Abstract
The initial isolation of Helcococcus ovis from a valvular thrombus prompted us to investigate the prevalence of this bacterium in bovine valvular endocarditis. Specimens from 55 affected hearts were examined by culture using Columbia blood agar and cross streaking the inoculated plate with a Staphylococcus aureus strain. As confirmed by 16S rRNA gene sequencing, H. ovis was isolated with an unexpectedly high frequency of 33%, predominantly as heavy growth and pure culture. The majority of H. ovis isolates showed distinct satellitism around S. aureus and pyridoxal dependency, resembling “nutritionally variant streptococci” (now assigned to the genera Abiotrophia and Granulicatella). Using the API rapid ID 32 Strep, API ZYM, and Rosco Diatabs systems, incongruent results were obtained for alkaline phosphatase, β-galactosidase, β-glucuronidase, and leucine aminopeptidase activities. Based on the satellitism/pyridoxal dependency; hemolysis on blood agar; the API rapid ID 32 Strep results for arginine dihydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, and pyroglutamic acid arylamidase activities; hippurate hydrolysis; and acidification of sucrose, a scheme for the identification of H. ovis and its differentiation from other members of the Helcococcus genus and the pyridoxal-dependent species Abiotrophia defectiva, Granulicatella adiacens, and Granulicatella elegans is proposed. By establishing specific fluorescence in situ hybridization, large H. ovis aggregates were specifically detected within the fibrinous exudate of the valvular thrombi. Our results demonstrate for the first time that H. ovis represents an emerging pathogen in bovine valvular endocarditis that is frequently isolated if appropriate culture conditions are used.
Helcococcus ovis, belonging to the family Peptostreptococcaceae, is a catalase-negative, facultatively anaerobic, gram-positive coccus first described by Collins et al. in 1999 (4). Initially recovered from lung, liver, and spleen of a sheep together with Arcanobacterium pyogenes and from a case of subclinical ovine mastitis together with a Staphylococcus species, its clinical significance was unclear (4). Later on, H. ovis was isolated from equine and bovine pulmonary abscesses and two cases of bovine valvular endocarditis, indicating that H. ovis might be etiologically involved in infections of different mammalian hosts and organ systems (10, 12, 15, 18).
To the best of our knowledge, so far only four H. ovis strains have been characterized phenotypically (4, 15, 18). On blood agar, colonies were described as pinpoint, nonpigmented, and nonhemolytic at 24 h but slightly alpha-hemolytic after 72 h of incubation. Initially growth restricted to the periphery of a Staphylococcus species was observed for the H. ovis strain (CCUG 39041) from sheep and the equine isolate. However, after repeated subculture on blood agar, the strains lost their dependency on the Staphylococcus species (4, 18). Biochemical tests were performed using the API rapid ID 32 Strep and API ZYM kits (bioMérieux) (4), the API 20 Strep kit (bioMérieux) (18), or without further specification (15), leading to varying results for alkaline phosphatase, leucine aminopeptidase, and pyrrolidonyl arylamidase activities.
During routine bacteriology of a bovine heart showing valvular endocarditis, we isolated H. ovis in pure culture, pointing toward an etiological significance of this bacterium in that particular case. We therefore initiated the present study, investigating the prevalence of H. ovis in this important disease by examining a considerable number of bovine endocarditis samples.
MATERIALS AND METHODS
Collection of specimens.
In a 1.5-year period, 54 samples from bovine hearts with valvular endocarditis were collected during meat inspection at two abattoirs located in the Federal State of Brandenburg, Germany. The specimens were refrigerated immediately at 4°C and transported to the laboratory on the same day. One additional specimen was obtained during a necropsy carried out in the pathology department of the Landeslabor Brandenburg, Frankfurt (Oder), Germany.
Pathological examination.
All heart samples were examined macroscopically, and affected parts were determined. From six hearts with positive H. ovis culture results, representative tissue sections were fixed in 10% (vol/vol) neutral buffered formalin, embedded in paraffin wax, cut at 4-μm thickness, and stained with hematoxylin and eosin for histological investigation. In addition, Gram staining was used to characterize bacterial aggregates present in the slides.
Bacteriology.
Primary culture was carried out by transferring valvular plaque material on Columbia agar containing 5% sheep blood (CBA) (Oxoid, Wesel, Germany) and overlaying the inoculated area with a single streak of Staphylococcus aureus. CBA was incubated at 36°C in ambient air for 48 h. Isolates showing satellitism were examined phenotypically and by 16S rRNA gene sequencing to verify H. ovis. All other isolates were identified by conventional bacteriological methods described elsewhere (20). For storage, H. ovis colony material was suspended in a cryovial containing 1.8 ml brain heart infusion broth and 0.2 ml sterile glycerol and kept at −80°C.
Phenotypic characteristics.
The H. ovis isolates were retrieved from storage, grown on Columbia blood pyridoxal agar (CBPA) (Columbia agar base [Merck, Darmstadt, Germany] supplemented with 10% [vol/vol] defibrinated sheep blood and 0.002% [wt/vol] pyridoxal HCl [Sigma-Aldrich, Deisenhofen, Germany]) at 36°C in an atmosphere of 6% CO2 for 48 h, and subcultured once for 24 h using the same conditions. The isolates were examined and compared to the H. ovis strains CCUG 37441T and CCUG 39041 for the following characteristics: Gram stain reaction, catalase reaction, vancomycin resistance, anaerobic growth on CBPA, satellitism, pyridoxal dependency, and various biochemical reactions. For testing satellitism and pyridoxal dependency, a saline suspension equivalent to a turbidity of a 0.5 McFarland standard was plated onto CBA. Thereafter, an S. aureus streak was placed on one half and a pyridoxal HCl (0.01% [wt/vol] aqueous solution)-impregnated 6-mm paper disk on the other half of the agar plate. Incubation was performed at 36°C in an atmosphere of 6% CO2 for 24 and 48 h. The strains Abiotrophia defectiva DSM 9849T, Granulicatella adiacens DSM 9848T, and Granulicatella elegans DSM 11693T served as positive controls and the strain Helcococcus kunzii DSM 10548T as a negative control.
Biochemical tests were performed in duplicate using the API rapid ID 32 Strep and API ZYM kits according to the manufacturer's instructions (bioMérieux, Nürtingen, Germany). In addition, alkaline phosphatase, arginine dihydrolase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, leucine aminopeptidase, and pyrrolidonyl arylamidase activities; hippurate hydrolysis; and acid production from maltose and sucrose were examined using Rosco Diatabs (A/S Rosco, Taastrup, Denmark). Diatabs tests for alkaline phosphatase activity and hippurate hydrolysis were analyzed at 4 h and the remaining reactions after overnight incubation.
16S rRNA gene sequencing analysis.
DNA extraction from H. ovis isolates was performed using the Qiagen tissue kit according to the bacterial support protocol. For PCR amplification, approximately 1,490 bp of the 16S rRNA gene was amplified using forward primer Seq-fw (5′-TGGCTCAGGACGAACGCT-3′, positions 20 to 37 in the Escherichia coli 16S rRNA gene) and Seq-rev (5′-CTTCGGGTATTGCCAACTC-3′, positions 1416 to 1434 in the Escherichia coli 16S rRNA gene). Amplification of the 16S rRNA gene was conducted in a final volume of 50 μl containing 2 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 10 pmol of each primer, and 2.5 U Taq polymerase. Samples were preheated at 94°C for 5 min, followed by amplification at 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s. A total of 30 cycles were carried out, with a final elongation step at 72°C for 10 min. Successful amplification was verified by agarose gel electrophoresis.
Sequencing of the 16S rRNA gene was conducted commercially (AGOWA, Berlin, Germany), resulting in at least 1,290 bp for each strain. Phylogenetic analyses were performed by comparative 16S rRNA gene sequence analysis using the Lasergene software package (DNASTAR Inc., Madison, WI).
Fluorescence in situ hybridization (FISH).
The H. ovis-specific probe HOVIS (5′-ATAGTATAGTTTCTTCGGAAAC-3′), corresponding to positions 91 to 104 in E. coli 16S rRNA, was designed after comparative analysis of the 16Sr RNA gene sequences obtained for the H. ovis endocarditis isolates and the sequences of H. ovis CCUG 37441T, H. kunzii DSM 10548T, and Helcococcus sueciensis CCUG 47334T as deposited in the GenBank database. The specificity of the probe HOVIS was checked against all 16S rRNA gene sequence entries available in GenBank and the Ribosomal Database Project II using the software tool “Probe Match,” which is part of the Ribosomal Database Project environment (11).
The oligonucleotide probe HOVIS used for the FISH technique was synthesized commercially (Thermo Hybaid, Ulm, Germany) and 5′ labeled with Cy3 (indocarbocyanine). Representative tissue samples were prepared as described for histological investigations and deparaffanized in xylene prior to FISH. To test the specificity of the probe and to adjust stringency, fixed control strains were used at formamide concentrations of 10, 20, 30, and 40% (vol/vol).
FISH was performed at 48°C in a humid chamber for 18 h using 20 μl hybridization buffer (30% [vol/vol] deionized formamide, 0.9 M NaCl, 20 mM Tris-HCl [pH 7.4], 0.01% [wt/vol] sodium dodecyl sulfate) and 100 ng of HOVIS for each section. After incubation, sections were washed with distilled water at 48°C for 10 min and mounted with ProLong antifade reagent (Molecular Probes, Leiden, The Netherlands). Fluorescence microscopy was performed using a Leica DMBL microscope (Leica, Wetzlar, Germany).
To assess specific hybridization conditions, the following Helcococcus strains as well as strains of unrelated bacterial species were included in FISH experiments to serve as positive and negative controls: H. ovis CCUG 37441T, H. ovis CCUG 39041, H. kunzii DSM 10548T, H. sueciensis CCUG 47334T, A. pyogenes IMT 8803, Clostridium perfringens ATCC 13124T, Enterococcus dispar DSM 6630T, Enterococcus faecium ATCC 6057, S. aureus ATCC 25923, and Streptococcus sp. strain ATCC 9932.
RESULTS
Pathology and bacteriology.
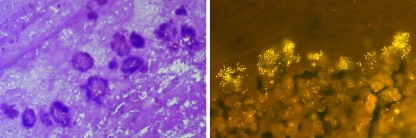
All 55 hearts investigated showed typical lesions of chronic valvular endocarditis. The valvular thrombi were of variable size and composed of a luminal zone containing unorganized fibrinous exudate. Deeper parts of the thrombi were organized by granulation tissue maturing into avascular, collagen-rich scar tissue. The type and quantity of affected valves were determined as follows: tricuspic, 39; tricuspic and bicuspic with myocard abscesses, 1; tricuspic and pulmonic, 1; pulmonic, 6; bicuspic, 6; aortic, 1; and atrioventricular but not further identifiable, 1. In H. ovis culture-positive thrombi, histology revealed large aggregates of gram-positive cocci within the luminal fibrinous exudate (Fig. 1).
FIG. 1.
Gram staining (left) (magnification, ×100) and FISH (right) (magnification, ×400) showing multifocal aggregates within the fibrinous exudate of an H. ovis culture-positive valvular thrombus.
From all valvular plaque specimens, bacteria were recovered as heavy growth, mainly in pure culture or accompanied by just a few colonies of unspecific flora, e.g., coliform bacteria, enterococci, and/or coagulase-negative staphylococci. Mixed infections were found in only three cases (Table 1). H. ovis was obtained from 17 cows and 1 bull originating from 17 different farms. The mean age of the animals was 47 months (range, 20 to 75 months).
TABLE 1.
Bacteriological findings in 55 cases of bovine valvular endocarditis
| Predominant bacterial species | Coisolate(s) | No. of cases |
|---|---|---|
| A. pyogenes | None | 33 |
| S. aureus | 1 | |
| H. ovis | None | 16 |
| Pseudomonas aeruginosa, Enterococcus faecalis | 1 | |
| Streptococcus dysgalactiae subsp. dysgalactiae | 1 | |
| S. dysgalactiae subsp. dysgalactiae | None | 1 |
| Streptococcus sp. | None | 1 |
| Member of the family Neisseriaceae | None | 1 |
In primary culture, the H. ovis isolates demonstrated distinct satellitism around the S. aureus streak. For only one isolate, a slight growth outside the S. aureus hemolysis zone could be observed. Colony polymorphism, ranging from pinpoint, grayish colonies to tiny, translucent colonies, was common and remained stable after subculture on CBPA. All H. ovis isolates were nonhemolytic on CBA and CBPA after 48 h of incubation.
Phenotypic characteristics.
All H. ovis strains were gram positive to gram labile, with cocci arranged in clusters and pairs. They were catalase negative and vancomycin sensitive and grew facultatively or anaerobically. After subculture and storage, 14 H. ovis endocarditis isolates (78%) showed satellitism and pyridoxal dependency, whereas the H. ovis type strain (CCUG 37441T) did not. H. ovis strain CCUG 39041 yielded an ambiguous result, characterized by marked growth restricted to the zones of S. aureus hemolysis and pyridoxal disc diffusion at 24 h but sparse growth even outside these zones after 48 h of incubation.
Details of the biochemical characterization are given in Tables 2 to 4. The results for some enzyme activities differed depending on the test system used. Alkaline phosphatase activity was always positive in the API ZYM system but negative in the API rapid ID 32 Strep (color shift to very pale yellow) and the Rosco Diatabs systems. β-Galactosidase activity was continuously demonstrated for the substrate resorufin-β-d-galactopyranoside (API rapid ID 32 Strep) but not for 2-naphtyl-β-d-galactopyranoside (API rapid ID 32 Strep and API ZYM). Using the Rosco o-nitrophenyl-β-d-galactopyranoside Diatabs, predominantly weak positive reactions were observed. All strains except one that were positive for β-glucuronidase in the API rapid ID 32 Strep yielded positive results with the respective Rosco Diatabs, but all strains were negative in the API ZYM. All strains displayed strong leucine aminopeptidase activity in the API ZYM, but four endocarditis isolates and H. ovis CCUG 39041 were negative using the Rosco Diatabs, exhibiting a yellow or yellow-orange color.
TABLE 2.
API rapid ID 32 Strep results for 18 H. ovis endocarditis isolates compared to results obtained for H. ovis control strains
| Phenotypic characteristica | Test result for H. ovis strain:
|
Aggregated test resultb (no. of positive isolates) for H. ovis endocarditis isolates | |
|---|---|---|---|
| CCUG 37441T | CCUG 39041 | ||
| Arginine dihydrolase | − | − | − (0) |
| β-Glucosidase | − | − | − (0) |
| β-Galactosidasec | + | + | + (18) |
| β-Glucuronidase | + | + | V (11) |
| α-Galactosidase | − | − | − (0) |
| Alkaline phosphatase | − | − | − (0) |
| Ribose | − | − | − (0) |
| Mannitol | − | − | − (0) |
| Sorbitol | − | − | − (0) |
| Lactose | − | − | V (4) |
| Trehalose | − | − | V (5) |
| Raffinose | − | − | − (0) |
| Acetoin | − | − | − (0) |
| Alanyl-phenylalanyl-proline arylamidase | − | − | − (0) |
| β-Galactosidased | − | − | − (1) |
| Pyroglutamic acid arylamidase | − | − | − (0) |
| N-Acetyl-β-Glucosaminidase | − | − | − (0) |
| Glycyl-tryptophan arylamidase | − | − | − (0) |
| Hippurate hydrolysis | − | − | − (0) |
| Glycogen | − | − | − (0) |
| Pullulan | − | − | V (11) |
| Maltose | + | + | + (18) |
| Melibiose | − | − | − (0) |
| Melezitose | − | − | − (0) |
| Sucrose | − | − | − (0) |
| l-Arabinose | − | − | − (0) |
| d-Arabitol | − | − | − (0) |
| Methyl-β-d-glucopyranoside | − | − | − (0) |
| Tagatose | − | − | − (1) |
| β-Mannosidase | − | − | − (0) |
| Cyclodextrin | − | + | + (16) |
| Urease | − | − | − (0) |
In order of the arrangement in the kit.
+, V, and −, ≥85%, 16 to 84%, and ≤15% of strains were positive, respectively.
With resorufin-β-d-galactopyranoside as the substrate.
With 2-naphtyl-β-d-galactopyranoside as the substrate.
TABLE 4.
Rosco Diatabs results for 18 H. ovis endocarditis isolates compared to the results obtained for H. ovis control strains
| Phenotypic characteristic | Test result for H. ovis strain:
|
Aggregated test resulta (no. of positive isolates) for H. ovis endocarditis isolates | |
|---|---|---|---|
| CCUG 37441T | CCUG 39041 | ||
| Alkaline phosphatase | − | − | − (0) |
| Arginine dihydrolase | − | − | − (0) |
| α-Galactosidase | − | − | − (0) |
| β-Galactosidaseb | + | + | + (16) |
| β-Glucosidase | − | − | − (0) |
| β-Glucuronidase | + | + | V (10) |
| Leucine aminopeptidase | + | − | V (14) |
| Pyrrolidonyl arylamidase | − | − | − (0) |
| Hippurate hydrolysis | − | − | − (0) |
| Maltose | + | + | + (18) |
| Sucrose | − | − | − (0) |
+, V, and −, ≥85%, 16 to 84%, and ≤15% of strains were positive, respectively.
Predominantly weak reaction.
16S rRNA sequence analysis.
All H. ovis endocarditis isolates shared an almost-identical 16S rRNA gene sequence, with at least 99.6% sequence identity. By comparing their 16S rRNA gene sequences to all available entries in GenBank, every strain could be assigned to the species H. ovis, showing at least 99.7% sequence identity with H. ovis CCUG 37441T.
FISH.
Using 30% (vol/vol) formamide, only H. ovis was detected by FISH with the HOVIS probe, whereas no signals were obtained from the negative control strains. When FISH was applied to the tissue sections, H. ovis was demonstrated in large amounts, predominantly as multifocal aggregates within the fibrinous exudate in luminal parts of the valvular thrombi (Fig. 1).
DISCUSSION
Valvular endocarditis is an important cause of illness and death in adult cattle, predominantly cows, with an incidence ranging from 0.12% at slaughter to up to 9% in necropsy cases (13, 21). A. pyogenes is known to be the most important bacterial agent in bovine endocarditis (13, 14, 16, 19, 21) and was also found in the majority of cases (62%) in this study. Remarkably, as confirmed by 16S rRNA gene sequence analysis, H. ovis (33%) represented the second most common isolate, recovered mainly in pure culture.
Histologically, the H. ovis-induced valvular lesions were characterized by large bacterial aggregates, thus corresponding to the results of Post et al. (15). In addition, we applied FISH in the postmortem examination and specifically detected H. ovis in the superficial parts of the lesions. Due to the fastidious character of some bacteria capable of causing endocarditis, culture often remains difficult. Therefore, FISH has already been proven to be an important diagnostic tool for the detection of human endocarditis pathogens (8), and here we also demonstrate its usefulness in bovine endocarditis.
Satellitism and pyridoxal dependency were found to be major characteristics of H. ovis endocarditis isolates, resembling the case for the so-called “nutritionally variant streptococci” (7), which are now designated A. defectiva, G. adiacens, and G. elegans (5, 9, 17). As reported for Abiotrophia and Granulicatella species (1, 6), H. ovis may also adapt to growth without a Staphylococcus strain or pyridoxal supplement (4, 18), which might complicate the phenotypic identification. In the present study, the first two H. ovis isolates obtained demonstrated satellitism in primary culture. Prior to storage, they were transferred three or four times on CBA with an S. aureus streak. After recovery, growth on CBA did not require S. aureus or pyridoxal. Therefore, all other isolates were subcultured only once on CBPA before storage at −80°C, but despite this measure, two of them also lost satellitism and pyridoxal dependency.
Examination of biochemical reactions using different test systems revealed incongruent results for previously defined key characteristics, i.e., alkaline phosphatase, β-glucuronidase, and leucine aminopeptidase activities (4), and for β-galactosidase activity. This might be due to the various substrates contained in the systems and indicates the need for precise method specification when presenting biochemical data. If commercially available test kits are used, the manufacturer's instructions should be strictly followed to achieve comparable results. In contrast to the work of Collins et al. (4), production of esterase C4 and esterase lipase C8 were consistently observed using the API ZYM. Moreover, the large majority of H. ovis strains displayed weak valine arylamidase activity.
Based on evaluation of the phenotypic characteristics examined during this work, an API rapid ID 32 Strep-based scheme for the identification of H. ovis and its differentiation from other Helcococcus species and the pyridoxal-dependent species A. defectiva, G. adiacens, and G. elegans is proposed (Table 5). The predicted results for H. kunzii, H. sueciensis, A. defectiva, G. adiacens, and G. elegans (1-3, 5, 9, 17) were verified for the API rapid ID 32 Strep kit using the type strains of the respective species. In contrast to previous findings (1), β-galactosidase activity was negative for the type strain of G. adiacens (DSM 9848T).
TABLE 5.
Phenotypic characteristics, including API rapid ID 32 Strep-based tests, potentially useful in the identification of H. ovis and its discrimination from other Helcococcus species and the pyridoxal-dependent species A. defectiva, G. adiacens, and G. elegans
| Phenotypic characteristic | Result for speciesa (source)
|
|||||
|---|---|---|---|---|---|---|
| H. ovis (animal) | H. kunzii (human) | H. sueciensis (human) | A. defectiva (human) | G. adiacens (human) | G. elegans (human) | |
| Satellitism/pyridoxal dependency | + | − | − | + | + | + |
| Hemolysis on sheep blood agar | γ | γ | γ | α | α | α |
| Arginine dihydrolase | − | − | − | − | − | + |
| α-Galactosidase | − | − | − | + | − | − |
| β-Galactosidaseb | + | + | + | + | − | − |
| β-Galactosidasec | − | − | + | + | − | − |
| β-Glucuronidase | V | − | − | − | + | − |
| Hippurate hydrolysis | − | − | − | − | − | + |
| Pyroglutamic acid arylamidase | − | + | − | + | + | + |
| Sucrose | − | − | − | + | + | + |
+, V, and −, ≥85%, 16 to 84%, and ≤15% of strains were positive, respectively.
With resorufin-β-d-galactopyranoside as the substrate.
With 2-naphtyl-β-d-galactopyranoside as the substrate.
The high culture prevalence and FISH results presented here demonstrate that H. ovis is an emerging pathogen in bovine valvular endocarditis. For the future, attention should be paid to this bacterium in veterinary microbiology, ensuring appropriate culture conditions crucial for its isolation, particularly coincubation of specimens with a Staphylococcus strain and use of pyridoxal-supplemented media.
Although one report assumes the skin to be the source of H. ovis (18), there is no valid information concerning the natural habitat. Thus, it remains to be investigated how H. ovis enters the bloodstream and how this pathogen circumvents serum complement activity.
TABLE 3.
API ZYM results for 18 H. ovis endocarditis isolates compared to the results obtained for H. ovis control strains
| Phenotypic characteristica | Test result for H. ovis strain:
|
Aggregated test resultb (no. of positive isolates) for H. ovis endocarditis isolates | |
|---|---|---|---|
| CCUG 37441T | CCUG 39041 | ||
| Alkaline phosphatase | + | + | + (18) |
| Esterase (C4) | + | + | + (18) |
| Esterase lipase (C8) | + | + | + (18) |
| Lipase (C14) | − | − | − (0) |
| Leucine arylamidase | + | + | + (18) |
| Valine arylamidasec | + | + | + (16) |
| Cystine arylamidase | + | + | + (18) |
| Trypsin | − | − | − (0) |
| α-Chymotrypsin | − | − | − (0) |
| Acid phosphatase | + | + | + (18) |
| Naphtol-AS-BI-phosphohydrolase | + | + | + (18) |
| α-Galactosidase | − | − | − (0) |
| β-Galactosidase | − | − | − (0) |
| β-Glucuronidase | − | − | − (0) |
| α-Glucosidasec | + | + | V (14) |
| β-Glucosidase | − | − | − (0) |
| N-Acetyl-β-glucosaminidase | − | − | − (0) |
| α-Mannosidase | − | − | − (0) |
| α-Fucosidase | − | − | − (0) |
In order of the arrangement in the kit.
+, V, and −, ≥85%, 16 to 84%, and ≤15% of strains were positive, respectively.
Frequently weak reaction.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Christensen, J. J., and R. R. Facklam. 2001. Granulicatella and Abiotrophia species from human clinical specimens. J. Clin. Microbiol. 393520-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins, M. D., R. R. Facklam, U. M. Rodrigues, and K. L. Ruoff. 1993. Phylogenetic analysis of some Aerococcus-like organisms from clinical sources: description of Helcococcus kunzii gen. nov., sp. nov. Int. J. Syst. Bacteriol. 43425-429. [DOI] [PubMed] [Google Scholar]
- 3.Collins, M. D., E. Falsen, K. Brownlee, and P. A. Lawson. 2004. Helcococcus sueciensis sp. nov., isolated from a human wound. Int. J. Syst. Evol. Microbiol. 541557-1560. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. D., E. Falsen, G. Foster, L. R. Monasterio, L. Dominguez, and J. F. Fernandez-Garazabal. 1999. Helcococcus ovis sp. nov., a gram-positive organism from sheep. Int. J. Syst. Bacteriol. 491429-1432. [DOI] [PubMed] [Google Scholar]
- 5.Collins, M. D., and P. A. Lawson. 2000. The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov. and Granulicatella balaenopterae comb. nov. Int. J. Syst. Evol. Microbiol. 50365-369. [DOI] [PubMed] [Google Scholar]
- 6.Facklam, R. R. 2001. Newly described, difficult-to-identify, catalase-negative, Gram-positive cocci. Clin. Microbiol. Newsl. 231-7. [Google Scholar]
- 7.Frenkel, A., and W. Hirsch. 1961. Spontaneous development of L forms of streptococci requiring secretions of other bacteria or sulphydryl compounds for normal growth. Nature 191728-730. [DOI] [PubMed] [Google Scholar]
- 8.Gescher, D. M., C. Mallmann, D. Kovacevic, D. Schmiedel, A. C. Borges, B. Schweickert, U. B. Gobel, and A. Moter. 2008. A view on Bartonella quintana endocarditis-confirming the molecular diagnosis by specific fluorescence in situ hybridization. Diagn. Microbiol. Infect. Dis. 6099-103. [DOI] [PubMed] [Google Scholar]
- 9.Kawamura, Y., X. G. Hou, F. Sultana, S. Liu, H. Yamamoto, and T. Ezaki. 1995. Transfer of Streptococcus adjacens and Streptococcus defectivus to Abiotrophia gen. nov. as Abiotrophia adiacens comb. nov. and Abiotrophia defectiva comb. nov., respectively. Int. J. Syst. Bacteriol. 45798-803. [DOI] [PubMed] [Google Scholar]
- 10.Kubota, Y., and K. Morimoto. 2006. Association between cases of bovine abscess during the previous 20-year period in Hiroshima prefecture and Helcococcus ovis. Hiroshima J. Vet. Med. 2124-27. [Google Scholar]
- 11.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morimoto, K., Y. Kubota, A. Fujita, C. Kawamoto, and Y. Ibaraki. 2006. A case of bovine valvular endocarditis in which Helcococcus ovis is isolated. J. Jpn. Vet. Med. Assoc. 59325-328. [Google Scholar]
- 13.Muller, M., S. Platz, J. Ehrlein, T. Ewringmann, G. Molle, and A. Weber. 2005. Bacterially conditioned thromboembolism in dairy cows—a retrospective study of 31 necropsy cases with special consideration of the causative complex. Berl. Munch. Tierarztl. Wochenschr. 118121-127. [PubMed] [Google Scholar]
- 14.Narucka, U., J. van den Berg, J. F. Nouws, B. D. Okma, J. P. Peelen, and A. E. Soethout. 1985. Disorders in slaughtering animals. V. Endocarditis in slaughtering pigs, sows and cattle. Tijdschr. Diergeneeskd. 110776-779. [PubMed] [Google Scholar]
- 15.Post, K. W., S. D. Rushton, and S. J. Billington. 2003. Valvular endocarditis associated with Helcococcus ovis infection in a bovine. J. Vet. Diagn. Investig. 15473-475. [DOI] [PubMed] [Google Scholar]
- 16.Power, H. T., and W. C. Rebhun. 1983. Bacterial endocarditis in adult dairy cattle. J. Am. Vet. Med. Assoc. 182806-808. [PubMed] [Google Scholar]
- 17.Roggenkamp, A., M. Abele-Horn, K. H. Trebesius, U. Tretter, I. B. Autenrieth, and J. Heesemann. 1998. Abiotrophia elegans sp. nov., a possible pathogen in patients with culture-negative endocarditis. J. Clin. Microbiol. 36100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothschild, C. M., J. L. Oaks, J. K. Schaupp, F. R. Rurangirwa, D. C. Sellon, and M. T. Hines. 2004. Helcococcus ovis isolated from a pulmonary abscess in a horse. J. Clin. Microbiol. 422224-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiele, R., G. Amtsberg, and C. Meier. 1984. Diagnosis of bacterial endocarditis in cattle using intra vitam cultured blood samples. Dtsch. Tierarztl. Wochenschr. 9115-18. [PubMed] [Google Scholar]
- 20.Songer, J. G., and K. W. Post. 2005. Veterinary microbiology: bacterial and fungal agents of animal disease. Elsevier Saunders, St. Louis, MO.
- 21.Strehle, U., and N. Welbers. 1987. The pyogenes-endocarditis of cattle. Tierärztliche Umschau. 42869-878. [Google Scholar]