Abstract
The detection and successful typing of dengue virus (DENV) from patients with suspected dengue fever is important both for the diagnosis of the disease and for the implementation of epidemiologic control measures. A technique for the multiplex detection and typing of DENV serotypes 1 to 4 (DENV-1 to DENV-4) from clinical samples by PCR-ligase detection reaction (LDR) has been developed. A serotype-specific PCR amplifies the regions of genes C and E simultaneously. The two amplicons are targeted in a multiplex LDR, and the resultant fluorescently labeled ligation products are detected on a universal array. The assay was optimized using 38 DENV strains and was evaluated with 350 archived acute-phase serum samples. The sensitivity of the assay was 98.7%, and its specificity was 98.4%, relative to the results of real-time PCR. The detection threshold was 0.017 PFU for DENV-1, 0.004 PFU for DENV-2, 0.8 PFU for DENV-3, and 0.7 PFU for DENV-4. The assay is specific; it does not cross-react with the other flaviviruses tested (West Nile virus, St. Louis encephalitis virus, Japanese encephalitis virus, Kunjin virus, Murray Valley virus, Powassan virus, and yellow fever virus). All but 1 of 26 genotypic variants of DENV serotypes in a global DENV panel from different geographic regions were successfully identified. The PCR-LDR assay is a rapid, sensitive, specific, and high-throughput technique for the simultaneous detection of all four serotypes of DENV.
The dengue virus (DENV), a mosquito-borne flavivirus, consists of four closely related but genetically distinct antigenic serotypes: DENV serotype 1 (DENV-1), DENV-2, DENV-3, and DENV-4. It is tropical and subtropical in distribution and is prevalent in Asia, Africa, and Central and South America (45). Infection with any of the four serotypes of DENV may cause a mild febrile illness, dengue fever (DF). In some cases, however, more-severe manifestations, such as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), occur; these may prove fatal without proper early intervention (15).
Geographic spread of both the mosquito vector and the virus over the past 25 years has led to the increased occurrence of epidemic DF/DHF/DSS, making dengue a major global health problem. The disease is endemic in more than 100 countries, with an estimated 2.5 billion people at risk of infection. It is estimated that 50 million DENV infections occur each year, with 500,000 cases of DHF and at least 22,000 deaths, mainly in children (31, 32, 45; WHO/WPRO/SEARO meeting on DengueNet implementation in Southeast Asia and the Western Pacific, Kuala Lumpur, Malaysia, 11 to 13 December 2003).
DENV infection confers lifelong serotype-specific immunity. Multiple infections with different DENV serotypes occur in regions of hyperendemicity (31, 35). Secondary infections with a different DENV serotype are major risk factors for DHF and DSS (13, 14, 39) due to antibody-dependent enhancement of disease (35). Serotype identification and the differentiation of primary and secondary infections are therefore important both for patient management and for the implementation of public health measures (26, 33).
The diagnosis of DENV infection and the typing of DENV serotypes can be confirmed using viral isolation techniques, serology, or molecular methods. Virus isolation is the gold standard for detection but requires 7 to 10 days and is often insensitive (26). Serological tests for the detection of viral antibodies, such as immunoglobulin M and immunoglobulin G antibody capture enzyme-linked immunosorbent assays, require the demonstration of a rise in antibody titer from an acute-phase to a convalescent-phase serum sample and therefore have little impact on patient management (24, 41). Additionally, the extensive antigenic cross-reactivity in serological assays, both among flaviviruses and between DENV serotypes, further complicates definitive diagnosis and the interpretation of the assays (18, 20, 41).
Molecular techniques based on the detection of genomic sequences by reverse transcription-PCR (RT-PCR), nested PCR, and real-time PCR are rapid and sensitive and have replaced virus isolation as the new standard method for the detection of DENV in acute-phase serum samples (15). These methods identify the four different serotypes by using genus- or serotype-specific primers or a combination of both. A two-step nested RT-PCR approach is routinely employed in laboratories worldwide (27).
Although most molecular techniques have the advantage of being rapid and sensitive, there are limitations. Real-time assays are limited by the large number of reactions required, as in Sybr green-based assays, and the manipulation of samples necessary for serotype identification is a limitation of fluorogenic dye-based assays (20, 26, 29, 44). In addition, the genetic diversity among DENV isolates raises concerns regarding false-negative PCR results due to mismatches in sequences resulting from the continual evolution of variant viral sequences. For example, recent studies indicate the presence of well-defined phylogenetic groups within each serotype of DENV. The genotypes described within the different serotypes are based on sequence variations in gene E and NS1. The number of genotypes varies, ranging from three (for DENV-4) to five (for DENV-1, -2, and -3), depending on the region sequenced (19, 31).
The present study was conducted on clinical samples from Puerto Rico, which is regarded as a prototype of the urban establishment of DENV. During the past 2 decades, Puerto Rico has experienced increasingly severe DENV epidemics (4). All four serotypes and multiple genotypes have been in circulation on this island. This situation is considered ideal for the genetic evolution of the virus, which has been demonstrated by molecular analysis of DENV-2 and -4 (3, 4). The situation is expected to be further complicated by the recent introduction of West Nile virus, which may clinically mimic DF and is difficult to distinguish from DENV by serologic tests, due to cross-reactive antibodies (7, 25).
In this study, we combine multiplex PCR using multiple degenerate primers with a ligase detection reaction (LDR) and a universal zip-code array for the simultaneous identification and serotyping of DENV from viral cultures and clinical samples from Puerto Rico. Originally developed for discriminating single-base mutations or polymorphisms in cancer genes, LDR uses a thermostable DNA ligase that ligates two adjacent oligonucleotides annealed to a complementary target only if the nucleotides are perfectly matched at the junction (1, 2). This method has subsequently been used in detecting mutations, insertions, and deletions in cancer genes (8-10, 21, 22). More recently this assay has been adapted for the detection of bacterial pathogens in blood cultures (34) and of West Nile virus in serum and mosquito pools (38). Since even a single base mismatch at the ligation junction prevents successful ligation, the technique is highly specific (21). Furthermore, such assays are ideal for multiplexing, since several primer sets can ligate along a DNA template without the interference encountered in purely polymerase-based assays (10, 22).
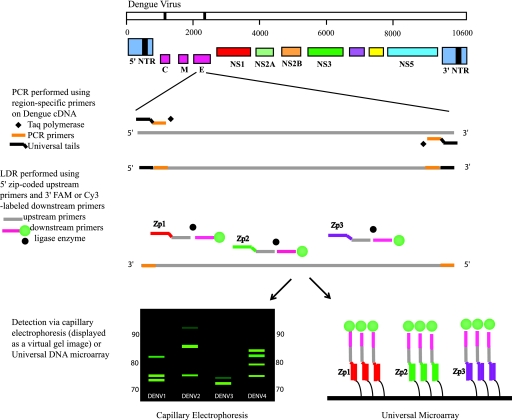
The LDR primers are designed to produce ligation products that are fluorescently labeled at their 3′ ends and have zip-code complements (complementary to zip-code addresses in a universal array) appended to their 5′ ends. The specific zip-code address spotted onto the array hybridizes only to the complementary sequence included on the LDR product (9, 10). A schematic representation of the assay is shown in Fig. 1. The universal array is a powerful technique that permits the simultaneous detection of a large number of genes or gene products, making it ideal for use in multiplex, high-throughput assays.
FIG. 1.
Schematic of the PCR-LDR assay for the identification of DENV serotypes. Serotype-specific PCR primers amplify one region of gene C and one region of gene E (for clarity, only the gene E amplicon is shown). Within each PCR amplicon, LDR primers are designed to identify and differentiate the four different DENV serotypes. The LDR primers target two locations in gene C and three locations in gene E. The 5′ upstream LDR primers bear zip-code complements, while the 3′ downstream LDR primers have either a FAM or a Cy3 fluorescent label. Ligation of the LDR primers results in fluorescently labeled products of different lengths that are then detected either by CE or on a universal array. NTR, nontranslated region.
Thus, the unique specificity and sensitivity of PCR-LDR coupled to the specificity of the universal array enables the detection and differentiation of all four serotypes of DENV in a single assay.
MATERIALS AND METHODS
Viral cultures and RNA from viruses and clinical samples.
Viral culture supernatants were obtained from the Dengue Branch, Centers for Disease Control and Prevention (CDC), San Juan, Puerto Rico (n = 38). These included 10 isolates each of DENV-1 and -3 and 9 isolates each of DENV-2 and -4.
DENV strains selected from the global DENV panel (6) maintained at the Division of Vector-Borne Infectious Diseases (DVBID), CDC, Fort Collins, CO, were used (Table 1). The panel consists of unique strains of DENV-1 to -4 representing the latest isolates of each serotype from different geographic regions. Isolates of other flaviviruses (Table 2) were kindly provided by Robert Lanciotti of the DVBID, CDC, the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch in Galveston, M. Niedrig from the Robert Koch Institute, Berlin, Germany (European Network for Diagnostics of Imported Viral Diseases), and the New York City Department of Health (38).
TABLE 1.
Global DENV panel used in assay validation
| Sample ID | Origin | Yr of isolation | Source | Serotype | Genotype |
|---|---|---|---|---|---|
| 276RKI | India | 1997 | Human | DENV-1 | Genotype I |
| 498 RKI | Thailand | 1998 | Human | DENV-1 | Genotype I |
| 1266 | Indonesia | 1978 | Mosquito | DENV-1 | Genotype II |
| 12150 | Philippines | 1984 | Human | DENV-1 | Genotype II |
| 228690 | Jamaica | 1977 | Human | DENV-1 | Genotype III |
| BC89/94 | Costa Rica | 1994 | Human | DENV-1 | Genotype III |
| 10674 | Dakar | 1970 | Human | DENV-2 | Sylvatic genotype |
| BC102/94 | Saudi Arabia | 1994 | Human | DENV-2 | Cosmopolitan/genotype III |
| P8-1407MS | Malaysia | 1968 | Human | DENV-2 | Cosmopolitan/genotype III |
| S-40921 | Myanmar | 1976 | Human | DENV-2 | Asian genotype I |
| BC27/96 | Vietnam | 1995 | Unknown | DENV-2 | Asian genotype II |
| BC171/96 | Philippines | 1996 | Human | DENV-2 | Asian genotype II |
| BC100/98 | Bolivia | 1998 | Unknown | DENV-2 | American/Asian |
| BC141/96 | Puerto Rico | 1994 | Unknown | DENV-2 | American/Asian |
| S-14635 | Tonga | 1974 | Unknown | DENV-2 | American |
| BC182/96 | Philippines | 1997 | Human | DENV-3 | Genotype I |
| BC14/97 | Malaysia | 1997 | Human | DENV-3 | Genotype I |
| MK-594-87 | Thailand | 1987 | Unknown | DENV-3 | Genotype II |
| S-40580 | Myanmar | 1976 | Unknown | DENV-3 | Genotype II |
| BC188/97 | Mexico | 1997 | Human | DENV-3 | Genotype III |
| 271242 | Sri Lanka | 1991 | Human | DENV-3 | Genotype III |
| BC123/97 | Malaysia | Unknown | Monkey | DENV-4 | Sylvatic genotype |
| D85-019 | Thailand | 1985 | Human | DENV-4 | Genotype I |
| BC13/97 | Malaysia | 1997 | Human | DENV-4 | Genotype I |
| BC287/97 | Mexico | 1997 | Human | DENV-4 | Genotype II |
| BC258/97 | Puerto Rico | 1994 | Human | DENV-4 | Genotype II |
TABLE 2.
Details of other flavivirus strains tested
| Flavivirus | Strain | Origin/yr |
|---|---|---|
| St. Louis encephalitis virus | MSI-7 | Mississippi/1977 |
| Murray Valley fever virus | OR2 | Victoria, Australia/1951 |
| Kunjin virus | MRM 16 | Australia/1960 |
| Powassan encephalitis virus | M11665 | Ontario, Canada/1965 |
| Yellow fever virus | 17D | Ghana/1927 |
| Japanese encephalitis virus | SA14-14-2 | China/1954 |
| West Nile virus | Unknowna | New York City/1999, 2000 |
Unidentified strain collected from mosquito pools (38).
A total of 350 clinical serum samples tested in this study were obtained from the repository at the Dengue Branch, CDC, San Juan, Puerto Rico, where they were confirmed as either positive or negative for DENV by real-time PCR (6). These samples were collected from patients with suspected DF during epidemics and in the intraepidemic periods corresponding with the years when the different serotypes circulated in the region: 1994 to 1995 for DENV-1, 2002 to 2005 for DENV-2, 2000 to 2004 for DENV-3, and 1995 to 1998 for DENV-4. The serum specimens were collected from 25 different municipalities in Puerto Rico.
RNA extraction.
Total RNA was extracted from 140 μl of human serum samples or virus-infected tissue culture supernatants by using the QIAamp viral RNA kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions. The RNA was extracted in 60 μl of elution buffer and was used immediately to synthesize the first-strand cDNA with the Superscript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Briefly, a master mix consisting of 0.1 mM deoxynucleoside triphosphates, 150 ng of random hexamers, 5 mM MgCl2, 2 μl of 10× buffer, 10 mM dithiothreitol, 40 U of RNaseOUT recombinant RNase inhibitor, and 50 U of Superscript II reverse transcriptase enzyme was prepared. A 6-μl aliquot of extracted RNA was added to this mixture, and the sample was incubated at 42°C for 50 min, followed by 15 min at 70°C to terminate the reaction. An additional incubation of 20 min at 37°C with 1 μl of RNase H eliminated any residual RNA in the reaction product. The resultant cDNA was stored at −20°C for future use.
Primer design and assay development.
PCR primers were designed using Oligo 6.0 software (Molecular Biology Insights, Cascade, CO) and were targeted to the capsid protein (C) and envelope protein (E) regions of the DENV genome. In the original version of the assay, nested forward and reverse primers were designed for the amplification of one region in gene C and two regions in gene E. Relatively conserved regions of the genome were chosen after alignment of known DENV sequences accessed from GenBank for primer design, but due to the extensive sequence variation in the DENV genome, different sets of primers had to be designed for each serotype, and degenerate bases were included to accommodate variation within each serotype. Primers had melting temperatures of approximately 72 to 75°C and were designed such that there were no more than three degenerate positions in each primer. Universal tail sequences were appended to the 5′ ends of forward and reverse PCR primers to prevent the formation of primer dimers. A total of 73 PCR primers were designed for all four serotypes of DENV (see Tables S1 and S2 in the supplemental material).
LDR primers were designed at three positions within each of the gene E amplicons and two positions within the gene C amplicon. The upstream LDR primers had unique oligonucleotides (zip-code complements), 20 bases long, attached at the 5′ end; the downstream primers had a fluorescent label, either 6-carboxyfluorescein (FAM) or cyanine 3 (Cy3), at the 3′ end. A total of 128 LDR primers were designed (see Tables S3 and S4 in the supplemental material), with melting temperatures of 75 to 80°C; degenerate bases (no more than three in each primer) were introduced where required to account for sequence variations among different strains of the virus. Primers were obtained from Integrated DNA Technologies (Coralville, IA).
PCR-LDR assay and detection of products.
Gene C and gene E were amplified using serotype-specific primers. The PCR mixture consisted of 10 mM Tris-HCl buffer containing 50 mM KCl (pH 8.0), 2.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates, and 1.25 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA).
PCRs were optimized by performing serotype-specific uniplex reactions using DENV culture supernatants with 5 to 10 pmol of each primer per reaction. To this mixture, 1 μl of template cDNA was added to make a final volume of 25 μl. Amplification was performed using a GeneAmp 9700 thermocycler (Applied Biosystems, Foster City, CA). Initial denaturation of template DNA was achieved by heating at 95°C for 5 min. This was followed by 40 cycles of 30 s at 95°C, 30 s at 58°C, and 1 min at 72°C. A final extension step was conducted for 7 min at 72°C, followed by a termination step at 99°C for 30 min.
Multiplex PCRs were performed using the same method, except that all primers for all serotypes were used in a single reaction. Multiplex reactions were optimized by varying the concentrations of the primer (10 to 0.5 μM) and MgCl2 (1.5 to 2.5 mM). Ultimately, the optimum reaction conditions were found to be similar to those described above, except that the final concentration of each primer used was 2 μM and the MgCl2 concentration was 2.5 mM.
Ligation reactions were conducted in a solution (20 μl) containing LDR buffer (20 mM Tris-HCl buffer [pH 7.6], 100 mM KCl, 10 mM MgCl2), 1 mM NAD+, 1 mM dithiothreitol, 12.5 nM (250 fmol) each LDR primer, 2 μl of each PCR product, and 0.01 μM AK 16D ligase (expressed from Thermus species AK 16D) (42). Reaction mixtures were initially heated at 94°C for 1 min, followed by 20 thermal cycles at 94°C for 30 s (denaturation) and 64°C for 4 min (annealing/ligation). The ligation products were analyzed by two methods: capillary electrophoresis (CE) and a universal zip-code array.
CE.
A 0.5-μl aliquot of each LDR product was added to 9.2 μl of Hi-Di formamide and 0.3 μl of a LIZ-500 DNA size standard (Applied Biosystems, Foster City, CA). The samples were denatured by heating to 95°C for 3 min and were cooled rapidly to 4°C before being loaded onto the ABI 3730 DNA analyzer for CE. The data generated were analyzed using Gene Mapper software (version 3.5; Applied Biosystems, Foster City, CA). The fragment size and peak area data of the different ligation products were exported and used to generate a 2-dimensional virtual gel image using the Gelrender software program (34). All reactions were performed in duplicate, and the experiment was run twice.
Universal zip-code array.
Unique 20-bp oligonucleotides (zip-code addresses) were double spotted onto polymer-coated slides (CodeLink slides; GE Healthcare, Piscataway, NJ). Zip-code addresses for spotting were prepared in 50 mM sodium phosphate (pH 8.5) at a final concentration of 25 μM in a 384-well plate. A fiducial oligonucleotide (1 μM) was added to the printing mix in each well and cospotted with each zip-code address. Arrays were printed using a QArrayMini robotic array printer (Genetix, Boston, MA) at 10°C and 50 to 60% humidity. Printed slides were incubated in a saturated NaCl chamber overnight and then treated with a blocking solution (0.1 M Tris, 50 mM ethanolamine [pH 9.0]) to block residual reactive carboxyl groups. The slides were washed with 4× standard sodium citrate (SSC) buffer (20× SSC is 3 M sodium chloride and 0.3 M sodium citrate; pH 7.0) and 0.1% sodium dodecyl sulfate (SDS) and were dried by spinning. A 6-carboxy-X-rhodamine-labeled fiducial complement included in the hybridization mixture served as an internal positive control to determine the position and quality of each address. Printed slides were randomly selected for quality control by hybridizing fluorescent-labeled zip-code complements; a batch of slides was used only if the quality control produced a specific fluorescent signal in the absence of extraneous signals on adjacent addresses. Printed slides were stored in a desiccator at room temperature until use.
Ligation products were diluted in a hybridization buffer (5× SSC buffer) containing 0.1% SDS, 0.1 mg/ml salmon sperm DNA (Fisher Scientific), and 5 nM fiducial complement in a total volume of 30 μl. A Pro-Plate multiarray slide chamber (Grace Bio-Labs, Bend, OR) was attached to universal array slides, and the entire amount of the hybridization mixture was added to the chambers. Hybridization was carried out at 60°C for 2 h in the dark in a hybridization oven (Lab-Line; VWR, West Chester, PA). Following hybridization, the slides were rinsed with 5× SSC and washed with 1× SSC-0.1% SDS at 60°C for 15 min. After two more wash steps of 1 min each with 0.2× SSC and 0.1× SSC, respectively at room temperature, the slides were spin-dried and scanned using a Pro-Scan array (Perkin-Elmer, Wellesley, MA). Positive signals were detected and quantified with ScanArray Express (version 3.0; Perkin-Elmer, Wellesley, MA) and were manually inspected when necessary. The signal intensity data obtained were transferred as text files, and only those addresses where the signal intensity was ≥10-fold higher than background were considered positive. Reactions were performed twice and identification confirmed for both experiments.
LOD of the assay.
To determine the limit of detection (LOD), 10-fold serial dilutions of viral culture stocks were prepared for all four DENV serotypes from standard stock cultures, with starting concentrations of 2,500,000 PFU/ml for DENV-1 (strain Hawaii), 290,000 PFU/ml for DENV-2 (strain New Guinea C), 1,200,000 PFU/ml for DENV-3 (strain Philippines H87), and 10,000,000 PFU/ml for DENV-4 (strain Philippines H241). The concentration ranges tested for the different serotypes were as follows: 2.5 × 106 to 0.0025 PFU/ml (1.75 × 104 to 1.75 × 10−5 PFU/reaction) for DENV-1, 2.9 × 105 to 0.0029 PFU/ml (2 × 104 to 2.03 × 10−5 PFU/reaction) for DENV-2, 1.2 × 106 to 0.0012 PFU/ml (8.4 × 103 to 0.84 × 10−5 PFU/reaction) for DENV-3, and 1 × 107 to 0.01 PFU/ml (7 × 104 to 0.7 × 10−4 PFU/reaction) for DENV-4. Dilutions were prepared in Dulbecco's minimum essential medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). RNA was extracted from 140 μl of each dilution, and RT-PCR-LDR with a universal array was used to determine the lower limit of detection.
RESULTS
Assay design and initial optimization.
Initial optimization and validation of the multiplex PCR-LDR assay for the detection and identification of DENV serotypes were performed on 38 culture supernatants from stock virus cultures. Products of PCR amplification were analyzed by electrophoresis on a 2% agarose gel. Both uniplex (each serotype amplified separately for each gene target) and multiplex (all serotypes amplified together for each gene target) reactions were individually validated, and PCR products were observed at 300 bp and 400 bp, respectively, for gene C and gene E (data not shown). LDRs were then performed on the PCR products, and the LDR products were analyzed by CE. The assay was able to successfully identify 37 out of the 38 samples and to type them correctly compared to standard detection by real-time PCR at the CDC in Puerto Rico (data not shown). The sample that failed identification was found to have insufficient nucleic acid. Our initial optimization and validation experiments for the detection of DENV serotypes were carried out successfully by analyzing LDR products from two amplicons (one each in gene E and gene C) (data not shown). Analysis of the third amplicon did not provide any additional information for the detection or differentiation of DENV serotypes. All further experiments were therefore performed using primers for only two amplicons: 46 PCR primers and 75 LDR primers in all (see Tables S1 to S4 in the supplemental material).
Assessment of the multiplex capability of the PCR-LDR assay.
A multiplex PCR amplification of both target regions used all 46 PCR primers in a single reaction. Subsequently, amplicons were subjected to a multiplex ligation reaction using the 75 primers for all four DENV serotypes in a single reaction. For the initial validation, LDR products were analyzed by CE. Results from representative samples are shown in Fig. 2. The sizes and number of LDR products at a single position may differ depending on the target viral sequence in that particular ligation position. Table 3 shows the observed lengths of the LDR products for each serotype, which differed by 1 to 3 bases from those expected. A given DENV serotype can produce four or five different ligation products of approximately 73 to 92 bases. Due to variations in product length and the comigration of products of similar length, CE is not an optimal method for the interpretation of the PCR-LDR assay (Table 3 and Fig. 2). In a similar study, we have observed that LDR products of identical lengths but different sequences (due to the use of degenerate oligonucleotides) can migrate at separate positions in CE, resulting in broadened peaks that may overlap with nearby peaks (38). Other studies have also observed that it is difficult to predict CE results based on the lengths of the primers, because the migration pattern cannot be deduced from the length of the product (28). Therefore, after successful optimization of the PCR-LDR, the universal array was used for the analysis of LDR products.
FIG. 2.
Reconstructed CE images of representative DENV isolates. Ligation products were labeled with FAM, separated by CE on a 3730 DNA analyzer, and sized using a LIZ-labeled internal standard. Each serotype produces a distinct pattern depending on the number and sizes of the ligation products obtained. The approximate sizes of the ligation products, which ranged from 73 to 92 bases, are indicated on each side. The CE data shown are reconstructed images produced by Gelrender software (34). Negative samples (n = 7) and samples from other viruses yielded no observable products. For other flaviviruses, lanes are as follows: 1, West Nile virus; 2, St. Louis encephalitis virus; 3, Japanese encephalitis virus; 4, Kunjin virus; 5, Murray Valley virus; 6, Powassan virus; 7, yellow fever virus.
TABLE 3.
Comparison of the expected sizes of the ligation products and the sizes observed by CE analysis
| Serotype | Target detected | Sizes of LDR products (bp)
|
|
|---|---|---|---|
| Predicted | Observed | ||
| DENV-1 | Gene C | 76, 83 | 74.5, 76-76.2, 82.3-82.6 |
| Gene E | 76, 78, 88 | 73.6-74, 75.9-76.2, 88.6-88.8 | |
| DENV-2 | Gene C | 76, 86 | 76.2-76.4, 85.9-86.0 |
| Gene E | 85, 86, 92 | 82.4-82.6, 84.7-84.8, 91.7-91.9 | |
| DENV-3 | Gene C | 74, 74 | 73.7-73.9 |
| Gene E | 75, 78, 78 | 75.2-75.4, 77.4-77.6 | |
| DENV-4 | Gene C | 79, 84 | 79.8-80.0, 81.3-81.4, 83.8-84.1 |
| Gene E | 76, 83, 86 | 75.5-75.7, 82.6-82.8, 84.0-84.3 | |
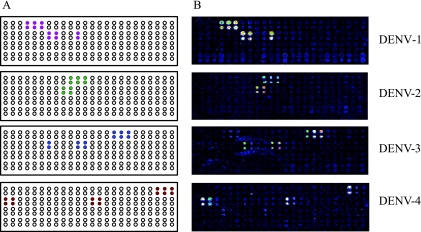
The detection of LDR products on the universal array is independent of their size. The serotype-specific products hybridize to unique complementary zip-code addresses on the array. The LDR products are detected by fluorescent signals appended to their 3′ ends. A typical universal array layout is shown in Fig. 3A. All zip-code addresses were spotted in duplicate. Two positive signals were required, one from each amplicon or both from a single amplicon, for identification (Fig. 3B). Two additional zip-code addresses were spotted for DENV-4 due to the use of additional LDR primers designed for the detection of gene E; however, for any given DENV-4 serotype, only five positive signals were observed on the array. Two addresses on the array produced positive signals for both DENV-1 and DENV-3 due to similarities in the primer sequences. This did not, however, interfere with the identification of the two serotypes, because the other addresses clearly distinguished them (Fig. 3B).
FIG. 3.
Detection of DENV by the universal array. (A) Universal microarray layout of expected positive signals (filled circles) for each DENV serotype. Zip-codes were spotted in duplicate. Open circles indicate addresses available for the detection of other viruses as the assay is further developed. (B) Representative universal array detection of DENV-1 to -4 using the RT-PCR-LDR assay. Each serotype generates at least four or five unique signals that permit identification. DENV-1 and DENV-3 produce two common signals, as described in the text.
Identification and typing of DENV from clinical samples.
Serum specimens from 161 cases of DF confirmed at the Dengue Branch, CDC, Puerto Rico, were analyzed by the PCR-LDR assay. The results are shown in Table 4. It was possible to identify 159 of the 161 positive samples correctly (sensitivity, 98.7%). Serotype identification was concordant for all 159 samples in which DENV was detected. The two samples that tested positive at the CDC but could not be detected by PCR-LDR were archived samples; material could have been lost due to prolonged storage or multiple freeze-thaw cycles. Specificity was evaluated using a panel of 189 serum samples that tested negative for the presence of DENV. Three of the negative samples were identified as DENV-2 by our assay (specificity, 98.4%). Two of these samples came from patients presenting with acute symptoms of DF but were found to be DENV negative when tested by RT-PCR and an enzyme-linked immunosorbent assay at the CDC; no second, paired serum sample, which might have been tested for seroconversion, was ever received from either patient. No clinical data were available for the third patient, but the sample was found to be negative upon repeated analysis by real-time PCR.
TABLE 4.
Detection and serotyping of DENV from clinical serum samples using a multiplex RT-PCR-LDR-universal array
| CDC serotype determination | No. of samples tested | No. of samples with the following serotype by the RT-PCR-LDR-universal array:
|
||||
|---|---|---|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | Negative | ||
| DENV-1 | 20 | 20 | 0 | 0 | 0 | 0 |
| DENV-2 | 62 | 0 | 62 | 0 | 0 | 0 |
| DENV-3 | 59 | 0 | 0 | 57 | 0 | 2 |
| DENV-4 | 20 | 0 | 0 | 0 | 20 | 0 |
| Negative | 189 | 0 | 3 | 0 | 0 | 186 |
Performance of the PCR-LDR against the global DENV panel and other flaviviruses.
A global DENV panel consisting of six strains each of DENV-1 and -3, nine strains of DENV-2, and five strains of DENV-4 was tested using PCR-LDR with the universal array for detection and serotype identification. The strains belonged to different genotypes and came from different geographic regions (Table 1) (6). All except one of the strains (strain identification [ID] 10674; Dakar, 1970) were detected and correctly serotyped by PCR-LDR. Sequence analysis of gene E and gene C confirmed that the isolate had significant genetic variation at the PCR primer binding sites. Two additional PCR primers were subsequently designed and added to the PCR primer mix in order to accommodate the sequence difference and permit the amplification of this isolate (see Tables S1 and S2 in the supplemental material).
A panel of seven other flaviviruses (West Nile virus, Kunjin virus, Powassan virus, Japanese encephalitis virus, St. Louis encephalitis virus, Murray Valley encephalitis virus, and yellow fever virus) was tested with the assay, and CE results are shown in Fig. 2. The PCR-LDR primers were specific for DENV serotypes, and no signals were detected for any of the other flaviviruses by use of CE or the universal array.
Determination of LOD.
Using serial dilutions of the virus stock cultures for RNA extraction and subsequent PCR-LDR, detection limits were found to be 0.017, 0.004, 0.8, and 0.7 equivalent PFU of virus for DENV-1, DENV-2, DENV-3, and DENV-4, respectively. Figure 4 shows the universal array data for the DENV-1 detection limit. Detection limits were found to be the same by using CE or the universal array and were calculated by taking the dilution factors into account; i.e., RNA was extracted from 140 μl of each dilution and was subsequently eluted out in 60 μl of buffer, from which 6 μl was used for cDNA synthesis.
FIG. 4.
LOD of the PCR-LDR assay. Microarray images show the LOD for DENV-1. The number of PFU used in each PCR is given. Fluorescent signals ≥10-fold higher than the background negative signals were deemed positive.
DISCUSSION
In this study, we report the development and evaluation of a nucleic acid detection assay, based on RT-PCR followed by LDR with specific primers, for the simultaneous identification of all four serotypes of DENV in a single reaction.
We used the PCR-LDR approach to overcome the high degree of sequence variation between DENV serotypes. The wide distribution of DENV serotypes in the tropics and subtropics allows for considerable genotypic heterogeneity among the circulating serotypes and has also been linked to the virulence of the circulating strains (5, 12, 17, 23, 30). The four different serotypes of DENV are thought to be more dissimilar than different “species” of Flavivirus (17). Such viral genetic heterogeneity has implications for the design of molecular assays. False-negative PCR results due to sequence mismatches between DENV RNA and the primers used in real-time PCR assays have been reported, necessitating assay revisions (6, 16, 36). In the design of our assay, we used several primer pairs with degeneracies to accommodate possible failures of amplification due to sequence mismatches. The envelope (E) and capsid (C) genes were chosen as targets, because they were found to have sufficient variation to be useful for serotype differentiation. However, due to the inherent heterogeneity in the E region, a large number of primers had to be used to account for all anticipated sequence variations. Primers were designed for two regions in order to provide a certain amount of redundancy that would circumvent any dropouts in PCR amplification due to sequence variation.
A multiplex PCR-LDR assay was optimized such that all primers could be used in a single reaction for amplification of all four DENV serotypes. This approach was found to be >98% sensitive and specific in the detection and serotype identification of DENV from 350 archived acute-phase serum samples and to compare favorably to techniques described previously (11, 24, 40). The assay has been developed as a prototype for the multiplex detection of RNA viruses and will be expanded for the detection of other hemorrhagic fever viruses. Therefore, a multiplex format was adopted to define the feasibility of the assay, and it was possible to multiplex 46 PCR primers and 75 LDR primers in a single reaction, with significant signal intensity in the CE and microarray formats.
The primers were very specific for DENV serotypes and did not cross-react with any of the other flaviviruses tested. Only 3 of 189 samples were positive by PCR-LDR but negative by the real-time PCR assay performed at the CDC. Upon analysis of clinical findings, it was discovered that the serum samples from two of these patients were collected within 1 to 5 days of the onset of symptoms, suggesting that they may have had acute infections. Seroconversion could not be demonstrated, because convalescent-phase serum samples were never obtained. It is therefore possible that these patients indeed had viremia that could not be detected by real-time PCR. No clinical history was available for the third patient whose sample was retrieved from the CDC repository archives.
Current reports suggest that several lineages of DENV-2 and DENV-4 circulate in Puerto Rico and that DENV strains exhibit complex patterns of lineage turnover and extensions (3, 4). Thus, it is likely that several lineages of each DENV serotype could have been circulating within the time period encompassing the collection of specimens included in the panel of clinical samples used for this study, which were detected by our assay with high sensitivity and specificity.
In order to verify the ability of the PCR-LDR assay to detect different genotypes of DENV, we used a global DENV panel. Our assay was able to detect all but one isolate (strain ID 10674), a sylvatic strain from Africa (G.-J. J. Chang, personal communication). The analysis of complete genome sequences suggests that sylvatic DENV-2 isolates are evolutionarily distinct from endemic DENV-2 isolates and supports the classification of DENV-2 into two discrete ecotypes (4, 43). In another study, real-time PCR primers for DENV targeted to the C-PrM region had to be modified to detect variant sylvatic virus (6; G.-J. J. Chang, personal communication). The sequence of sylvatic isolate 10674 was not available when our primers were initially designed; subsequently, two new PCR primers were added, and it was possible to amplify this isolate. The addition of two new primers did not adversely affect the efficiency of the PCR, indicating the flexibility of our assay in the identification of new genotypes. The evolutionary genetics of DENV suggest that its population is becoming increasingly diverse, and the presence of sylvatic reservoirs of the virus may allow the introduction and emergence of sylvatic virus in human populations. It has also been suggested that these conditions could lead to the development of new pathogenic DENV strains (17). With the PCR-LDR strategy, it would be possible to incorporate new primers for the detection of these variant genotypes as they evolve.
The detection limit of our assay ranged from 0.004 to 0.7 equivalent PFU/reaction. The lower LOD differed for the different serotypes; the assay was 100 times more sensitive for DENV-2 and DENV-1 (LOD, 0.004 and 0.017 equivalent PFU, respectively) than for DENV-3 and DENV-4 (LOD, 0.8 and 0.7 equivalent PFU, respectively). Variable LOD have been reported for DENV serotypes in several different studies. Johnson et al. have reported an assay 100 times more sensitive for DENV-2, -3, and -4 (0.0016 to 0.008 equivalent PFU) than for DENV-1 (0.5 equivalent PFU) and postulated that the difference could be due to differences in the proportion of noninfectious RNA transcripts to infectious particles (PFU) between DENV-1 and the other serotypes (20). In a comparable study, Lai et al. have reported variable LOD of 0.1 PFU for DENV-1 and -2, 1 PFU for DENV-3, and 0.01 PFU for DENV-4 (26). In yet another real-time PCR-based assay evaluated recently, serotype-specific primers were 10 times more sensitive for DENV-2, -3, and -4 (LOD, 0.1 PFU) than for DENV-1 (LOD, 1 PFU) (33). The LOD observed in the present study is comparable to those for the other techniques reported (20, 26, 40).
The use of zip-code addresses for spotting the array and its unique potential to recognize pathogen-specific zip-code complements appended to the LDR primers obviate the use of actual genetic sequence for pathogen detection. This is especially useful for multiplexing: a large number of genomic targets can be detected using this array, since the zip-code addresses are synthetic oligonucleotides. Additionally, the same array can be used for the simultaneous detection of different organisms, since positive hybridization is dependent on the chemistry of the synthesized zip-code oligonucleotides spotted onto the array and their complements appended to the primers.
The current study was undertaken to prove the feasibility of using the PCR-LDR technique for the multiplexed detection of RNA viruses. Since viral infections often present with nonspecific clinical symptoms, and arboviruses have similar geographical distributions, a multiplexed approach to the detection of several arboviruses in a single sample could be extremely beneficial. In addition to serotype identification, a critical element of epidemic control measures is the early identification of emerging genotypes and the replacement of a genotype(s) in a given geographic region (37). Since some DENV serotypes have more than four genotypes, PCR-LDR may be useful in genotype detection and differentiation without the involvement of nucleotide sequence analysis and phylogenetic studies.
We envision extending the scope of the assay to permit the multiplexed identification of a panel of hemorrhagic fever viruses. Our group has already reported the use of the PCR-LDR assay for the identification of a panel of 20 different bacteria in blood cultures (34) and has developed an assay for the detection of West Nile virus (38). The DENV detection assay has been developed using similar principles, and the technique is currently being developed for use in the simultaneous identification of hemorrhagic fever viruses. Ultimately the technique may prove useful in a comprehensive assay for blood-borne infectious agents.
Supplementary Material
Acknowledgments
We acknowledge Pius Brzoska at Applied Biosystems for providing genomic sequence alignments for DENV serotypes and Robert Lanciotti at the CDC, Fort Collins, CO, the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch in Galveston, M. Niedrig from the Robert Koch Institute, Berlin, Germany (European Network for Diagnostics of Imported Viral Diseases), and the New York City Department of Health for viruses other than DENV used in this study.
This work was supported by Public Health Service grant UC1-AI062579 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 6 August 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Barany, F. 1991. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl. Acad. Sci. USA 88189-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barany, F. 1991. The ligase chain reaction in a PCR world. PCR Methods Appl. 15-16. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, S. N., E. C. Holmes, M. Chirivella, D. M. Rodriguez, M. Beltran, V. Vorndam, D. J. Gubler, and W. O. McMillan. 2006. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J. Gen. Virol. 87885-893. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, S. N., E. C. Holmes, M. Chirivella, D. M. Rodriguez, M. Beltran, V. Vorndam, D. J. Gubler, and W. O. McMillan. 2003. Selection-driven evolution of emergent dengue virus. Mol. Biol. Evol. 201650-1658. [DOI] [PubMed] [Google Scholar]
- 5.Bray, M., R. Men, I. Tokimatsu, and C. J. Lai. 1998. Genetic determinants responsible for acquisition of dengue type 2 virus mouse neurovirulence. J. Virol. 721647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien, L. J., T. L. Liao, P. Y. Shu, J. H. Huang, D. J. Gubler, and G. J. Chang. 2006. Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. J. Clin. Microbiol. 441295-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuis, A. P., II, P. P. Marra, R. Reitsma, M. J. Jones, K. L. Louie, and L. D. Kramer. 2005. Serologic evidence for West Nile virus transmission in Puerto Rico and Cuba. Am. J. Trop. Med. Hyg. 73474-476. [PubMed] [Google Scholar]
- 8.Favis, R., and F. Barany. 2000. Mutation detection in K-ras, BRCA1, BRCA2, and p53 using PCR/LDR and a universal DNA microarray. Ann. N. Y. Acad. Sci. 90639-43. [DOI] [PubMed] [Google Scholar]
- 9.Favis, R., J. P. Day, N. P. Gerry, C. Phelan, S. Narod, and F. Barany. 2000. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat. Biotechnol. 18561-564. [DOI] [PubMed] [Google Scholar]
- 10.Gerry, N. P., N. E. Witowski, J. Day, R. P. Hammer, G. Barany, and F. Barany. 1999. Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol. 292251-262. [DOI] [PubMed] [Google Scholar]
- 11.Gomes, A. L., A. M. Silva, M. T. Cordeiro, G. F. Guimaraes, E. T. Marques, Jr., and F. G. Abath. 2007. Single-tube nested PCR using immobilized internal primers for the identification of dengue virus serotypes. J. Virol. Methods 14576-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gualano, R. C., M. J. Pryor, M. R. Cauchi, P. J. Wright, and A. D. Davidson. 1998. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J. Gen. Virol. 79437-446. [DOI] [PubMed] [Google Scholar]
- 13.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gubler, D. J. 1998. The global pandemic of dengue/dengue haemorrhagic fever: current status and prospects for the future. Ann. Acad. Med. Singapore 27227-234. [PubMed] [Google Scholar]
- 15.Guzmán, M. G., and G. Kouri. 2004. Dengue diagnosis, advances and challenges. Int. J. Infect. Dis. 869-80. [DOI] [PubMed] [Google Scholar]
- 16.Harris, E., T. G. Roberts, L. Smith, J. Selle, L. D. Kramer, S. Valle, E. Sandoval, and A. Balmaseda. 1998. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J. Clin. Microbiol. 362634-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes, E. C., and S. S. Burch. 2000. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 874-77. [DOI] [PubMed] [Google Scholar]
- 18.Ito, M., T. Takasaki, K. Yamada, R. Nerome, S. Tajima, and I. Kurane. 2004. Development and evaluation of fluorogenic TaqMan reverse transcriptase PCR assays for detection of dengue virus types 1 to 4. J. Clin. Microbiol. 425935-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, M., K. Yamada, T. Takasaki, B. Pandey, R. Nerome, S. Tajima, K. Morita, and I. Kurane. 2007. Phylogenetic analysis of dengue viruses isolated from imported dengue patients: possible aid for determining the countries where infections occurred. J. Travel Med. 14233-244. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, B. W., B. J. Russell, and R. S. Lanciotti. 2005. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 434977-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanna, M., W. Cao, M. Zirvi, P. Paty, and F. Barany. 1999. Ligase detection reaction for identification of low abundance mutations. Clin. Biochem. 32287-290. [DOI] [PubMed] [Google Scholar]
- 22.Khanna, M., P. Park, M. Zirvi, W. Cao, A. Picon, J. Day, P. Paty, and F. Barany. 1999. Multiplex PCR/LDR for detection of K-ras mutations in primary colon tumors. Oncogene 1827-38. [DOI] [PubMed] [Google Scholar]
- 23.Kinney, R. M., S. Butrapet, G. J. Chang, K. R. Tsuchiya, J. T. Roehrig, N. Bhamarapravati, and D. J. Gubler. 1997. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230300-308. [DOI] [PubMed] [Google Scholar]
- 24.Klungthong, C., R. V. Gibbons, B. Thaisomboonsuk, A. Nisalak, S. Kalayanarooj, V. Thirawuth, N. Nutkumhang, M. P. Mammen, Jr., and R. G. Jarman. 2007. Dengue virus detection using whole blood for reverse transcriptase PCR and virus isolation. J. Clin. Microbiol. 452480-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komar, N., and G. G. Clark. 2006. West Nile virus activity in Latin America and the Caribbean. Rev. Panam. Salud Pública 19112-117. [DOI] [PubMed] [Google Scholar]
- 26.Lai, Y. L., Y. K. Chung, H. C. Tan, H. F. Yap, G. Yap, E. E. Ooi, and L. C. Ng. 2007. Cost-effective real-time reverse transcriptase PCR (RT-PCR) to screen for dengue virus followed by rapid single-tube multiplex RT-PCR for serotyping of the virus. J. Clin. Microbiol. 45935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanciotti, R. S., C. H. Calisher, D. J. Gubler, G. J. Chang, and A. V. Vorndam. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen, L. A., C. Jespersgaard, and P. S. Andersen. 2007. Single-strand conformation polymorphism analysis using capillary array electrophoresis for large-scale mutation detection. Nat. Protoc. 21458-1466. [DOI] [PubMed] [Google Scholar]
- 29.Laue, T., P. Emmerich, and H. Schmitz. 1999. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. J. Clin. Microbiol. 372543-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leitmeyer, K. C., D. W. Vaughn, D. M. Watts, R. Salas, I. Villalobos de Chacon, C. Ramos, and R. Rico-Hesse. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 734738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackenzie, J. S., D. J. Gubler, and L. R. Petersen. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10S98-S109. [DOI] [PubMed] [Google Scholar]
- 32.Morens, D. M., and A. S. Fauci. 2008. Dengue and hemorrhagic fever: a potential threat to public health in the United States. JAMA 299214-216. [DOI] [PubMed] [Google Scholar]
- 33.Parida, M., K. Horioke, H. Ishida, P. K. Dash, P. Saxena, A. M. Jana, M. A. Islam, S. Inoue, N. Hosaka, and K. Morita. 2005. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 432895-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pingle, M. R., K. Granger, P. Feinberg, R. Shatsky, B. Sterling, M. Rundell, E. Spitzer, D. Larone, L. Golightly, and F. Barany. 2007. Multiplexed identification of blood-borne bacterial pathogens by use of a novel 16S rRNA gene PCR-ligase detection reaction-capillary electrophoresis assay. J. Clin. Microbiol. 451927-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porterfield, J. S. 1986. Antibody-dependent enhancement of viral infectivity. Adv. Virus Res. 31335-355. [DOI] [PubMed] [Google Scholar]
- 36.Reynes, J. M., S. Ong, C. Mey, C. Ngan, S. Hoyer, and A. A. Sall. 2003. Improved molecular detection of dengue virus serotype 1 variants. J. Clin. Microbiol. 413864-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rico-Hesse, R., L. M. Harrison, R. A. Salas, D. Tovar, A. Nisalak, C. Ramos, J. Boshell, M. T. de Mesa, R. M. Nogueira, and A. T. da Rosa. 1997. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230244-251. [DOI] [PubMed] [Google Scholar]
- 38.Rondini, S., M. R. Pingle, S. Das, R. Tesh, M. S. Rundell, J. Hom, S. Stramer, K. Turner, S. N. Rossmann, R. Lanciotti, E. G. Spier, J. Muñoz-Jordán, D. Larone, E. Spitzer, F. Barany, and L. M. Golightly. 2008. Development of multiplex PCR-ligase detection reaction assay for detection of West Nile virus. J. Clin. Microbiol. 462269-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothman, A. L. 2004. Dengue: defining protective versus pathologic immunity. J. Clin. Investig. 113946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu, P. Y., S. F. Chang, Y. C. Kuo, Y. Y. Yueh, L. J. Chien, C. L. Sue, T. H. Lin, and J. H. Huang. 2003. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J. Clin. Microbiol. 412408-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavakoli, N. P., E. H. Tobin, S. J. Wong, A. P. Dupuis II, B. Glasheen, L. D. Kramer, and K. A. Bernard. 2007. Identification of dengue virus in respiratory specimens from a patient who had recently traveled from a region where dengue virus infection is endemic. J. Clin. Microbiol. 451523-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong, J., W. Cao, and F. Barany. 1999. Biochemical properties of a high fidelity DNA ligase from Thermus species AK16D. Nucleic Acids Res. 27788-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasilakis, N., E. B. Fokam, C. T. Hanson, E. Weinberg, A. A. Sall, S. S. Whitehead, K. A. Hanley, and S. C. Weaver. 2008. Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains. Virology 377296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warrilow, D., J. A. Northill, A. Pyke, and G. A. Smith. 2002. Single rapid TaqMan fluorogenic probe based PCR assay that detects all four dengue serotypes. J. Med. Virol. 66524-528. [DOI] [PubMed] [Google Scholar]
- 45.World Health Organization. 2002. Dengue and dengue hemorrhagic fever. WHO fact sheet 117. World Health Organization, Geneva, Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.