Abstract
The KI and WU polyomaviruses were found in 11 (2.7%) and 17 (4.2%) of 406 nasopharyngeal aspirates, respectively, from children with acute respiratory tract infection (ARTI). The phylogenetic analysis indicates that they are all in the same cluster as the prototype strains. Our findings suggest that they are common in children with ARTI in China.
In 1971, two human polyomavirus species named BK and JC were first isolated (7, 12). These two viruses are ubiquitous in most human populations and can cause persistent subclinical infections in healthy individuals and then reactivate under immunocompromised conditions. Recently, two novel human polyomaviruses were found, KI polyomavirus (KIPyV) in 6 (1%) of 637 nasopharyngeal aspirates (NPA) and 1 (0.5%) of 192 fecal samples in Sweden (2) and WU polyomavirus (WUPyV) in 43 (2%) of 2,135 patients with acute respiratory tract infection (ARTI) in Australia and the United States (8). Subsequently, these viruses were detected in Australia (6), South Korea (9), and Canada (1). In this study, 406 children with ARTI in Gansu Province, China, were analyzed for the presence of KIPyV and WUPyV with the associated clinical presentations and epidemiological characteristics.
From 1 December 2006 to 31 November 2007, 406 nasopharyngeal aspirates were collected from 406 children with acute respiratory infections in the First Hospital of Lanzhou University, Gansu Province, China. The mean age of the patients in our study was 2 years, and 88% of the specimens were collected from children 5 years of age or younger. Sixty percent were from males and 40% from females. Demographic data and data on the clinical findings were recorded. Informed consent was obtained from the parents of the children. The specimens were collected and transported immediately to a laboratory at the National Institute for Viral Disease Control and Prevention, China Center for Disease Control, and stored at −80°C until further processing. DNA and RNA were extracted from 0.2 ml of each NPA specimen using a QIAamp viral DNA mini kit and QIAamp viral RNA mini kit (Qiagen, Beijing, China). Nested PCR assays were used to detect KIPyV and WUPyV using the primers originally described by Allander et al. (2) and Gaynor et al. (8). In addition, we screened KIPyV- and WUPyV-positive specimens for human metapneumovirus, respiratory syncytial virus (RSV), influenza virus A and B, parainfluenza virus types 1 to 3 (PIV1 to -3), human rhinoviruses, and human coronaviruses (229E, OC43, NL63, and HKU1) using a standard reverse transcription-PCR technique (4, 5, 13, 14) and for adenovirus and human bocavirus (HBoV) using traditional PCR methods (3, 10). All of the PCR products were sequenced to confirm the specificity of each virus. The cloned sequences were determined and analyzed using the DNASTAR software package. A neighbor-joining tree was constructed using the program MEGA 3.1.
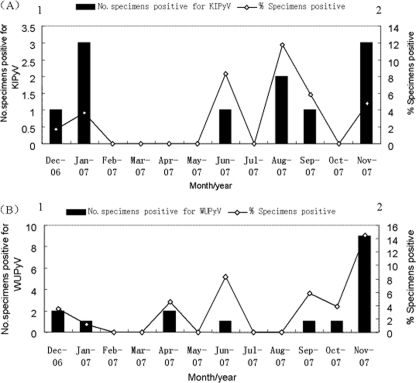
KIPyV was detected in 11 (2.7%) of 406 NPA specimens, and WUPyV was detected in 17 (4.2%) NPA specimens. KIPyV were detected in December, January, June, August, September, and November. WUPyV were detected in December, January, April, June, and September to November (Fig. 1). The maximum number of positive cases was detected in November for WUPyV (9 [52.9%] of 17 positive specimens) and in January and November for KIPyV (3 [27.3%] of 11 positive specimens). Patients who tested positive for KIPyV and WUPyV ranged in age from 3 days to 9 years, and 64% (16/25) of the patients were age 3 years and under. The median age of the patients infected with KIPyV was 7 months, and that of the patients infected with WUPyV was 24 months. No significant difference was observed in the ages of patients infected with KIPyV and WUPyV (P = 0.509; Wilcoxon two-sample test). When tested for other respiratory viruses, another respiratory virus was detected in 8 (72.7%) of the 11 KIPyV-positive specimens and 12 (70.5%) of the 17 WUPyV-positive specimens. For the KIPyV-positive specimens, PIV3 was the most common additional respiratory virus detected, accounting for 4 (50%) of 8 coinfections, while for coinfections among the WUPyV-positive cases, the most prevalent copathogen was RSV (4/17 [23.5%]) (Table 1). KIPyV (three cases) or WUPyV (five cases) was the sole virus detected in eight patients with clear clinical evidence of respiratory tract infection.
FIG. 1.
Seasonal distribution of KIPyV (A) and WUPyV (B) in children with ARTI from December 2006 until approximately November 2007. The percentage of specimens positive for KIPyV (A) and WUPyV (B) by month are shown.
TABLE 1.
Clinical presentation and demographic information for the 25 patients who tested positive for KIPyV or WUPyV
| Polyomavirus and patient | Age | Sexb | Coinfectionc | Diagnosisd | Signs/symptoms |
|---|---|---|---|---|---|
| KIPyV | |||||
| Lz42 | 9 yr | M | PIV3 | AURI | Fever, cough |
| Lz61 | 8 mo | F | PIV3 + IFVA | AURI | Fever, cough |
| Lz64 | 38 mo | M | PIV3 + IFVA | Acute laryngitis | Fever, cough, hoarseness, pharyngeal obstruction |
| Lz65 | 6 mo | M | PIV3 + IFVA hMPV + HBoV | Bronchopneumonia | Fever, cough, wheezing, diarrhea, crackles |
| Lz285 | 6 mo | F | PIV1 + HBoV | Bronchopneumonia | Cough, wheezing, crackles |
| Lz302 | 9 yr | M | None | Bronchopneumonia | Cyanosis, shortness of breath, nausea, vomiting, respiratory failure |
| Lz303a | 17 days | F | HBoV + WUPyV | Bronchopneumonia | Fever, cough |
| Lz312 | 8 mo | F | None | Asthma | Fever, diarrhea |
| Lz345 | 10 mo | M | None | Bronchopneumonia | Fever, cough, wheezing, diarrhea, crackles |
| Lz384a | 3 days | M | WUPyV | Bronchopneumonia | Difficulty in feeding, cyanosis |
| Lz386a | 6 mo | F | RSV + WUPyV | Bronchiolitis | Fever, cough, wheezing, cyanosis, crackles |
| WUPyV | |||||
| Lz41 | 4 yr | M | IFVA | AURI | Fever, cough |
| Lz121 | 3 yr | F | None | Bronchopneumonia | Cough, crackles |
| Lz128 | 11 mo | M | hMPV | Bronchopneumonia | Fever, cough, wheezing, crackles, diarrhea |
| Lz162 | 26 mo | M | None | Bronchopneumonia | Fever, cough, wheezing, crackles, diarrhea |
| Lz170 | 8 yr | F | None | Bronchiolitis | Fever, croup, crackles |
| Lz299 | 6 yr | M | None | Asthma | Cough, wheezing, croup |
| Lz338 | 3 yr | M | RSV | Bronchopneumonia | Fever, cough |
| Lz354 | 2.5 yr | M | RSV | Bronchopneumonia | Fever, cough, crackles |
| Lz368 | 2 yr | F | RSV | Bronchopneumonia | Cough, crackles |
| Lz385 | 7 yr | F | None | AURI | Fever, convulsion |
| Lz387 | 16 days | F | hRV + HKU1 | AURI | Fever, cough |
| Lz389 | 8 yr | M | RSV | Bronchiolitis | Cough, wheezing, croup, crackles |
| Lz390 | 1 mo | M | hRV | Bronchopneumonia | Cough, cyanosis, crackles |
| Lz400 | 7 mo | M | AdV | Bronchiolitis | Fever, cough, wheezing, diarrhea |
Specimens Lz303, Lz384, and Lz386 were infected with both KIPyV and WUPyV.
M, male; F, female.
IFVA, influenza virus A; hMPV, human metapneumovirus; hRV, human rhinovirus; HKU1, human coronavirus HKU1; AdV, adenovirus.
AURI, acute upper respiratory tract infection.
Of the polyomavirus-positive children, 76% (19/25) were inpatients with a median stay of 9 days. Their diagnoses included ARTI, acute laryngitis, bronchiolitis, bronchopneumonia, and asthma (Table 1). The common clinical presentations of these children included cough (n = 21; 84%), fever (n = 17; 68%), crackles (n = 12; 48%), wheezing (n = 9; 36%), diarrhea (n = 6; 24%), and influenza-like illnesses (n = 5; 20%). Of the six children who had chest radiography performed, five had abnormal findings and four children had evidence of lobar infiltrates. Complete blood counts were performed for 20 of the 25 children. The mean white blood cell count was 86,500 cells/μl (range, 31,000 to 134,000 cells/μl). Bacterial cultures of NPAs were obtained for five patients, three of which were positive for Haemophilus influenzae (Lz121), Streptococcus pneumoniae (Lz338), and Escherichia coli (Lz285) and two of which had normal flora. In addition, three patients (Lz303, Lz384, and Lz386) were infected with both KIPyV and WUPyV.
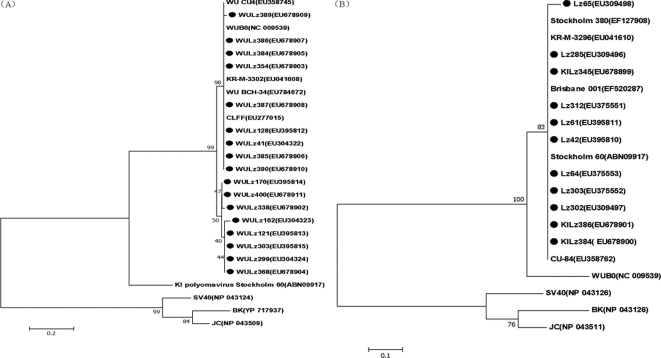
The partial sequences of theVP1 gene from KIPyV and the VP2 gene from WUPyV obtained from children in China were aligned with the reference strains which are shown in Fig. 2. Based on the VP1 gene sequences (204 bp), all of the KIPyV strains found in our study were in the same cluster as the KIPyV prototype strains, with over 99% DNA sequence homology. All of the WUPyV strains in our study also showed 98 to 100% nucleotide identity with the VP2 region (250 bp) of the WUPyV reference strains (Fig. 2).
FIG. 2.
Phylogenetic analysis of the deduced amino acid sequences of the WUPyV VP2 gene (250 bp) (A) and KIPyV VP1 gene (204 bp) (B) with reference strains. Phylogenetic trees were constructed by the neighbor-joining method by using MEGA 3.1, and bootstrap values were determined by 1,000 replicates. Viral sequences in marks were generated from the present study; representative WUPyV and KIPyV sequences and other polyomaviruses from GenBank are indicated by isolate name. The GenBank accession numbers of each strain are given in parentheses.
The respective incidence of KIPyV in our sample population was 2.7%, which is higher than the 1% reported by Allander et al. (2) and Han et al. (9) but consistent with the 2.5% detection rate for Australian children (6). The detection rate of WUPyV for respiratory tract infections has been reported to be 0.4% to 7% (9, 11), and most were between 2% to 3%, which is similar to our data (4.2%). In South Korea and Canada, the detection rates of WUPyV in asymptomatic children were 4.2% (3/72) and 6.4% (5/78), respectively (1, 9). It is not yet known whether WUPyV and KIPyV are infectious or whether they are capable of replication in the respiratory tract. One possibility is that they can cause subclinical infections in healthy individuals and then reactivate under immunocompromised conditions. Allander et al. found that the ages of the six subjects positive for KIPyV in the nasopharynx ranged from 1 month to 26 years (median, 2 years) (2) and Gaynor et al. reported that 89% of the cases positive for WUPyV were children 3 years of age and under (8). Similar results were reported by Bialasiewicz et al. in Australia (6) and Abed et al. from Canada (1). Our data show that 82% (9/11) KIPyV-infected and 53% (9/17) WUPyV-infected children were less than 5 years of age. Notably, a 3-day-old neonate was infected with WUPyV (Lz384), and a 17-day-old patient (Lz303) was infected with both KIPyV and WUPyV. It is indicated that primarily the two polyomavirus infections might occur early in life. Further serological study is needed to validate this hypothesis.
In America, WUPyV were detected throughout the year, with small peaks in April, May, and July (8), while the majority of the positive detections of KIPyV were made in the Australian winter months (May to July) (6). Our data showed that the majority of the positive cases of WUPyV were detected in autumn and winter with a high peak in November, while KIPyV were detected throughout the year with small peaks in January and November. These seasonal detection variations need to be investigated by additional epidemiological studies in more regions. The rates of coinfection with KIPyV and WUPyV were 72.7% and 70.5% in this study, respectively. Similar high percentages were found in preliminary studies, which reported coinfection rates of 25% (6) to 83.3% (2, 6, 8). Moreover, the two cases positive for both KIPyV and WUPyV were coinfected with HBoV (Lz303) and RSV (Lz386). In one sample (Lz65), a total of five viruses (KIPyV, HBoV, human metapneumovirus, influenza virus A, and PIV3) were detected. WUPyV or KIPyV was the only virus detected in 5 and 3 patients, respectively. The frequent codetection of KIPyV and WUPyV with other respiratory viruses raises concern over their causative roles in human respiratory tract infection. Clinical findings for KIPyV- and WUPyV-positive patients are indistinguishable from those for patients with other respiratory viruses. The most frequent symptoms in patients infected with KIPyV and WUPyV included cough, fever, wheezing, diarrhea, and influenza-like symptoms. In these children, the common clinical diagnoses were bronchopneumonia (52% [13/25]) and upper respiratory tract infection (20% [5/25]). Given the lack of testing for bacterial pathogens for every specimen or a control population, the relationship between these two new polyomaviruses and respiratory tract infection is not clear.
In conclusion, we found in this study that KIPyV and WUPyV were prevalent in children, especially infants and young children with ARTI in China. Coinfection of KIPyV and WUPyV with other respiratory viruses was common. The clinical symptoms of KIPyV- and WUPyV-positive patients are not distinguishable from those of patients infected with other respiratory viruses. A single genetic lineage of the KIPyV and WUPyV strain is circulating in China. To clarify the role of these two new polyomaviruses in respiratory tract infection, future priorities should include case-control studies using larger cohorts of cases and the establishment of a serology examination method to monitor the infection in patients.
Nucleotide sequence accession numbers.
The sequences generated from the present study were deposited in GenBank, and the accession numbers are shown in Fig. 2.
Acknowledgments
We thank the First Hospital of Lanzhou University, which routinely submits specimens to China CDC.
Footnotes
Published ahead of print on 30 July 2008.
REFERENCES
- 1.Abed, Y., D. Wang, and G. Boivin. 2007. WU polyomavirus in children, Canada. Emerg. Infect. Dis. 131939-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., K. Andreasson, S. Gupta, A. Bjerkner, G. Bogdanovic, M. A. Persson, T. Dalianis, T. Ramqvist, and B. Andersson. 2007. Identification of a third human polyomavirus. J. Virol. 814130-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 10212891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastien, N., K. Anderson, L. Hart, P. Van Caeseele, K. Brandt, D. Milley, T. Hatchette, E. C. Weiss, and Y. Li. 2005. Human coronavirus NL63 infection in Canada. J. Infect. Dis. 191503-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellau-Pujol, S., A. Vabret, L. Legrand, J. Dina, S. Gouarin, J. Petitjean-Lecherbonnier, B. Pozzetto, C. Ginevra, and F. Freymuth. 2005. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 12653-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bialasiewicz, S., D. M. Whiley, S. B. Lambert, D. Wang, M. D. Nissen, and T. P. Sloots. 2007. A newly reported human polyomavirus, KI virus, is present in the respiratory tract of Australian children. J. Clin. Virol. 4015-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner, S. D., A. M. Field, D. V. Coleman, and B. Hulme. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i1253-1257. [DOI] [PubMed] [Google Scholar]
- 8.Gaynor, A. M., M. D. Nissen, D. M. Whiley, I. M. Mackay, S. B. Lambert, G. Wu, D. C. Brennan, G. A. Storch, T. P. Sloots, and D. Wang. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, T. H., J. Y. Chung, J. W. Koo, S. W. Kim, and E. S. Hwang. 2007. WU polyomavirus in children with acute lower respiratory tract infections, South Korea. Emerg. Infect. Dis. 131766-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hierholzer, J. C., P. E. Halonen, P. O. Dahlen, P. G. Bingham, and M. M. McDonough. 1993. Detection of adenovirus in clinical specimens by polymerase chain reaction and liquid-phase hybridization quantitated by time-resolved fluorometry. J. Clin. Microbiol. 311886-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, F., M. Zheng, H. Li, C. Zheng, X. Li, G. Rao, M. Zheng, F. Wu, and A. Zeng. 2008. WU polyomavirus in children with acute lower respiratory tract infections, China. J. Clin. Virol. 4294-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padgett, B. L., D. L. Walker, G. M. ZuRhein, R. J. Eckroade, and B. H. Dessel. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i1257-1260. [DOI] [PubMed] [Google Scholar]
- 13.Vabret, A., J. Dina, S. Gouarin, J. Petitjean, S. Corbet, and F. Freymuth. 2006. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis. 42634-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vabret, A., F. Mouthon, T. Mourez, S. Gouarin, J. Petitjean, and F. Freymuth. 2001. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J. Virol. Methods 9759-66. [DOI] [PMC free article] [PubMed] [Google Scholar]