Abstract
The colonic mucosa-associated flora (MAF) in patients with active ulcerative colitis (UC) (n = 13) was investigated by examining 16S rRNA gene signatures during remission and relapse against levels for controls (n = 5). Baseline reduction, temporal instability, and decrease of bacterial richness toward relapse were observed for UC patients, whereas the MAF for controls was stable over time.
Microbial studies have shown a persistent reduction in biodiversity of the mucosa-associated flora (MAF) in patients with active inflammatory bowel disease compared with the MAF in normal and non-inflammatory bowel disease inflammatory controls (2). A loss of normal anaerobic components (Bacteroides, Eubacterium, and Lactobacillus spp.) has contributed to this difference (7, 11). A shift to an increased representation of gram-negative bacteria appears to accompany a reduced bacterial diversity in Crohn's disease patients during remission, suggesting that these abnormalities may be primary and not secondary to the inflammatory process (4). We have conducted a prospective study of the MAF in patients with ulcerative colitis (UC) during remission to investigate temporal changes in bacterial richness and diversity. Monthly biopsy specimens were obtained during a 1-year follow-up. Follow-up was terminated if patients developed a clinical relapse.
The 1-year prospective follow-up protocol was conducted at St. Mark's Hospital, Harrow, United Kingdom. The patients included had established UC confirmed by clinical, endoscopic, and histological features and had frequently relapsing disease (defined as at least two episodes requiring medical treatment in the past 12 months). Clinical characteristics are given in Table 1. Inclusion criteria allowed concurrent medication with oral 5-aminosalicylates (5-ASA) at a stable dose for at least 4 weeks and azathioprine or 6-mercaptopurine at a stable dose for at least 3 months prior to screening. Treatments with steroids, antibiotics, anti-tumor necrosis factor agents, and rectal 5-ASA were not permitted for a minimum of 4 weeks before or at any time during the study. The UC disease activity index (UCDAI) was used for assessment of disease activity (8). At enrollment, patients were in remission, defined as a UCDAI of 1 or 0 and an endoscopic subscore of 0 (normal mucosal appearance). Following screening, UC patients attended monthly study visits until relapse or for a maximum of 12 months. Relapse was defined as a UCDAI of greater than 3, including an endoscopic score of at least 1 (mucosal friability). Five healthy volunteers were included for benchmarking the assessments (Table 2). Specimens from these controls were collected at two time points (0 and 6 months) from the same location. No colonic cleansing with oral agents or enemas was used. All laboratory investigations were carried out at the Institute for Clinical Molecular Biology in Kiel, Germany. The research protocol was approved by the ethics committee of the Medical Faculty of Christian Albrechts University in Kiel. PCR, single-stranded conformation polymorphism analysis, cloning, and sequencing of bands, including sequence analysis, were performed as described earlier (5, 6, 7). For comparison of remission status and relapse status in UC, a total of 43 bands were included in the statistical analysis. Standard statistical tests were conducted using SPSS (10.0; Chicago, IL). To investigate the regularities of the bacterial dynamics in individuals with UC, information on the intensity levels of 43 bacterial bands for the 13 patients were used. These multidimensional data were treated using methods of the analysis of neural networks (9) (see the supplemental material). These measurements are based on community fingerprint profiles of bacterial consortia of monthly biopsy specimens taken after initial induction of remission until the moment of relapse. The time to relapse, in months, is given in Table 1.
TABLE 1.
Patient characteristics
| Patient no. | Age (yr) | Gendera | Disease duration (yr) | Disease extent | No. of relapses per yr | Medication(s) | EIMc | Smoking statusd | Time to relapse (mo) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 66 | M | 3 | Total | 2 | 5-ASA, Azab | Non | 2 | |
| 2 | 26 | F | 4 | Left sided | 2 | 5-ASA, Aza | Non | 2 | |
| 3 | 49 | M | 6 | Total | 3 | 5-ASA | Non | 3 | |
| 4 | 46 | M | 18 | Left sided | 2 | 5-ASA | Non | 10 | |
| 5 | 59 | F | 3 | Left sided | 2 | 5-ASA | Arthritis | Non | 3 |
| 6 | 51 | M | 16 | Total | 4 | 5-ASA | Arthritis | Ex | 3 |
| 7 | 35 | M | 7 | Total | 2 | 5-ASA | Non | 3 | |
| 8 | 27 | M | 7 | Total | 2 | 5-ASA, Aza | Non | 3 | |
| 9 | 62 | F | 30 | Total | 2 | 5-ASA | Ex | 3 | |
| 10 | 53 | F | 5 | Total | 2 | 5-ASA | Ex | 3 | |
| 11 | 30 | M | 22 | Total | 5 | 5-ASA, Aza | Non | 3 | |
| 12 | 45 | M | 5 | Total | 3 | 5-ASA, Aza | Non | 4 | |
| 13 | 45 | F | 10 | Left sided | 2 | 5ASA | Non | >3e |
M, male; F, female.
Aza, azathioprine.
EIM, extraintestinal manifestations.
Non, nonsmoker; Ex, former smoker.
Lost to follow-up.
TABLE 2.
Controls
| Control IDa no. | Age (yr) at time of first sampling | Genderb | Description | Endoscopic and histological diagnosis | Localization |
|---|---|---|---|---|---|
| NC3 | 24 | M | Normal healthy volunteer | Normal | Sigmoid colon |
| NC8 | 20 | F | Normal healthy volunteer | Normal | Sigmoid colon |
| NC13 | 24 | F | Normal healthy volunteer | Normal | Sigmoid colon |
| NC24 | 32 | F | Normal healthy volunteer | Normal | Sigmoid colon |
| NC23 | 40 | M | Normal healthy volunteer | Normal | Sigmoid colon |
ID, identification.
M, male; F, female.
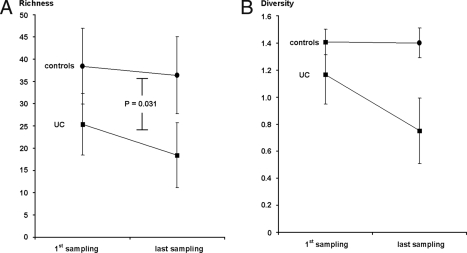
A total of 43 different band classes were identified for UC patients and 61 band classes for controls, corresponding to a baseline reduction in bacterial richness by 15% in UC patients at study start in comparison to levels for normal controls. One band of each class was excised, cloned, and sequenced. Sequencing of the 61 band classes resulted in taxonomic results from 53 bands, while 8 gave no reproducible results. The overall richness of bacterial operational taxonomic units (OTUs) as revealed by BLAST analysis is shown in Table 3. A total of 91 different OTUs were identified from the 53 band classes, including 17 (18.7%) sequences assigned to uncultured bacteria. As indicated in Fig. 1A, a significant reduction of bands was seen for UC patients between remission and relapse in comparison with repeated sampling of controls at the two different time points (25.31 versus 18.38 for UC patients, 38.4 versus 36.4 for the control sample, P = 0.031). A similar change in diversity scores was seen (Fig. 1B). No specific OTUs that would predict the time point of relapse were found during remission (i.e., at study start). Densitometric intensities of matched bands for the UC patients were pairwise compared between enrollment and time of relapse by use of a Wilcoxon matched-paired signed-rank test. For five bands, a significant difference was seen (P = 0.011), due to a reduction of the bacterial OTUs listed in Table 4. As shown, the decrease in bacterial complexity at relapse that was observed was due to a loss of normal intestinal bacterial taxa, such as Bacteroides, Escherichia, Eubacterium, Lactobacillus, and Ruminococcus spp. In comparison with the repeat biopsy specimens from normal controls, bacterial communities in specimens from patients with UC during remission showed variation in richness and diversity over time. Unfortunately, the relapse biopsy specimen endpoint was reached at month 2 for four patients and at month 3 for six patients (one patient lost to follow-up at month 3). One patient stayed in remission until month 4.
TABLE 3.
Taxonomic correlates of overall bacterial richness (alphabetical order)
| Bacterial species (according to BLAST analysis)a |
|---|
| Bacteroides acidifaciens |
| Bacteroides caccae |
| Bacteroides distasonis |
| Bacteroides fragilis |
| Bacteroides merdae |
| Bacteroides sp. (different strains) |
| Bacteroides stercoris |
| Bacteroides thetaiotaomicron |
| Bacteroides uniformis |
| Bacteroides vulgatus |
| Bifidobacterium adolescentis |
| Bifidobacterium breve |
| Bifidobacterium lo ngum subsp. infantis |
| Bifidobacterium longum |
| Bilophila wadsworthia |
| Burkholderia sp. |
| Burkholderiales bacterium |
| Citrobacter freundii |
| Citrobacter sp. |
| Citrobacter youngae |
| Clostridium amygdalinum |
| Clostridium indolis |
| Clostridium leptum |
| Clostridium methoxybenzovorans |
| Clostridium ramosum |
| Clostridium sp. (different strains) |
| Desulfovibrio piger |
| Dialister invisus |
| Escherichia coli |
| Escherichia fergusonii |
| Eubacterium eligens |
| Eubacterium rectale |
| Faecalibacterium prausnitzii |
| Finegoldia magna |
| Gemella sanguinis |
| Hafnia alvei |
| Lactobacillus gasseri |
| Lactobacillus johnsonii |
| Lactobacillus sp. |
| Morganella morganii |
| Obesumbacterium proteus |
| Peptoniphilus ivorii |
| Peptoniphilus sp. |
| Peptostreptococcus anaerobius |
| Peptostreptococcus micros |
| Peptostreptococcus sp. |
| Photorhabdus luminescens |
| Prevotella bivia |
| Prevotella bryantii |
| Prevotella melaninogenica |
| Prevotella sp. oral clone |
| Prevotella zoogleoformans |
| Rahnella aquatilis |
| Ruminococcus gnavus |
| Ruminococcus lactaris |
| Ruminococcus luti |
| Ruminococcus obeum |
| Serratia fonticola |
| Serratia marcescens |
| Streptococcus anginosus |
| Streptococcus constellatus |
| Streptococcus mitis |
| Sutterella wadsworthensis |
| Veillonella atypica |
| Veillonella ratti |
| Veillonella sp. |
| Yersinia frederiksenii |
| Uncultured bacteria |
| Bacterium 84634 |
| Bacterium mpn-isolate group 1 |
| Bacterium mpn-isolate group 20 |
| Butyrate-producing bacterium L2-21 |
| Butyrate-producing bacterium M21/2 |
| Butyrate-producing bacterium SM4/1 |
| Butyrate-producing bacterium SR1/5 |
| Butyrate-producing bacterium T2-132 |
| Uncultured bacterium |
| Uncultured bacterium adhufec151 |
| Uncultured bacterium adhufec405 |
| Uncultured eubacterium |
| Uncultured eubacterium E1-K8 |
| Uncultured gammaproteobacterium |
| Uncultured human fecal bacterium |
| Uncultured rumen bacterium 2C28d-8 |
| Unidentified rumen bacterium RFN27 |
Nomenclature according to reference 3.
FIG. 1.
Comparison of indicators (±standard deviations) for bacterial richness (number of bands in community fingerprinting gels [10]) (A) and bacterial diversity (weighted diversity scores calculated according to the method of Shannon and Weaver, as described earlier [7]) (B) at the points of first and last (i.e., the second for the controls) sampling, carried out using a nonparametric Wilcoxon test. As indicated, richness and diversity decrease in association with relapse in UC patients, whereas the same parameters were relatively stable for controls. The test indicators for richness in UC patients reached statistical significance compared to levels for controls (P = 0.031).
TABLE 4.
Intestinal bacteria downregulated at relapse in UC patients
| Bacterial species (according to BLAST analysis)a |
|---|
| Bacteroides distasonis |
| Bacteroides merdae |
| Bacteroides sp. |
| Butyrate-producing bacterium |
| Escherichia coli |
| Escherichia fergusonii |
| Eubacterium rectale |
| Lactobacillus gasseri |
| Lactobacillus johnsonii |
| Lactobacillus sp. |
| Morganella morganii |
| Peptostreptococcus anaerobius |
| Ruminococcus obeum |
| Streptococcus gordonii |
| Streptococcus mitis |
| Uncultured bacterium |
| Uncultured eubacterium |
| Uncultured gammaproteobacterium |
Nomenclature according to reference 3.
The loss of commensal organisms could profoundly modify gut mucosal homeostasis through a loss of essential micronutrients and redox potential (e.g., short-chain fatty acids) (1). A further decrease of microbial diversity therefore not only could be a marker of relapse in UC but might be a causative factor driving the inflammatory change. Gut microbiota in normal individuals are remarkably stable over time (12, 13). Although the study was hampered by the early relapse that most patients developed, it appears that UC is associated with a high degree of instability in richness and diversity of the MAF. The inability to find specific changes in the microbial composition to predict relapse or to detect a temporal interaction between different species of the MAF may result from a lack of power due to the limited observation time with the small number of patients. Larger consecutive series, including long-term remission studies, are necessary to investigate this question. There is a significant decrease in diversity and richness of the MAF of patients with UC in remission compared to that of controls, and a further decrease in diversity was observed at relapse. In particular, the decrease in bacterial complexity at relapse that was observed was due to a loss of normal intestinal bacterial taxa, such as Bacteroides, Escherichia, Eubacterium, Lactobacillus, and Ruminococcus spp. The composition of MAF in UC patients appears to be remarkably unstable. Further studies will investigate treatment outcome and long-term courses of remission to examine a putative link between MAF and the pathophysiology of active disease for UC.
Supplementary Material
Acknowledgments
We acknowledge funding through the BMBF (Competence Network IBD, National Genome Research Network, and Infectious Disease Network), DFG, and MFG.
S.S. declares competing financial interests as a speaker for SigmaTau (makers of VSL#3) and a member of the scientific advisory board for Applied Biosystems, an Applera corporation.
Footnotes
Published ahead of print on 13 August 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Blaut, M., and T. Clavel. 2007. Metabolic diversity of the intestinal microbiota: implications for health and disease. J. Nutr. 137751S-755S. [DOI] [PubMed] [Google Scholar]
- 2.Hart, A. L., A. J. Stagg, M. Frame, H. Graffner, H. Glise, P. Falk, and M. A. Kamm. 2002. The role of the gut flora in health and disease, and its modification as therapy. Aliment. Pharmacol. Ther. 161383-1389. [DOI] [PubMed] [Google Scholar]
- 3.Holt, J. G. (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, MD.
- 4.Manichahn, C., L. Rigottier-Gois, E. Bonnaud, K. Gloux, E. Pelletier, L. Frangeul, R. Nalin, C. Jarrin, P. Chardon, P. Marteau, J. Roca, and J. Dore. 2006. Reduced diversity of faecal microbiota in Crohn's disease revealed by metagenomic approach. Gut 55205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ott, S. J., M. Musfeldt, K. N. Timmis, J. Hampe, D. F. Wenderoth, and S. Schreiber. 2004. In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn. Microbiol. Infect. Dis. 50237-245. [DOI] [PubMed] [Google Scholar]
- 6.Ott, S. J., M. Musfeldt, U. Ullmann, J. Hampe, and S. Schreiber. 2004. Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J. Clin. Microbiol. 422566-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott, S. J., M. Musfeldt, D. F. Wenderoth, J. Hampe, O. Brandt, U. R. Fölsch, K. N. Timmis, and S. Schreiber. 2004. Reduction in diversity of the colonic mucosa associated microflora in patients with active inflammatory bowel disease. Gut 53685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder, K. W., W. J. Tremaine, and D. M. Ilstrup. 1987. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N. Engl. J. Med. 3171625-1629. [DOI] [PubMed] [Google Scholar]
- 9.Schryver, J. C., C. C. Brandt, S. M. Pfiffner, A. V. Palumbo, A. D. Peacock, D. C. White, J. P. McKinley, and P. E. Long. 2006. Application of nonlinear analysis methods for identifying relationships between microbial community structure and groundwater geochemistry. Microb. Ecol. 51177-188. [DOI] [PubMed] [Google Scholar]
- 10.Seksik, P., L. Rigottier-Gois, G. Gramet, M. Sutren, P. Pochart, P. Marteau, R. Jian, and J. Dore. 2003. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sepehri, S., R. Kotlowski, C. N. Bernstein, and D. O. Krause. 2007. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm. Bowel Dis. 13675-683. [DOI] [PubMed] [Google Scholar]
- 12.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 622273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 683401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.