Abstract
One hundred four bacterial strains mediating urinary tract infections in separate individuals from a Uruguayan community were isolated. Forty-six strains conferred a multidrug resistance phenotype. All 104 strains were examined for the presence of class 1, 2, and 3 integrons. Class 1 integrons were found in 21 isolates across four distinct bacterial genera. A large class 1 integron in a Klebsiella pneumoniae strain was fully sequenced and was 29,093 bp in length. This integron probably arose by homologous recombination since it was embedded in a hybrid Tn21-like transposon backbone which comprised a Tn5036-like tnp transposition module at the IRi integron end and a Tn21 mer module at the IRt integron end. The parent integron/transposon that contributed the Tn5036 module was not related to Tn1696 since the integron insertion points in the transposon backbones were 16 bases apart. Examination of the other 20 class 1 integron-containing strains revealed further evidence of genetic exchange. This included a strain that possessed a Tn5036 module at the IRt end but not at the IRi end and another that possessed a tnp module beyond IRi that was a hybrid of Tn21 and Tn5051 and that is presumed to have arisen by site-specific recombination. This study highlights the ability of different genetic elements to act cooperatively to spread and rearrange antibiotic resistance in a community.
Community-acquired urinary tract infection (UTI) is one of the most common infectious diseases and a frequent cause of presentation for outpatient treatment. While mortality rates are not usually high, the cost to the global economy is substantial (1, 11). Generally, UTIs are mediated by gram-negative bacteria, with the most common of these being Escherichia coli and Klebsiella pneumoniae, but can also include Acinetobacter and Enterobacter spp. (1). Management of the infection is becoming progressively complicated by the ongoing increase in resistance to antibiotics manifested by UTI-causing organisms.
Lateral gene transfer (LGT) is a significant driver of the evolution of multiple-drug-resistant pathogenic bacteria (29). In gram-negative bacteria, the elements that facilitate LGT include plasmids, transposons, and integrons, along with their associated mobile gene cassettes. These elements do not act in isolation, however, and it is common that, for example, conjugative plasmids can carry various transposable elements which, in turn, may harbor class 1 integrons with a diverse combination of associated resistance cassettes (51). Such combinations of elements that collectively contain conjugation, transposition, and site-specific recombination systems can infectiously spread resistance genes through a variety of disparate bacteria very quickly and rearrange them in a number of ways in the new host after transfer. It is this cooperative approach that has led to the rapid spread of antibiotic resistance genes in pathogens and a major reason why managing resistance is proving so difficult.
Epidemiological studies to better manage infections such as UTIs generally involve surveys of isolates from a defined location or locations for the purpose of identifying common resistance profiles (10, 20, 47). These studies attempt to survey resistance to clinically important antibiotics generally or focus on particular families of antibiotics and the corresponding genes that encode resistance to them. Such surveys can lead to treatment strategies that make use of antibiotics to which strains are likely to be sensitive and can be useful in the short term for the development of detection and management strategies (14, 48). However, they do not usually take into account the genetic contexts in which resistance genes are found, a shortcoming considering that transposons in which integrons are embedded also contribute to the spread of associated resistance gene cassettes (25).
One group of elements that have been an increasing focus of study in the context of UTIs is integrons. In particular, integrons belonging to classes 1, 2, and 3, the classes that are most commonly associated with the spread of antibiotic resistance in pathogens. While integrons are a problem generally in managing the spread of resistance, they are especially common in UTIs (5). Surveys that examine the prevalence of various genetic elements in multidrug-resistant UTI-causing strains invariably show a high correlation between the presence of a class 1 integron and particular antimicrobial resistance profiles. Most notable among these are resistance to ampicillin (AMP) and streptomycin but especially trimethoprim and sulfamethoxazole (SMX) (5, 39, 40). In the case of class 1 integrons, SMX resistance is normally derived from the sul1 gene that is contained within a region downstream of the integron called the 3′ conserved segment (3′-CS), and this region is present in the majority of class 1 integrons from clinical environments. Consequently, SMX resistance is a common feature of strains that carry class 1 integrons. In addition, the use of cotrimoxazole (SXT) to treat UTIs and many other community-acquired infections is a likely additional reason why a variety of dfrA gene cassettes are found in class 1 integrons from UTI-mediating organisms, thus leading to SXT resistance being very common (13, 40). The ongoing use of antibiotics is associated with more complex multiresistance patterns attributable to the influx of more resistance genes into clinical isolates (52).
The class 1 integrons that carry drug resistance genes in clinical isolates have a relatively conserved structure. This structure commonly comprises two conserved DNA sequences—the 5′-CS and 3′-CS—separating a variable region where mobile gene cassettes are located (43). This arrangement has been used as a PCR tool for the simple recovery of cassette arrays irrespective of knowledge of the many and varied cassettes that may be present (24, 36). As well as being able to recover and analyze cassettes from specific isolates, this method has also been used as a tool in broader epidemiological studies (19, 38). The use of PCR for array analysis in the latter type of studies, however, has limitations. Firstly, some class 1 integrons from clinical isolates do not have part or all of the 3′-CS, or alternatively, they may carry a very large array of cassettes that prevents detectable amplification (34). Either outcome can generate a false negative. In addition, PCR array length analysis alone underestimates cassette diversity since some array combinations may be similar or even identical in length (34). Also, given that LGT is mediated by a number of different types of elements, the combinatorial exchange and spread of integron cassette arrays can occur in a variety of ways that can include homologous recombination, transposition, and even non-integron-mediated site-specific recombination (32, 33). Investigation of integron cassette arrays in the absence of context therefore may not give a wholly accurate picture of the processes influencing resistance gene spread in pathogenic bacteria.
In this study, 104 organisms that mediated UTIs in a South American community over 6 months were typed and examined for the presence of class 1, 2, and 3 integrons. Class 1 integrons were the most common and were found in 21 isolates derived from at least four bacterial genera. These isolates collectively contained a number of different resistance arrays, with the combination dfrA17-aadA5 being the most common. One class 1 integron was examined in detail and was found to be 29,093 bp long. With this transposon/integron as a starting point, the molecular context of the other class 1 integrons circulating in this cohort of strains was examined. It was found that most of the integrons were related but showed evidence of exchange of their respective transposon backbones. These exchange events included the generation of hybrid transposons probably derived from homologous recombination of common sequence within them and resolution-type site-specific recombination between different integron-containing transposons.
MATERIALS AND METHODS
Bacterial isolates and antimicrobial susceptibility testing.
Isolates were collected from early March 2005 to the end of August 2005 from outpatients with UTIs attending the Central Hospital of the Armed Forces, a public health center in Montevideo, Uruguay. Identification of enterobacteria was performed by the simplified identification system SYS9E (42); when confirmation was needed, the API 20E (BioMérieux) was used. Standard laboratory procedures were used for identification of the Staphylococcus, Streptococcus, and Enterococcus genera (28). Antimicrobial susceptibility testing was performed by the disc diffusion method following CLSI recommendations (7, 8). The antimicrobials tested were AMP, ciprofloxacin, norfloxacin, SXT, trimethoprim, SMX, gentamicin, and nitrofurantoin (BBL, Becton Dickinson, Le Pont de Claix, France). Multidrug resistance was defined as resistance of an isolate to more than two unrelated drugs.
DNA extraction, sequencing, and PCR analysis.
The DNA sequencing and PCR primers used are listed in Table 1. Total DNA was extracted with the Perfect DNA gDNA Blood Mini Isolation kit (Eppendorf), and all DNA templates were analyzed by 16S rRNA gene amplification with primers f27 and r1492 as previously described (22) to confirm DNA viability for PCR amplification. PCR analysis were performed with a Perkin-Elmer DNA thermal cycler 2400 and corresponding primers synthesized by Alpha DNA (Montreal, Quebec, Canada).
TABLE 1.
Primers used in this study
| Primer | Sequence 5′ → 3′ | Target | Application (reference) |
|---|---|---|---|
| HS458 | GTTTGATGTTATGGAGCAGCAACG | 5′-CS attI1 end | Amplify class 1 cassette arrays (18) |
| HS459 | GCAAAAAGGCAGCAATTATGAGCC | 3′-CS | Amplify class 1 cassette arrays (18) |
| HS915 | CGTGCCGTGATCGAAATCCAG | intI1 | PCR screen for presence of intI |
| HS916 | TTCGTGCCTTCATCCGTTTCC | intI1 | PCR screen for presence of intI |
| HS501 | CACGGATATGCGACAAAAAGGT | intI2 | PCR screen for presence of intI2 |
| HS502 | GTAGCAAACGAGTGACGAAATG | intI2 | PCR screen for presence of intI2 |
| HS503 | GCCTCCGGCAGCGACTTTCAG | intI3 | PCR screen for presence of intI3 |
| HS504 | ACGGATCTGCCAAACCTGACT | intI3 | PCR screen for presence of intI3 |
| HS549 | ACTAAGCTTGCCCCTTCCGC | sul1 | PCR screen for presence of sulI |
| HS550 | CTAGGCATGATCTAACCCTCG | sul1 | PCR screen for presence of sulI |
| HS817 | CCTCCAATTGCCGTTCC | Tn5036-like tnpR genea | PCR screen for Tn5036 at IRi |
| HS818 | TCCTGGCGGATTCACTACC | 5′-CS adjacent IRi | PCR screen for IRi insertion point |
| HS819 | GGGCCAGGTCTTGAGTATCG | orf513 | PCR screen for presence of ISCR1 |
| HS820 | GCTTCGGCCATCACACC | orf513 | PCR screen for presence of ISCR1 |
| HS825 | TGTTTTCGGAATCGTAGTCGC | Tn402-like tniA gene | PCR screen tniA and IRt insertion point |
| HS826 | CTGACCGGCTTGTTCGTTC | Tn21 merE gene | PCR screen for Tn21 at IRt |
| HS841 | GAAGGGTTACGCCAGTACCAG | IS26 | PCR screen for IS26 adjacent to IRi |
| HS842 | ATGCTCAATACTCGTGTGCACC | Tn402-like tniA gene | PCR screen for presence of tniA gene |
| HS845 | CCAAACCTAGAAACGC | cmlA1 | Targeted PCR screen for 6-cassette array identified in strain 8157 |
| HS853 | GTGCAAGGTATCCTCCTGAGG | Tn5051 res region | PCR screen for presence of Tn5051 res region beyond IRi |
| HS856 | CATTCGCCGCTCAATGTTAAC | blaCTX-M-2 gene | PCR screen for presence of blaCTX-M-2 |
| HS857 | GAAGGTCTCATCACCCAACG | blaCTX-M-2 gene | PCR screen for presence of blaCTX-M-2 |
| HS911 | GCGCGGAATACGTCGAAC | Tn5036-like merE gene | PCR screen for presence of Tn5036 at IRt |
Also for tnpR in Tn21 (see text).
PCR was preformed with the Promega Taq Mastermix (Promega). For most reactions, the conditions were 1 cycle of 94°C for 3 min; 35 cycles of 94°C for 30 s, 60 or 65°C for 30 s, and 72°C for 90 s; and 1 cycle of 72°C for 5 min. For PCR with primer HS845, the annealing temperature was reduced to 52°C.
Fosmid libraries were constructed from genomic DNA (gDNA) with the CopyControl fosmid library production kit (Epicentre). gDNA was extracted from overnight cultures by the XS buffer protocol (45), treated with RNase A (Sigma), and further purified by phenol-chloroform extraction. Fosmid library production relies on gDNA to be sheared into approximately 40-kb fragments due to the size restriction of the packaging phage capsids. Post gDNA purification, pulsed-field gel electrophoresis was undertaken with a 1% agarose gel to ensure that the gDNA was sufficiently sheared to the 40-kb range. Where necessary, shearing was achieved by passage through a 26-gauge needle. Sheared DNA was then end repaired, ligated into the fosmid vector, packaged into phage capsids, and used to infect E. coli EPI300-T1R as instructed in the CopyControl fosmid library production kit manual. E. coli clones carrying fosmids were plated on LB plates containing chloramphenicol. Screening of the fosmid library for intI1 was carried out as previously described. intI2-positive clones were identified by the same method but with the use of HS501 and HS502 as PCR primers. Fosmids from positive clones were extracted with the Wizard plus SV Minipreps kit (Promega). Terminal ends of some inserts were sequenced with the FP and RP vector primers. Complete fosmid clone sequencing was performed by Macrogen (Korea).
Nucleotide sequence accession numbers.
The sequence of fosmid clone 4E6 from K. pneumoniae strain 12836 has been submitted to GenBank under accession no. EU780013. The sequence of the IRt junction of strain 8180 can be found under GenBank accession no. EU918721, and that of the resolution region of strain 5293 can be found under GenBank accession no. EU930720.
RESULTS
Multidrug resistance is common among organisms mediating community-acquired UTIs.
Bacteria were isolated from 104 individuals treated at an outpatient clinic for UTIs. The clinic is part of a public health care facility serving the needs of past and present members of the Uruguayan armed forces and their families. It meets the medical needs of about 160,000 people (5% of the population of Uruguay) of all ages and can be considered representative of the population as a whole. Of the 104 isolates, 83 were identified as E. coli, 6 were Proteus mirabilis, 4 were Enterococcus sp., 4 were K. pneumoniae, 3 were Staphylococcus saprophyticus, and there was 1 each of Enterobacter cloacae, E. aerogenes, Streptococcus agalactiae, and Citrobacter diversus. The resistance profiles varied significantly between strains, but 46 were resistant to two or more of the agents tested. All 104 isolates were screened by PCR for the presence of class 1, 2, and 3 integrons with primer pairs that targeted regions internal or adjacent to, and specific for, the respective intI genes. Of the 104 strains, 21 were positive for intI1 (Table 2), 12 were positive for intI2, and none were positive for intI3. Three isolates were positive for both intI1 and intI2 (Table 2). Despite the variable resistance profiles overall, there was a strong correlation between the presence of intI1 and resistance to SXT and AMP. Of the 25 strains resistant to these compounds, 19 were also intI1 positive. Of the 21 intI1-positive isolates, the 2 not meeting this profile were strain 3843 (AMPs SXTr) and strain 8175 (AMPr SXTs). All 21 intI1-containing strains were part of the cohort of 46 isolates conferring a multidrug resistance phenotype.
TABLE 2.
Characteristics of intI1-positive strains
| Strain | Isolation dateb | Organism | Cassette array | IRi junction | IRt junction | Sul1f |
|---|---|---|---|---|---|---|
| 3020 | 2 March | E. coli | dfrA17-aadA5 | UKd | NPe | + |
| 3843a | 5 April | E. coli | NAc | Tn21 > IS26 | NP | − |
| 3665 | 5 April | E. coli | dfrA17-aadA5 | Tn21g | Tn21h | + |
| 3879 | 15 April | E. coli | dfrA17-aadA5 | UK | Tn21h | + |
| 5293 | 10 May | E. coli | aadA1a | Tn21 > Tn5051g | NP | + |
| 8047 | 27 May | E. coli | dfrA1-aadA1a | UK | NP | − |
| 5302 | 27 May | E. coli | dfrA17-aadA5 | UK | NP | + |
| 8157a | 12 June | E. coli | NA | Tn21 > IS26g | NP | − |
| 8167 | 12 June | E. coli | dfrA17-aadA5 | Tn21g | Tn21h | + |
| 8175 | 12 June | E. coli | aadA1a | Tn21g | Tn21h | + |
| 8221 | 20 June | E. cloacae | blaoxa101-aacA4 | Tn21 | NP | + |
| 8180 | 20 June | K. pneumoniae | dfrA22 | UK | Tn5036h | + |
| 8392 | 30 June | E. coli | dfrA17-aadA5 | Tn21g | Tn21 | + |
| 8393 | 30 June | E. coli | NA | Tn21 | Tn21 | + |
| 9445 | 30 June | E. coli | NA | Tn21 > IS26 | NP | − |
| 13014 | 20 August | E. coli | dfrA17-aadA5 | Tn21 | Tn21 | + |
| 12674/1 | 20 August | E. coli | dfrA17-aadA5 | UK | NP | + |
| 12928 | 20 August | E. coli | dfrA17-aadA5 | UK | NP | + |
| 12950 | 20 August | P. mirabilis | aadB-aadA1a | Tn21g | NP | + |
| 12836a | 20 August | K. pneumoniae | dfrA12-orfF-aadA2 | Tn5036g | Tn21h | + |
| 4012/2 | 30 August | E. coli | dfrA17-aadA5 | Tn21 | NP | + |
Also positive for intI2.
All dates 2005.
NA, no amplicon generated by PCR with HS458 and HS459.
UK, unknown. No PCR product was generated with any PCR primer pair.
NP, not present. IRt is inferred to be absent based on the failure to generate a PCR product with tniA-specific primers.
A plus or a minus sign indicates the presence or absence of a PCR product with primers HS549/HS550.
Junction confirmed by DNA sequencing. For the remainder, the junction was inferred by PCR fragment length.
Junction confirmed by DNA sequencing. For the remainder, the junction was inferred by PCR fragment length.
The method of amplifying class 1 cassette arrays by targeting adjacent conserved regions (23) was applied here with primers HS458 and HS459 and was carried out with all intI1-positive strains. As shown in Table 2, 17 of the 21 strains generated a product. All 17 of these were sequenced to determine the gene cassette array content (Table 2). The most common was a two-cassette array comprising dfrA17-aadA5. This array was found 10 times, all in E. coli isolates. One PCR product of about the same size, derived from E. coli isolate 8047, contained a dfrA1-aadA1a array. Two additional E. coli arrays comprised a single aadA1a cassette. Four intI1-positive E. coli strains failed to generate a product with HS458 and HS459. Three of these four also failed to generate a product with sul1-specific primers HS549/HS550, suggesting that the 3′-CS may be partly deleted or not present (Table 2). The four non-E. coli isolates were more variable in gene cassette array content (Table 2).
K. pneumoniae strain 12836.
K. pneumoniae strain 12836 was noteworthy for at least two reasons. It was resistant to all eight antimicrobial agents tested and carried both a class 1 integron and a class 2 integron, the former of which had a relatively long array (Table 2). Consequently, this strain was further examined via the construction of a fosmid genomic library. This library of 480 clones was screened by PCR with primer pairs specific for intI1 and intI2. One intI2-positive clone was recovered. The genomic location of this Tn7-like class 2 integron was determined with primers that sequence out from the left and right ends of the element. It was found that the integron was located in the chromosome immediately adjacent to the glmS gene, a region known to be an insertional hot spot for this family (17). Abutting the Tn7-like transposon were 5-bp direct repeats indicating insertion by transposition.
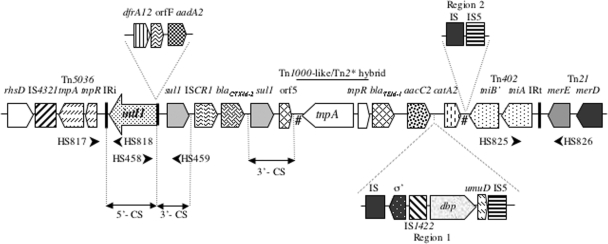
Six intI1-positive clones were recovered from the fosmid library. The insert of one of these (4E6) was sequenced completely, and a map of the relevant region is shown in Fig. 1. It was found that the class 1 integron, as measured between inverted repeats IRi and IRt (15, 43, 44), was 29,093 bp in length. This sequence included the presence of ISCR1 (46), which had a blaCTX-M-2 gene associated with it. The integron also had a partial duplication of the 3′-CS. The region extending from about the middle of intI1 through to the end of the second copy of sul1 was identical, with the exception of 10 bases, to the entire 9,017 bases determined from plasmid p04-9275-1 (accession no. EF592571). This plasmid was derived from a human fecal Salmonella enterica isolate from French Guiana in 2004 (12). The region beyond the second 3′-CS contained evidence of insertion of IS1326 at a point identical to that of In2 (30). However, most of this insertion sequence was missing and had been replaced with a region of more than 15 kb that included at least three other resistance determinants, blaTEM-1, aacC2, and catA2 (Fig. 1). The opposite boundary of IS1326 was located adjacent to the remnants of the Tn402-like tniB gene, again consistent with In2. Overall, this same region displayed evidence of acquisition of DNA sequences by transposition (Fig. 1). This included the presence of a transposon that was a hybrid in that it consisted of a Tn1000-like tnpA gene identical to a recently identified gene from a plasmid, pYE854, found in a strain of Yersinia enterocolitica (16) in association with the same tnpR gene found in Tn2* (31).
FIG. 1.
Physical map of the class 1 integron and surrounding sequence in K. pneumoniae strain 12836. Filled arrowheads indicate the relative positions of some PCR primers. The # symbols indicate the relative positions of the ends of IS1326. Regions not defined elsewhere are as follows: rhsD, identical to the rhsD gene from a Yersinia pestis plasmid (accession no. CP000603), the product of which is implicated in cell membrane biogenesis; region 1 IS, putative insertion sequence based on a weak match to insertion sequence elements from Rhizobium sp.; σ′, RNA polymerase sigma 70 factor (best match, an analogous protein from Myxococcus xanthus); dbp, putative DNA binding protein; umuD, DNA polymerase V subunit umuD (this gene is interrupted by IS5 but identical [DNA] to umuD from Acinetobacter baumannii strain 7030 over the 258 bases present); region 2 IS, insertion sequence with 99% DNA identity to ISPa12. The features shown are not necessarily to scale.
The above observations revealed that the class 1 integron was inserted into a Tn21-like transposon backbone. While such a general structure is common, the one found here was unusual since the backbone was a hybrid, compared to most other examples of this general structure. Specifically, in strain 12836, the tnp region beyond the IRi end of the integron is consistent with insertion into a Tn5036-like backbone (Fig. 2). In contrast, the transposon backbone beyond IRt, the mer module, is identical to Tn21 itself. Many clinical examples of insertion of an integron into a Tn5036-like backbone are known and are best exemplified by Tn1696. Comparison of the DNA sequence beyond IRi revealed only a 1-bp difference with Tn1696 across the region up to the end of tnpA. It is likely, therefore, that the integron in strain 12836 is a hybrid derived by homologous recombination between an integron inserted into a transposon of the Tn21 family and an integron inserted into a Tn5036 backbone.
FIG. 2.
Structural context of the integrons located in strains 12836 and 8180. (A) Diagrammatic representation of the transposon backbones (where known) at the IRi and IRt integron ends for the integrons in each of the two strains. (B) Sequence context of the IRi end of the strain 12836 integron and the IRt end of the strain 8180 integron in relation to the Tn5036 backbone. The common direct repeat (DR) is underlined. Numbered arrows indicate insertion points of integrons/transposons as follows: 1, accession no. AJ746361 and EU543272; 2, Tn6006 (21); 3, In4 (15). The location of the start codon (CAA) of tnpR is shown, with the value to the right indicating the number of intervening bases.
The hybrid backbone seen here is analogous to the context of In34, an integron with an In2-like integron structure and a member of the broader In5-like family of integrons (31, 33). The structure carrying In34 is presumed to have been generated by homologous recombination since, while it has a Tn21 mer region at its IRt end, it possesses a Tn5036-like tnp region at the IRi integron end with an insertion point at this latter end identical to that seen in Tn1696 (In4). The putative Tn5036-like integron parent that gave rise to the structure in strain 12836, however, was not Tn1696 since the integron insertion point in the transposon backbone is 16 bases away from that seen in Tn1696 (Fig. 2). Thus, the two integron acquisition events that created Tn1696 and the Tn5036-like parent of the integron in strain 12836 must have been independent. The insertion point at the IRt end of the 12836 element is identical to that seen in Tn21. The Pc promoter, P1, present in the 12836 integron is also noteworthy, as it is a “hybrid” with the −35 sequence comprising TGGACA and the −10 sequence comprising TAAACT. This promoter is therefore distinct from the strong and weak versions of P1 found in In4 and In2, respectively (9, 23), and is one of intermediate strength. The second promoter, P2, found in some class 1 integrons, including In2 (9), is not present in strain 12836.
Further examination of class 1-containing isolates.
A total of 21 isolates were positive for intI1. Given the unusual structure of the class 1 integron in strain 12836, we examined the other 20 isolates to determine if this same hybrid transposon/integron was circulating in this Uruguayan population. To do this, we designed two PCR primer pairs. One of these (HS817/HS818) was designed to amplify the IRi boundary as seen in strain 12836, and the other pair (HS825/HS826) was designed to amplify the IRt boundary (Fig. 1). The IRi pair was predicted to generate a product of 376 bases, and a product consistent with this length was found only in strain 12836. Twelve isolates failed to generate a product with HS817/HS818. Nine additional isolates (3665, 8167, 8175, 8221, 8392, 8393, 13014, 12950, and 4012/2 [Table 2]) generated a product of approximately 700 bases. Five of these products were sequenced, and all five were identical to the corresponding region in Tn21. The remaining four 700-bp PCR products were not sequenced but were assumed to be the same. Thus, these integrons have the same insertion point as In2. Examination of the Tn21 sequence at a region around the likely HS817 primer binding site revealed a sequence that matched this primer at 15 of 17 positions. Primer binding at this location would generate a product of 735 bases with HS818, consistent with the length of the PCR product observed.
The IRt primer pair HS825/HS826 generated a product of approximately 1,800 bases in eight isolates, including strain 12836 (Table 2). This was consistent with the predicted length of 1,847 bases for a Tn21-like insertion at this end. The remaining 13 strains did not generate a product with this primer pair. Since the IRi end of the integrons in this population can be Tn5036 like or Tn21 like, it was reasoned that the analogous scenario could apply at the IRt end. If so, it was likely that if a Tn5036-like sequence was present at the IRt end, HS826 would not bind to it. To investigate this possibility further, we designed another primer, HS911, that was used in conjunction with HS825 but which targeted the relevant region in a Tn5036-like backbone. Twenty isolates, including strain 12836, failed to generate a product with the HS825/HS911 primer pair. A product consistent with the predicted length of 1,284 bp was generated only with K. pneumoniae strain 8180 (Table 1). The sequence of this DNA fragment confirmed the presence of a Tn5036-like mer region and not Tn21. The insertion point in the Tn5036-like backbone for the IRt end of the integron in strain 8180 corresponded to the insertion point observed for the IRi end of the integron located in strain 12836 (Fig. 2). Surprisingly, sequence analysis of the tniA gene and adjacent DNA up to and including IRt (714 bp in total) revealed that this part of the transposition module was only 81% identical to the same region in Tn402 and other class 1 integrons/transposons. Instead, this region was 100% identical to the same region in the related mercury resistance transposon Tn5058 (accession number Y17897). Much more extensive sequence analysis is required to establish the relationship between the integrons in strains 12836 and 8180. It is possible that they are derived from unrelated recombination events or may be derived from two independent Tn5036 parents entirely, despite the common insertion point. Twelve isolates were negative for both Tn21 and Tn5036 sequences beyond IRt. IRt may not present in these 12 isolates since a PCR with primers internal to tniA, normally immediately adjacent to IRt, failed to generate a product (Table 2). In these 12 strains, the transposon backbone beyond IRt, if there is any, is unknown.
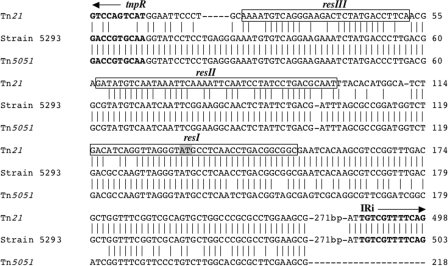
At the IRi integron end, 11 isolates failed to generate a product with HS817/HS818, implying the absence of both Tn21 and Tn5036 and suggesting that the spread of class 1 integrons associated with UTIs in this community was being facilitated by transposons additional to Tn21 and Tn5036 or by being inserted directly into a plasmid. To further investigate this, we selected two additional isolates, from which fosmid libraries were made. These isolates were E. coli strains 5293 and 8157. These were two of the seven strains that appeared to have no association with either Tn21 or Tn5036 (Table 2) in that they failed to produce a PCR product with primer pairs targeting these transposons at the IRi and IRt ends of the integron. Sequencing outward from IRi in an intI1-positive fosmid clone from strain 5293 revealed a junction identical to that in Tn21. Sequence identity continued for 363 bases beyond IRi, at which point the sequence diverged from Tn21 (Fig. 3). Analysis of the sequence immediately beyond this point revealed it to be identical to a sequence in another member of the Tn21-like transposon family, Tn5051 (27). The sequence identity between Tn21 and Tn5051 in this region is about 90%, and so the crossover point can only be localized to a region within 17 bases. This region, however, corresponds to resI of the Tn21 resolution site and includes the point at which cointegrate resolution occurs (Fig. 3). It is likely, therefore, that this transposon hybrid in strain 5293 arose by a site-specific recombination-mediated resolution event between Tn21 and a Tn5051-like transposon located on the same replicon. To test for the presence of this hybrid in other isolates, a PCR primer (HS853) was designed that would uniquely target Tn5051. This primer was used in a PCR with HS818 on all 21 class 1 integron-positive isolates with the result that only strain 5293 yielded a product, thereby suggesting that this IRi end structure was unique to this strain in the cohort sampled (Table 2). We also sequenced beyond the end of sul1 in strain 5293 toward the missing tniA gene for approximately 2 kb. This region was found to be identical (data not shown) to Tn21, including the presence of IS1326, which is found in Tn21 (25).
FIG. 3.
Alignment of sequences upstream of the tnpR gene in Tn21, Tn5051, and the transposon in E. coli strain 5293. The res sites of Tn21 are boxed and are as previously defined and numbered (37). The bases in resI between which site-specific mediated resolution occurs are shaded.
Sequencing outward from the IRi end of strain 8157 from an intI1-positive clone also revealed a boundary identical to that seen in Tn21. However, a copy of IS26 was found to be inserted at a position 32 bases beyond the insertion point, thus providing a likely explanation for the inability to generate a product with primer pair HS817/HS818. This location for IS26 has not been seen previously. Based on this information, a primer, HS841, based on the IS26 sequence was designed for use in a PCR with HS818. Application of this primer pair to all 21 class 1-positive strains generated a product of the expected length (491 bp) with strain 8157 and two other strains, 3843 and 9445 (Table 2). By sequencing through the attI1 site in a class 1 integron-containing fosmid clone, we found strain 8157 to possess a large cassette array and completely sequenced it. The array was 5,397 bp in length and comprised six cassettes in the order estX psp aadA2 cmlA1 aadA1 qacH. Clones that include this array have been recovered multiple times from Salmonella isolates from various sources in Portugal identified on the basis of the presence of the sul3 gene (2). Sequence generated beyond qacH in strain 8157 revealed a copy of IS440, which is also consistent with the organization of the Portuguese strains. While we did not sequence further, it is likely that strain 8157 also possesses sul3. This explained the observation that the strain is resistant to SXT, despite its not carrying the sul1 gene (Table 2). The fact that there is no 3′-CS is consistent with the failure to generate a PCR product with HS458 and HS459.
In total, four E. coli isolates failed to generate a PCR product with HS458 and HS459, and some of the features of these strains are compared in Table 3. To investigate whether the three strains additional to 8157 have the same array, a PCR was performed with HS458 and a second primer, HS845, which targets the cmlA1 gene. The three IS26-like strains generated a product of about 2,000 bp, implying that these three strains have the same array. It is possible that these isolates are clonal, although it is also noteworthy that strain 9445 does not possess a class 2 integron while strains 3843 and 8157 do. The fourth strain was also distinctive in that the integron backbone was different again, as was the array (Table 3), at least to the extent that it is probably lacking cmlA1. Thus, detailed examination of this subset of four strains demonstrated structural diversity despite the fact that all were identical in not generating an array product with HS458 and HS459.
TABLE 3.
Characteristics of E. coli strains failing to generate an array product with HS458 and HS459
| Strain | IRi junction | IRt junction | Size (bp) of PCR product obtained with HS458 and HS845 | Cassette array | Suld |
|---|---|---|---|---|---|
| 8157a | Tn21 > IS26 | NPb | 2,000 | estX-psp-aadA2-cmlA1-aadA1-qacH | − |
| 3843a | Tn21 > IS26 | NP | 2,000 | NSc | − |
| 9445 | Tn21 > IS26 | NP | 2,000 | NS | − |
| 8393 | Tn21 | Tn21 | None | NAe | + |
Also positive for intI2.
NP, not present. IRt is inferred to be absent based on the failure to generate a PCR product with tniA-specific primers.
NS, PCR product was not sequenced.
A plus or a minus sign indicates the presence or absence of a PCR product with primers HS549/HS550.
NA, not applicable. No array recovered.
The combination of blaCTX-M-2 linked to ISCR1 is common in clinical isolates from South America (3, 4, 35, 50), and this resistance gene encodes 69% of the extended-spectrum β-lactamases found in clinical isolates from Argentinean hospitals (3). Since a blaCTX-M-2 gene linked to ISCR1 was present in strain 12863, we designed primer pairs HS819/HS820, specific for ISCR1, and HS856/HS857, specific for blaCTX-M-2 (Table 1), and screened all 21 class 1-positive strains for their presence. Interestingly, it was found that neither of these genes was present in any of the strains, with the exception of 12836 (data not shown). Despite this observation, resistance to β-lactam antibiotics was very common, with only one of this same cohort of 21 strains being sensitive to AMP.
DISCUSSION
Class 1 integrons are a major clinical concern and are among the biggest contributors to the problem of multidrug-resistant infections due to gram-negative bacteria. Class 1 integrons from clinical isolates can vary, especially in relation to their cassette arrays but overall have several common, but not universal, characteristic features. These include the presence of two conserved segments that flank any inserted gene cassette array and association with at least the remnants of a Tn402-like transposon. The original linkage of a class 1 integron to a Tn402-like transposon is regarded as a key step in the mobilization of the former through populations of pathogens (44). Although most clinical type class 1 integrons are defective transposons, their transpositional spread is still ongoing for at least two reasons. The first of these is that many class 1 integrons that are defective transposons (6) but nonetheless retain the inverted repeats required for transposition. As a result, transposition can presumably still occur when missing proteins are provided in trans. The second reason is that clinically derived class 1 integrons are frequently embedded in other mobile elements—other transposons, plasmids, or both.
A detailed understanding of how class 1 integrons spread resistance genes in hospitals or communities requires an understanding of the contribution of other elements to the mobilization of gene cassettes. Thus, if the same array is recovered from independent isolates, does this mean that it is present in the same integron? Conversely, given that gene cassettes can be mobilized as independent units, does the recovery of different arrays imply cassette mobilization from the same integron or the presence of different integrons in the two isolates in question? If the numbers of isolates is very small, these questions can be relatively easily addressed, but they become more difficult to answer in large epidemiological studies.
Here we have attempted to take a holistic approach in looking at the interrelationships between class 1 integrons from different pathogenic isolates mediating UTIs in a community. To do this, we examined both for cassette array diversity and for the diversity of elements in which the class 1 integron was embedded. In total, 104 patients with UTIs were surveyed from a community in South America. Of the resulting 104 isolates, 21 carried class 1 integrons. Collectively, at least 11 distinct resistance cassettes were identified, spread across eight distinct array types. While all of these cassettes and arrays have been described previously, the cohort of strains contained a number of different array types. Most of the differences, however, were found in the small subset of non-E. coli-associated arrays. The arrays, where present, in the 17 E. coli strains were the most homogenous, with the two-cassette array dfrA17-aadA5 being recovered from 10 of these isolates, perhaps reinforcing the notion that LGT is more extensive within closely related organisms.
Investigation of the integron context proved to be particularly insightful and revealed another level of heterogeneity not obvious by inspection of cassette arrays alone. Two strains in particular highlight the disparate mechanisms by which class 1 integrons can be rearranged and spread in a community. One of these strains was K. pneumoniae isolate 12836. This strain possessed an unusually large integron, as measured between IRi and IRt, that was embedded in a hybrid transposon backbone that probably arose by homologous recombination. This hybrid comprised Tn5036 at the IRi end and Tn21 at the IRt end. While an analogous structure has been seen previously (33), the element described here was created independently of Tn1696 since the insertion point in Tn5036 observed in this study is different for that which is seen for In34 and has not been seen previously. The insertion point was also different from other identified integron insertion into a Tn5036 backbone (Fig. 2). In the strains studied here, we did not recover the Tn5036 parent that presumably generated the 12836 hybrid. We did, however, recover another likely hybrid in K. pneumoniae strain 8180. In this case, the Tn5036 mer region was located at the IRt integron end. It would not appear to be the reciprocal recombination product to 12836, however, since there was no evidence of the Tn21 tnp region being present adjacent to the IRi end of the integron and the tniA gene was related to but not identical to that of Tn402. Thus, we infer that it is the product of an independent recombination event.
Another feature of the 12836 integron was noteworthy and relevant to the origin of the Tn5036 backbone. This is the presence of ISCR1 linked to blaCTX-M-2. In our study, ISCR1/blaCTX-M-2 was only recovered once, despite being common in South America (3, 26, 35, 50), and this was in a context not described previously. Most other reports of ISCR1/blaCTX-M-2 do not provide a sequence context at IRi. Two database entries that do provide relevant sequence context information are those for integrons InS21 (GenBank accession no. AJ311891), from an S. enterica isolate, and InV117 and an integron from a Vibrio cholerae isolate, both recovered in Argentina (41). Both integrons are embedded in a conventional Tn1696-like element in that the insertion point is the same as in Tn1696. Also, the Pc promoter is the same strong promoter that is found in Tn1696. In contrast, one other integron of interest, In117, originates from an E. coli isolate from Spain (49). In117 is identical to Tn21 at the IRt end, but the sequence at the IRi end is not known. This integron is interesting in that it possesses the same hybrid Pc promoter seen in 12836 and not the weak promoter plus P2 normally associated with Tn21. The presence of this hybrid promoter therefore may imply that In117 possesses the same Tn5036 mer region at the IRi integron end seen in strain 12836. In any event, this reinforces the need to know more about integron/transposon context in understanding how integrons and their associated gene cassettes are mobilized. Specifically in this case, it may shed light on the origins of the unusual hybrid transposon found in 12836, as well as where it originated and what role it has in disseminating the ISCR1/blaCTX-M-2 combination.
E. coli strain 5293 demonstrated another aspect of the ability of different families of mobile DNA elements to cooperatively spread resistance genes in pathogens. In this case, two related transposons, Tn21 and Tn5051, have recombined, probably by site-specific recombination, to generate another type of hybrid element. Site-specific recombination is strongly implied since the crossover point can be localized to the resI sequence, the region in Tn21 known to be the point of cointegrate resolution for this transposon (37). In this instance, we would assume that the 5293 hybrid arose by a resolution event between transposons Tn21 and Tn5051, which were located in the same replicon. Such an event has a precedent, perhaps the most notable being the possible role of this process in the evolution of some blaTEM-1-containing transposons (32), but also includes the transposon backbone in which In34 is located (31). The hybrid observed here is noteworthy insomuch as Tn5051-like transposons are commonly associated with environmental bacteria (27). It represents an example that implies the ongoing intermingling of DNA between clinical isolates and bacteria from the general environment. The resulting increase in mobile DNA diversity has obvious ramifications for the evolution of multidrug resistance. In general, such transposon hybrids may be more common than previously thought since a number of these types of events have now been identified and include another example from this survey, namely, the transposon immediately adjacent to the blaTEM-1 gene in strain 12836 (Fig. 1).
This study has highlighted the extensive structural diversity that existed in a community of organisms that contain class 1 integrons that mediated UTI infections. This diversity of structure was despite the fact that many of these integrons had identical or similar cassette arrays and only manifested itself after relatively detailed examination of the backbone in which the integrons were embedded. The most general noteworthy finding was the previously unrealized extent of diversity that can be generated by a variety of mechanisms that facilitate resistance gene spread in a community. In the case of integrons, this includes not just mobilization of gene cassettes but also homologous recombination and site-specific recombination involving transposon backbones. It will be interesting to determine if the level of diversity seen here applies in other communities of pathogens. This survey involved bacteria mediating UTIs. The mediating organisms are predominantly those that are also human commensals and are found in the broader environment. Therefore, they are well placed to recruit mobile DNA from diverse sources and act as a conduit for the movement of resistance genes into the more specialized community of pathogens. The structural diversity seen here may reflect that role.
Acknowledgments
This work was supported in part by the National Health and Medical Research Council of Australia, PEDECIBA-Química, and grant FCE9029 from the Ministry of Education and Culture of Uruguay.
We thank the Clinical Microbiology Service of the Central Hospital of the Armed Forces of Uruguay for providing clinical strains. We thank Sally Partridge for critical assessment of the manuscript.
Footnotes
Published ahead of print on 27 August 2008.
REFERENCES
- 1.Akram, M., M. Shahid, and A. U. Khan. 2007. Etiology and antibiotic resistance patterns of community-acquired urinary tract infections in J N M C Hospital Aligarh, India. Ann. Clin. Microbiol. Antimicrob. 64. doi: 10.1186/1476-0711-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antunes, P., J. Machado, and L. Peixe. 2007. Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob. Agents Chemother. 511545-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arduino, S. M., M. Catalano, B. E. Orman, P. H. Roy, and D. Centron. 2003. Molecular epidemiology of orf513-bearing class 1 integrons in multiresistant clinical isolates from Argentinean hospitals. Antimicrob. Agents Chemother. 473945-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 462303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blahna, M. T., C. A. Zalewski, J. Reuer, G. Kahlmeter, B. Foxman, and C. F. Marrs. 2006. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J. Antimicrob. Chemother. 57666-672. [DOI] [PubMed] [Google Scholar]
- 6.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 1784429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2007. Performance standard for antimicrobial disk susceptibility testing (M100-S17). Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Clinical and Laboratory Standards Institute. 2006. Performance standard for antimicrobial disk susceptibility tests (M2-A9). Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell, D. J., I. Morrissey, D. De Rubeis, M. Robbins, and D. Felmingham. 2003. A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. J. Infect. 4694-100. [DOI] [PubMed] [Google Scholar]
- 11.Foxman, B. 2003. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 4953-70. [DOI] [PubMed] [Google Scholar]
- 12.García Fernández, A., A. Cloeckaert, A. Bertini, K. Praud, B. Doublet, F. X. Weill, and A. Carattoli. 2007. Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 514177-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grape, M., A. Farra, G. Kronvall, and L. Sundstrom. 2005. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant gram-negative bacteria. Clin. Microbiol. Infect. 11185-192. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, K., T. M. Hooton, and W. E. Stamm. 2001. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 13541-50. [DOI] [PubMed] [Google Scholar]
- 15.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 1766286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerl, J. A., I. Klein, E. Lanka, B. Appel, and S. Hertwig. 2008. Genetic and functional properties of the self-transmissible Yersinia enterocolitica plasmid pYE854, which mobilizes the virulence plasmid pYV. J. Bacteriol. 190991-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heikkilä, E. S., L, Skurnik, and M. P. Huovinen. 1991. Analysis of genetic localization of the type I trimethoprim resistance gene from Escherichia coli isolated in Finland. Antimicrob. Agents Chemother. 351562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes, A. J., M. R. Gillings, B. S. Nield, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2003. The gene cassette metagenome is a basic resource for bacterial genome evolution. Environ. Microbiol. 5383-394. [DOI] [PubMed] [Google Scholar]
- 19.Jones, L. A., C. J. McIver, W. D. Rawlinson, and P. A. White. 2003. Polymerase chain reaction screening for integrons can be used to complement resistance surveillance programs. Commun. Dis. Intell. 27(Suppl.)S103-S110. [DOI] [PubMed] [Google Scholar]
- 20.Kurutepe, S., S. Surucuoglu, C. Sezgin, H. Gazi, M. Gulay, and B. Ozbakkaloglu. 2005. Increasing antimicrobial resistance in Escherichia coli isolates from community-acquired urinary tract infections during 1998-2003 in Manisa, Turkey. Jpn. J. Infect. Dis. 58159-161. [PubMed] [Google Scholar]
- 21.Labbate, M., P. R. Chowdhury, and H. W. Stokes. 2008. A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments. J. Bacteriol. 1905318-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 23.Lévesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 14249-54. [DOI] [PubMed] [Google Scholar]
- 24.Lévesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melano, R., A. Corso, A. Petroni, D. Centron, B. Orman, A. Pereyra, N. Moreno, and M. Galas. 2003. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum β-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J. Antimicrob. Chemother. 5236-42. [DOI] [PubMed] [Google Scholar]
- 27.Mindlin, S., G. Kholodii, Z. Gorlenko, S. Minakhina, L. Minakhin, E. Kalyaeva, A. Kopteva, M. Petrova, O. Yurieva, and V. Nikiforov. 2001. Mercury resistance transposons of gram-negative environmental bacteria and their classification. Res. Microbiol. 152811-822. [DOI] [PubMed] [Google Scholar]
- 28.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.). 2003. Manual of clinical microbiology. American Society for Microbiology, Washington, DC.
- 29.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405299-304. [DOI] [PubMed] [Google Scholar]
- 30.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 451263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partridge, S. R., and R. M. Hall. 2004. Complex multiple antibiotic and mercury resistance region derived from the r-det of NR1 (R100). Antimicrob. Agents Chemother. 484250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Partridge, S. R., and R. M. Hall. 2005. Evolution of transposons containing blaTEM genes. Antimicrob. Agents Chemother. 491267-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Post, V., G. D. Recchia, and R. M. Hall. 2007. Detection of gene cassettes in Tn402-like class 1 integrons. Antimicrob. Agents Chemother. 513467-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Power, P., M. Galleni, J. Di Conza, J. A. Ayala, and G. Gutkind. 2005. Description of In116, the first blaCTX-M-2-containing complex class 1 integron found in Morganella morganii isolates from Buenos Aires, Argentina. J. Antimicrob. Chemother. 55461-465. [DOI] [PubMed] [Google Scholar]
- 36.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 1413015-3027. [DOI] [PubMed] [Google Scholar]
- 37.Rogowsky, P., S. E. Halford, and R. Schmitt. 1985. Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 42135-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Severino, P., and V. D. Magalhaes. 2004. Integrons as tools for epidemiological studies. Clin. Microbiol. Infect. 10156-162. [DOI] [PubMed] [Google Scholar]
- 39.Smith, S. P., A. R. Manges, and L. W. Riley. 2008. Temporal changes in the prevalence of community-acquired antimicrobial-resistant urinary tract infection affected by Escherichia coli clonal group composition. Clin. Infect. Dis. 46689-695. [DOI] [PubMed] [Google Scholar]
- 40.Solberg, O. D., R. M. Ajiboye, and L. W. Riley. 2006. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J. Clin. Microbiol. 441347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soler Bistué, A. J. C., F. A. Martín, A. Petroni, D. Faccone, M. Galas, M. E. Tolmasky, and A. Zorreguieta. 2006. Vibrio cholerae InV117, a class 1 integron harboring aac(6′)-Ib and blaCTX-M-2, is linked to transposition genes. Antimicrob. Agents Chemother. 501903-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soubes, M., S. Cobelli, G. Borthagaray, N. Bresciano, and P. Cerdeiras. 1990. Aplicación de un programa de computación para identificar los distintos géneros y especies de la familia Enterobacteriaceae en bacteriología clínica. Rev. Argent. Microbiol. 2273-78. [PubMed] [Google Scholar]
- 43.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 31669-1683. [DOI] [PubMed] [Google Scholar]
- 44.Stokes, H. W., C. L. Nesbo, M. Holley, M. I. Bahl, M. R. Gillings, and Y. Boucher. 2006. Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 1885722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tillett, D., and B. A. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36251-258. [Google Scholar]
- 46.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 581-6. [DOI] [PubMed] [Google Scholar]
- 47.Turnidge, J., J. Bell, D. J. Biedenbach, and R. N. Jones. 2002. Pathogen occurrence and antimicrobial resistance trends among urinary tract infection isolates in the Asia-Western Pacific region: report from the SENTRY Antimicrobial Surveillance Program, 1998-1999. Int. J. Antimicrob. Agents 2010-17. [DOI] [PubMed] [Google Scholar]
- 48.Turnidge, J., and D. L. Paterson. 2007. Setting and revising antibacterial susceptibility breakpoints. Clin. Microbiol. Rev. 20391-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valverde, A., R. Canton, J. C. Galan, P. Nordmann, F. Baquero, and T. M. Coque. 2006. In117, an unusual In0-like class 1 integron containing CR1 and blaCTX-M-2 and associated with a Tn21-like element. Antimicrob. Agents Chemother. 50799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vignoli, R., N. Cordeiro, V. Seija, F. Schelotto, M. Radice, J. Ayala, P. Power, and G. Gutkind. 2006. Entorno genetico de CTX-M2 en aislamientos de Klebsiella pneumoniae provenientes de pacientes hospitalizados en Uruguay. Rev. Argent. Microbiol. 3884-88. [PubMed] [Google Scholar]
- 51.Walsh, T. R. 2006. Combinatorial genetic evolution of multiresistance. Curr. Opin. Microbiol. 9476-482. [DOI] [PubMed] [Google Scholar]
- 52.Yu, H. S., J. C. Lee, H. Y. Kang, D. W. Ro, J. Y. Chung, Y. S. Jeong, S. H. Tae, C. H. Choi, E. Y. Lee, S. Y. Seol, Y. C. Lee, and D. T. Cho. 2003. Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J. Clin. Microbiol. 415429-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]