, K., and T. J. Inglis. 2003. Novel selective medium for isolation of Burkholderia pseudomallei. J. Clin. Microbiol. 413312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inglis, T. J., A. Merritt, G. Chidlow, M. Aravena-Roman, and G. Harnett. 2005. Comparison of diagnostic laboratory methods for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 432201-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglis, T. J., and J.-L. Sagripanti. 2006. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 726865-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inglis, T. J., D. B. Rolim, and J. L. Rodriguez. 2006. Clinical guideline for diagnosis and management of melioidosis. Rev. Inst. Med. Trop. São Paulo 481-4. [DOI] [PubMed] [Google Scholar]
- 7.Inglis, T. J., D. Chiang, G. S. Lee, and L. Chor-Kiang. 1998. Potential misidentification of Burkholderia pseudomallei by API 20NE. Pathology 3062-64. [DOI] [PubMed] [Google Scholar]
- 8.Jayasekara, K., S. Perera, and A. Wijesundere. 2006. Fatal Burkholderia pseudomallei septicaemia. Ceylon Med. J. 5169-70. [PubMed] [Google Scholar]
- 9.Kaestli, M., M. Mayo, G. Harrington, F. Watt, J. Hill, D. Gal, and B. J. Currie. 2007. Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Appl. Environ. Microbiol. 736891-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunakorn, M., and R. B. Markham. 1995. Clinically practical seminested PCR for Burkholderia pseudomallei quantitated by enzyme immunoassay with and without solution hybridization. J. Clin. Microbiol. 332131-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merritt, A., T. J. Inglis, G. Chidlow, and G. Harnett. 2006. PCR-based identification of Burkholderia pseudomallei. Rev. Inst. Med. Trop. São Paulo 48239-244. [DOI] [PubMed] [Google Scholar]
- 12.Meumann, E. M., R. T. Novak, D. Gal, M. E. Kaestli, M. Mayo, J. P. Hanson, E. Spencer, M. B. Glass, J. E. Gee, P. P. Wilkins, and B. J. Currie. 2006. Clinical evaluation of a type III secretion system real-time PCR assay for diagnosing melioidosis. J. Clin. Microbiol. 443028-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miralles, I. S., M. do Carmo Alves Maciel, M. R. F. Angelo, M. M. Gondini, L. H. F. Frota, C. M. F. dos Reis, and E. Hofer. 2004. Burkholderia pseudomallei: a case report of a human infection in Ceará, Brazil. Rev. Inst. Med. Trop. Sao Paulo 4651-54. [DOI] [PubMed] [Google Scholar]
- 14.Peetermans, W. E., E. van Wijngaerden, J. van Eldere, and J. Verhaegen. 1999. Melioidosis brain and lung abscess after travel to Sri Lanka. Clin. Infect. Dis. 28921-922. [DOI] [PubMed] [Google Scholar]
- 15.Rolim, D. B., D. C. F. L. Vilar, A. Q. Sousa, I. S. Miralles, D. C. A. de Oliveira, G. Harnett, L. O'Reilly, K. Howard, I. Sampson, and T. J. J. Inglis. 2005. Melioidosis, northeastern Brazil. Emerg. Infect. Dis. 111458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]