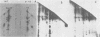Abstract
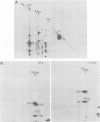
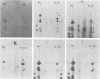
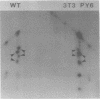
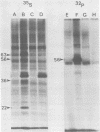
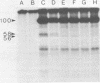
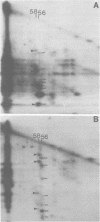
Two species of polyoma virus middle T antigen were detected in both lytically infected and transformed cells by in vitro kinase assay of immunoprecipitates. A minor species with an apparent molecular weight of 58,000 (58K) represented less than 10% of the total middle T protein. This species was roughly 10 times more active as a phosphate acceptor than was the predominant 56K form. Partial proteolytic mapping experiments showed that the same site was phosphorylated in both species. Mapping of the middle T antigens from a series of deletion mutants suggested that the major site of phosphorylation is tyrosine residue 315. Phosphorylation occurred on both middle T species in vivo, involving sites predominantly other than the tyrosine labeled in vitro. The 56K and 58K middle T forms differed from each other in their in vivo phosphorylation patterns. Some phosphate was incorporated into the 58K species in a region of the molecule to which at least part of the apparent molecular weight different could be mapped. hr-t mutant NG-59, which codes for a slightly altered middle T, produced only a single species (56K) which was inactive in the in vitro kinase reaction. Moreover, no 58K species appeared in vivo with this mutant. hr-t mutants are therefore defective in both aspects of phosphorylation. Phenotypically normal revertant cells of a polyoma transformed line failed to express any middle T antigens or associated kinase activity.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendig M. M., Thomas T., Folk W. R. Viable deletion mutant in the medium and large T-antigen-coding sequences of the polyoma virus genome. J Virol. 1980 Mar;33(3):1215–1220. doi: 10.1128/jvi.33.3.1215-1220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin T. L. Tumor viruses and cellular gene expression. Cancer Res. 1976 Nov;36(11 Pt 2):4289–4290. [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G., Benjamin T. L. Identification of DNA sequence changes leading to loss of transforming ability in polyoma virus. J Biol Chem. 1980 Jan 10;255(1):230–235. [PubMed] [Google Scholar]
- Collett M. S., Erikson E., Erikson R. L. Structural analysis of the avian sarcoma virus transforming protein: sites of phosphorylation. J Virol. 1979 Feb;29(2):770–781. doi: 10.1128/jvi.29.2.770-781.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S., Erikson R. L. Protein kinase activity associated with the avian sarcoma virus src gene product. Proc Natl Acad Sci U S A. 1978 Apr;75(4):2021–2024. doi: 10.1073/pnas.75.4.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W., Hutchinson M. A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979 Dec;18(4):925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- Feldman R. A., Hanafusa T., Hanafusa H. Characterization of protein kinase activity associated with the transforming gene product of Fujinami sarcoma virus. Cell. 1980 Dec;22(3):757–765. doi: 10.1016/0092-8674(80)90552-8. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Benjamin T. L. Comparisons of two early gene functions essential for transformation in polyoma virus and SV-40. Virology. 1979 Jul 15;96(1):205–228. doi: 10.1016/0042-6822(79)90185-5. [DOI] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Glynn I. M., Chappell J. B. A simple method for the preparation of 32-P-labelled adenosine triphosphate of high specific activity. Biochem J. 1964 Jan;90(1):147–149. doi: 10.1042/bj0900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman E., Benjamin T. L. Analysis of host range of nontransforming polyoma virus mutants. Virology. 1975 Aug;66(2):372–384. doi: 10.1016/0042-6822(75)90210-x. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Ito Y., Novak U., Spurr N., Dilworth S., Smolar N., Pollack R., Smith K., Rifkin D. B. Early mutants of polyoma virus (dl8 and dl23) with altered transformation properties: is polyoma virus middle T antigen a transforming gene product? Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):271–283. doi: 10.1101/sqb.1980.044.01.031. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori J., Carmichael G. G., Benjamin T. L. DNA sequence alterations in Hr-t deletion mutants of polyoma virus. Cell. 1979 Mar;16(3):505–513. doi: 10.1016/0092-8674(79)90025-4. [DOI] [PubMed] [Google Scholar]
- Hohmann P. Species-specific variations in H1 histone phosphopeptides. J Biol Chem. 1979 Sep 25;254(18):9022–9029. [PubMed] [Google Scholar]
- Hunter T., Hutchinson M. A., Eckhart W. Translation of polyoma virus T antigens in vitro. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5917–5921. doi: 10.1073/pnas.75.12.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., Sefton B. M. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Griffin B. E. Middle T antigen as primary inducer of full expression of the phenotype of transformation by polyoma virus. J Virol. 1980 Jul;35(1):219–232. doi: 10.1128/jvi.35.1.219-232.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Karess R. E., Hayward W. S., Hanafusa H. Cellular information in the genome of recovered avian sarcoma virus directs the synthesis of transforming protein. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3154–3158. doi: 10.1073/pnas.76.7.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S., Yoshida M., Segawa K., Sugiyama H., Ishizaki R., Toyoshima K. Characterization of Y73, an avian sarcoma virus: a unique transforming gene and its product, a phosphopolyprotein with protein kinase activity. Proc Natl Acad Sci U S A. 1980 Oct;77(10):6199–6203. doi: 10.1073/pnas.77.10.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs E. G., Beavo J. A. Phosphorylation-dephosphorylation of enzymes. Annu Rev Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levinson A. D., Oppermann H., Levintow L., Varmus H. E., Bishop J. M. Evidence that the transforming gene of avian sarcoma virus encodes a protein kinase associated with a phosphoprotein. Cell. 1978 Oct;15(2):561–572. doi: 10.1016/0092-8674(78)90024-7. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rübsamen H., Friis R. R., Bauer H. Src Gene product from different strains of avian sarcoma virus: Kinetics and possible mechanism of heat inactivation of protein kinase activity from cells infected by transformation-defective, temperature-sensitive mutant and wild-type virus. Proc Natl Acad Sci U S A. 1979 Feb;76(2):967–971. doi: 10.1073/pnas.76.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1092–1096. doi: 10.1073/pnas.73.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Phosphorylation of polyoma T antigens. Cell. 1979 Dec;18(4):935–946. doi: 10.1016/0092-8674(79)90206-x. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Silver J. E., Benjamin T. L. Tumor antigen(s) in cell productively infected by wild-type polyoma virus and mutant NG-18. Proc Natl Acad Sci U S A. 1978 Jan;75(1):79–83. doi: 10.1073/pnas.75.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R., Benjamin T. L. Cellular alterations dependent upon the polyoma virus Hr-t function: separation of mitogenic from transforming capacities. Cell. 1978 Jul;14(3):587–599. doi: 10.1016/0092-8674(78)90244-1. [DOI] [PubMed] [Google Scholar]
- Silver J., Schaffhausen B., Benjamin T. Tumor antigens induced by nontransforming mutants of polyoma virus. Cell. 1978 Oct;15(2):485–496. doi: 10.1016/0092-8674(78)90018-1. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Griffin B., Fried M. Protein kinase activity associated with polyoma virus middle T antigen in vitro. Cell. 1979 Dec;18(4):915–924. doi: 10.1016/0092-8674(79)90204-6. [DOI] [PubMed] [Google Scholar]
- Spector M., O'Neal S., Racker E. Regulation of phosphorylation of the beta-subunit of th Ehrlich ascites tumor Na+K+-ATPase by a protein kinase cascade. J Biol Chem. 1981 May 10;256(9):4219–4227. [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- WINOCOUR E. Purification of polyoma virus. Virology. 1963 Feb;19:158–168. doi: 10.1016/0042-6822(63)90005-9. [DOI] [PubMed] [Google Scholar]
- Willingham M. C., Jay G., Pastan I. Localization of the ASV src gene product to the plasma membrane of transformed cells by electron microscopic immunocytochemistry. Cell. 1979 Sep;18(1):125–134. doi: 10.1016/0092-8674(79)90361-1. [DOI] [PubMed] [Google Scholar]
- Witte O. N., Dasgupta A., Baltimore D. Abelson murine leukaemia virus protein is phosphorylated in vitro to form phosphotyrosine. Nature. 1980 Feb 28;283(5750):826–831. doi: 10.1038/283826a0. [DOI] [PubMed] [Google Scholar]
- Witte O. N., Rosenberg N., Baltimore D. Preparation of syngeneic tumor regressor serum reactive with the unique determinants of the Abelson murine leukemia virus-encoded P120 protein at the cell surface. J Virol. 1979 Sep;31(3):776–784. doi: 10.1128/jvi.31.3.776-784.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]