Abstract
The need for rapid methods to accurately detect methicillin-resistant Staphylococcus aureus (MRSA) is widely acknowledged, and a number of molecular assays are commercially available. This study evaluated the Xpert MRSA assay, which is run on the GeneXpert real-time PCR platform (Cepheid) for use in a clinical laboratory. The following parameters were investigated: (i) the limits of detection (LoDs) for four MRSA strains; (ii) the ability to detect isolates of MRSA from a collection representative of MRSA in Ireland since 1974 (n = 114) and the ability to detect control strains with staphylococcal cassette chromosome mec types IVa (IV.1.1.1), IVb (IV.2.1.1), IVc (IV.3.1.1), IVd (IV.4.1.1), V (V.1.1.1), VT, and VI; and (iii) performance in a clinical trial with swabs from nose, throat, and groin/perineum sites from 204 patients, where results were compared with those obtained by direct and enrichment cultures. The average LoD of the four test strains was 610 CFU/ml (equivalent to 58 CFU/swab). All 114 MRSA isolates and 7 control strains tested were detected. Sensitivity, specificity, and positive and negative predictive values for clinical specimens from all sites investigated were 90%, 97%, 86%, and 98%, respectively, but throat specimens yielded poor sensitivity (75%). Sensitivity, specificity, and positive and negative predictive values for nasal specimens were 95%, 98%, 90%, and 99%, respectively. Overall, the assay was rapid and easy to perform, but performance might be enhanced by the inclusion of an equivocal interpretive category based on analysis of all available amplification data.
The rapid detection of methicillin-resistant Staphylococcus aureus (MRSA) along with an early implementation of appropriate intervention has been reported to reduce the prevalence of MRSA, especially in critical care areas (1, 5, 8). In 2004, Huletsky et al. described a novel real-time PCR assay targeting DNA sequences in the region of the open reading frame orfX, where the staphylococcal cassette chromosome mec (SCCmec) integrates into the S. aureus chromosome (7). SCCmec carries the resistance determinant mecA, which encodes methicillin resistance and exhibits at least six different structural types and numerous subtypes (SCCmec; T. Ito, Department of Bacteriology, School of Medicine, Juntendo University, Japan. http://www.staphylococcus.net/ [10 November 2007, accession date]). To detect MRSA with all SCCmec types known in 2004, the assay used five forward primers designed to target sequences within SCCmec, while a sixth reverse primer and three probes were specific for orfX (7). Unlike earlier amplification techniques investigating mecA, nuc, and/or fem genes, this assay could distinguish between MRSA and mixtures of methicillin-susceptible S. aureus (MSSA) and methicillin-resistant coagulase-negative staphylococci (MR-CNS) (3, 7). The assay became commercially available as the IDI-MRSA assay and is currently marketed as the BD GeneOhm MRSA assay.
In 2007, a new real-time PCR MRSA assay also targeting DNA sequences in the chromosomal orfX-SCCmec junction became available. This assay is the Xpert MRSA kit (Cepheid, Sunnyvale, CA), and it is used in conjunction with the GeneXpert real-time PCR platform (Cepheid). In the Xpert MRSA assay, DNA from specimens is extracted, amplified, and detected in separate chambers of single-use disposable cartridges which contain freeze-dried beads with all reagents required for the real-time process. Sample preparation time is minimal and the PCR time is 75 min. The assay is validated for use with nasal specimens taken on Copan swabs with Stuart's liquid transport medium from patients at risk for colonization with MRSA and has a quoted limit of detection (LoD) of 80 CFU per swab (Xpert MRSA assay product insert; Cepheid, Sunnyvale, CA).
The present study evaluated the Xpert MRSA assay for use in a clinical laboratory. The following parameters were investigated: (i) LoD, (ii) ability to detect isolates of MRSA in a collection representative of MRSA in Ireland since 1974 and to detect isolates carrying recently described SCCmec elements, and (iii) performance in a clinical trial with swabs from nose, throat, and groin/perineum sites from 204 patients.
MATERIALS AND METHODS
LoD assays.
LoD assays of the Xpert MRSA kit were performed using four MRSA isolates. These isolates exhibited a range of oxacillin MICs and were representative of strains recovered in Ireland, as described previously (16). LoDs were determined for 10-fold dilutions of suspensions of each strain (from 105 to 101 CFU/ml) adsorbed onto Copan Stuart's liquid transport medium double swabs (Copan 139C swabs; Copan Italia SPA, Brescia, Italy). Colony counts (CFU/ml) of each dilution were determined by spiral plating onto brain heart infusion agar (Oxoid CM375; Oxoid Ltd., Basingstoke, United Kingdom) by use of a Whitley automatic spiral plater (WASP; Don Whitley Scientific Ltd., Shipley, England). LoDs were determined for all four isolates in pure culture and in the presence of a mixed cocktail of bacteria consisting of 10 MSSA isolates (which were mecA negative by in-house conventional PCR), five MR-CNS isolates, and single isolates of Moraxella catarrhalis, Escherichia coli, and Candida species (16). Each component of the cocktail was tested with the Xpert MRSA kit prior to use. The bacterial suspensions were also adsorbed onto Transwabs with Amies' clear transport medium (MW170 Transwabs; Medical Wire and Equipment Company, Corsham, England) for subsequent culture.
One Copan swab from each dilution was investigated for the presence of MRSA by use of the Xpert MRSA kit on a GeneXpert DX system, version 1.2 (Cepheid, Sunnyvale, CA), according to the manufacturer's instructions. The second Copan swab was reserved for repeat testing if required.
The corresponding Transwab was cultured onto Columbia agar (LabM Lab1; International Diagnostics Group plc, Bury, United Kingdom) containing 7% (wt/vol) horse blood (BA), onto MRSA-Select chromogenic agar (CA) (Bio-Rad 63747; Bio-Rad Life Science Group, Marnes-La-Coquette, France), and into 4 ml tryptic soy broth (BD 211825; Becton Dickinson) containing 6.5% NaCl (salt TSB). After 18 h of incubation at 35°C, enrichment broths (10-μl volumes) were subcultured onto BA and CA. All isolates from enriched and direct culture were identified as S. aureus and confirmed as MRSA as described previously (16).
To express the LoD value as CFU/swab, the volume adsorbed by either Copan swabs or Transwabs was determined by measuring the difference in weights of bijoux of saline before and after adsorbance for 10 s by each of 10 Copan swabs and 10 Transwabs. The average of the 10 replicate readings was taken as the average volume adsorbed by each swab type.
LoDs were recorded as the lowest concentration to give positive results by the Xpert MRSA kit or by culture. The Xpert MRSA assay defines a specimen as MRSA positive if the MRSA target has a cycle threshold (CT) value within the valid range (≤36 cycles). The LoDs for both culture methods were calculated on the basis of growth of >1 colony on a single culture medium or ≥1 colony on more than one medium and were expressed as averages of the counts obtained with the four isolates.
MRSA isolates and MRSA control strains.
To ensure that the kit could detect MRSA strains prevalent in Ireland, 114 MRSA isolates representative of MRSA recovered in Ireland were investigated (isolate details are shown in Table 1) (18). In addition, seven control MRSA strains representing isolates carrying SCCmec IV subtypes IVa (IV.1.1.1), IVb (IV.2.1.1), IVc (IV.3.1.1), IVd (IV.4.1.1), SCCmec V (V.1.1.1), VT, and VI were tested (2, 4, 13). Isolates were prepared in saline suspensions at concentrations of ∼102 CFU/ml above the LoD of the kit (determined in the present study) and adsorbed onto Copan swabs and Transwabs. One swab of each pair of Copan swabs was tested with the Xpert MRSA assay, and the Transwab was cultured onto BA and CA. Any isolate that was negative by use of the kit was retested in pure culture and at one 10-fold-higher concentration.
TABLE 1.
MRSA isolates (n = 114) used to determine whether the Xpert MRSA kit could detect all strains of MRSA prevalent in Ireland
| MLST (no. of isolates) | SCCmec typeb | AR type (no. of isolates)c |
|---|---|---|
| ST5d (10) | II | 07.3d (3); 07.4 (5); 11 (2) |
| ST5 (1) | IV | Unfamiliare (1) |
| ST8f (11) | IV or IVv | 43 (8); Unfamiliare (3f) |
| ST8 (26) | II or IIv | 13 (7); 14 (15); New03 (3); 05 (1) |
| ST12 (1) | IV | NT (1) |
| ST22 (14) | IV | 06 (13); NT (1) |
| ST30 (2) | IV | NT (2) |
| ST34 (1) | IV | NT (1) |
| ST36 (3) | II | 07.0 (2); 07.2 (1) |
| ST45 (1) | IV | NT (1) |
| ST239 (13) | III or IIIv | 01 (4); 09 (3); 15 (2); 44 (3); 23 (1) |
| ST247 (3) | IA | 22 (2); New02 (1) |
| ST250 (4) | I or Iv | 02 (4) |
| —a (6) | — | 07 (2); New02 (1); 06 (1); SCV (2) |
| — (18) | — | PVL-positive community-acquired MRSA (18) |
—, not done.
Antibiogram-resistogram (AR) types were determined from the patterns of resistance obtained when isolates were tested by disk diffusion against a panel of 23 antimicrobials as described previously (17). NT indicates AR patterns that were designated “no type” pending the results of DNA macrorestriction digestion analysis; PVL, Panton-Valentine leucocidin; SCV, small-colony variant.
One isolate was a double-locus variant of ST5.
Unfamiliar AR pattern.
Two isolates were single-locus variants of ST8.
Clinical trial.
The clinical trial was undertaken with specimens from patients attending St. James's Hospital (SJH), a large 936-bed tertiary-referral hospital catering to all specialties except maternity and pediatric services. A difficulty encountered when designing the clinical trial was that specimens processed by the Xpert MRSA assay could not be used for subsequent culture because the kit's elution reagent contains sodium hydroxide and quanidium thiocyanate. Ethical approval was obtained to take duplicate specimens from nose, throat, and groin/perineum sites from patients undergoing routine MRSA screening with Copan swabs after specimens on Transwabs had been obtained for routine diagnostic culture. Criteria for routine screening were as described previously (16).
Specimens collected with Copan swabs were tested using the Xpert MRSA kit according to the manufacturer's instructions as soon as possible after collection. Specimens were tested in batches of eight by use of two four-module GeneXpert machines. If not tested immediately, specimens were stored at 4°C. Following processing, all specimens were stored at 4°C until all molecular and culture testing was complete. Specimens yielding invalid results were retested using the second Copan swab.
Specimens collected with Transwabs were inoculated onto CA in the diagnostic laboratory and subsequently cultured into salt TSB. After incubation at 35°C for 18 h, 10-μl volumes of salt TSB were subcultured onto BA and CA. Growth was identified as MRSA as described previously, and the presence of mecA was confirmed by a conventional in-house end-point PCR assay (16). All MRSA isolates were stored at −70°C on cryoprotective beads (Protect beads; Technical Service Consultants Limited, Heywood, United Kingdom). Bacterial growth from BA plates from specimens yielding positive Xpert MRSA kit results and negative MRSA culture results were also preserved at −70°C on cryoprotective beads. Discrepancies between culture and kit results were investigated by salt enrichment culture of the second Copan swab (if available).
MRSA isolates recovered from specimens yielding kit-negative results were prepared in suspensions in saline at concentrations of 105 CFU/ml, adsorbed onto Copan swabs, and tested using the Xpert MRSA kit. Bacterial growth from BA plates from specimens which tested Xpert MRSA kit positive but from which MRSA was not recovered in culture was similarly tested to exclude the possibility that such positive results occurred as a result of MSSA or MR-CNS.
Control strains.
S. aureus ATCC 25923 and S. aureus ATCC 43300 were used as negative and positive controls, respectively. S. epidermidis ATCC 12228 was also used as a negative control as recommended by the manufacturer. Control isolates were prepared in saline suspensions containing 105 CFU/ml and adsorbed onto Copan swabs prior to testing. For determining LoDs, positive and negative controls were included with each batch of tests. When MRSA strains were tested, a negative control was included each day. During the clinical trial, a positive control was included once a week and after any batch of specimens where all specimens yielded kit-negative results. S. aureus ATCC 25923 and S. aureus ATCC 43300 were also included as negative and positive controls with each batch of CA plates.
Statistics.
In the clinical trial, the sensitivities, specificities, and positive and negative predictive values of the Xpert MRSA kit were calculated for all specimen types by comparison with direct culture and by comparison with enrichment culture. Because the Xpert MRSA kit detects DNA which may come from nonviable MRSA, whereas culture detects viable organisms only, Xpert MRSA kit-positive specimens that were culture negative were considered as giving possible true-positive results if the patient had been previously positive for MRSA within the last 2 years or was positive at another site. Sensitivities, specificities, and positive and negative predictive values were also calculated using these amended results for all specimen sites and for each specimen site individually. Further calculations were made when patients were grouped into those who were known to have been previously positive for MRSA and those who were not known to have been previously positive.
RESULTS
LoD assays.
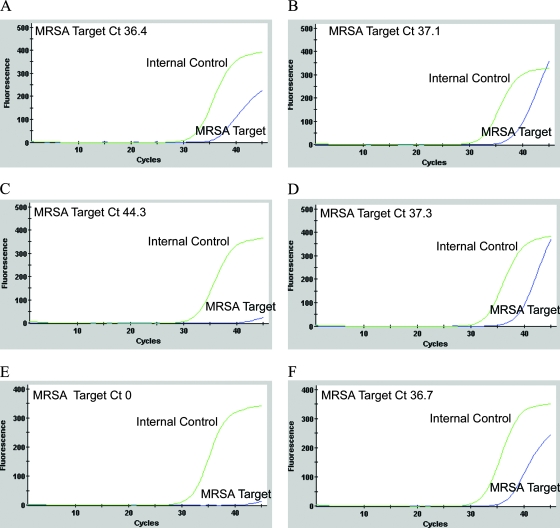
The average LoD value for the four MRSA test isolates in pure culture with the Xpert MRSA kit using Copan swabs was 610 CFU/ml. The average volume adsorbed by Copan swabs was 95 μl/swab; hence, the average LoD/swab was 58 CFU. The amplification data obtained with two of these isolates suggested that both isolates were positive at concentrations below 58 CFU/swab, but since CT values of 36.4 and 37.1 were obtained, the kit recorded negative results (Fig. 1A and B). The average LoD values of direct and enrichment cultures using Transwabs were 750 CFU/ml and 40 CFU/ml, respectively. The average volume adsorbed by Transwabs was 228 μl/swab; hence, the average LoDs of direct and enrichment cultures were 171 and 9 CFU/swab, respectively. The average LoD of the kit for the four test isolates in the presence of mixed flora was the same as the LoD obtained in pure culture (600 CFU/ml; 57 CFU/swab).
FIG. 1.
Amplification data obtained with the Xpert MRSA kit for two of the four MRSA isolates used in LoD assays (A and B), two replicates of S. aureus ATCC 25923 (C and D), and two replicates of an MRSA isolate (tested at 106 CFU/ml) recovered from a specimen yielding a kit-negative result (E and F). All isolates demonstrated kit-negative results with appropriate amplification of the internal control. The amplification plots in panels A and B demonstrate an exponential rise in fluorescence emission that is consistent with amplification of the MRSA target, suggesting false-negative kit results. The amplification data shown in panel C are representative of results obtained with most replicates of S. aureus ATCC 25923, but on some occasions this MSSA isolate yielded a plot suggestive of MRSA target amplification (panel D). The data shown in panels E and F were obtained from an MRSA isolate tested at a bacterial concentration of 106 CFU/ml on two separate occasions. While both replicates yielded kit-negative results, one replicate demonstrated evidence of MRSA target amplification (panel F).
During the preparation of the mixed-culture cocktail, the MSSA component yielded a kit-positive result. When the cocktail was prepared using fresh subcultures of the same 10 MSSA isolates, the cocktail tested negative and continued to test kit negative on the successive days when the LoDs of the kit were determined. However, when the cocktail was retested following completion of the LoD assays, the kit again recorded a positive result, although the accompanying amplification curve failed to demonstrate evidence of efficient amplification, suggesting that this could be a false-positive result. Conversely, although the Xpert assay always recorded negative results for the MSSA control strain S. aureus ATCC 25923 (Fig. 1C), on some occasions the amplification data demonstrated evidence of amplification of MRSA target DNA with CT values just exceeding the cutoff value of 36 cycles (for example, a CT value of 37.3) and accompanying amplification curves showed an exponential rise in fluorescence emission (Fig. 1D).
MRSA isolates and MRSA control strains.
When this study was designed, it was intended that the MRSA isolates and MRSA control strains would be tested in the presence of the mixed cocktail of bacteria to mimic normal flora, but the inconsistent results with the MSSA component of the cocktail suggested that this would be unwise. Isolate suspensions were therefore prepared in a pure culture in saline at a concentration of ∼104 CFU/ml (i.e., ∼102 CFU/ml above the LoD determined in the present study). All MRSA isolates except two yielded positive Xpert MRSA kit results; one negative isolate was positive upon repeat testing, and the second negative isolate was positive when retested at a higher concentration (105 CFU/ml). All seven control MRSA strains yielded positive results.
Clinical trial.
Six hundred twelve specimens from 204 patients were investigated. An overview of the numbers of positive specimens obtained with the Xpert MRSA kit, with direct culture, with enrichment culture, and with amended results is shown in Table 2. By use of amended results, Xpert MRSA kit-positive specimens that were culture negative were considered to be true-positive results if the patient had been previously positive for MRSA or was positive at another site. MRSA was detected in 99 specimens (16.2%; 99/612) from 58 patients (28.4%; 58/204) by the Xpert MRSA kit. Direct culture yielded positive results from 59 specimens (9.7%; 59/606) from 37 patients (18.3%; 37/202); direct culture results were unavailable for 6 specimens from 2 patients. Specimens from 89 specimens (14.5%; 89/612) from 45 patients (22.1%; 45/204) were positive by enrichment culture. No specimen was culture positive by direct culture and negative by salt enrichment culture. When amended results were calculated, 95 specimens (15.5%; 95/612) from 47 patients (23.0%; 47/204) were deemed positive. Sixteen specimens from 14 patients yielded invalid Xpert MRSA kit results upon initial testing, but upon repeat testing only four specimen results remained invalid (throat [n = 3]; groin [n = 1]).
TABLE 2.
Overview of results obtained with the Xpert MRSA kit and culture from 612 specimens
| Reference method and result | No. of specimens with Xpert MRSA kit result of:
|
|||
|---|---|---|---|---|
| Positive | Negative | Invalidb | Total | |
| Direct culturea | ||||
| Positive | 52 | 7 | 59 | |
| Negative | 46 | 497 | 4 | 547 |
| Total | 98 | 504 | 4b | 606a |
| Enrichment culture | ||||
| Positive | 70 | 19 | 89 | |
| Negative | 29 | 490 | 4 | 523 |
| Total | 99 | 509 | 4b | 612 |
| Amended resultsc | ||||
| Positive | 85 | 10 | 95 | |
| Negative | 14 | 499 | 4 | 517 |
| Total | 99 | 509 | 4b | 612c |
Direct culture results were unavailable for six specimens from two patients.
Four isolates were invalid by the Xpert MRSA kit.
Amended results include numbers of specimens that were culture positive and those that were Xpert MRSA kit positive but culture negative from previously positive patients or from patients from whom a specimen from another site was positive.
MRSA isolates from all but 2 of the 19 culture-positive kit-negative specimens were kit positive when tested in pure culture at a concentration of 105 CFU/ml. One of the two negative isolates tested positive when retested at 105 CFU/ml; the other remained kit negative when retested at both 105 and 106 CFU/ml (Fig. 1E). Repeat testing of the latter isolate at 106 CFU/ml also yielded a negative kit result, but evidence of amplification (with a CT value of 36.7) was noted on this occasion (Fig. 1F). This isolate was mecA gene positive by conventional in-house endpoint PCR.
Non-MRSA bacterial growths from BA plates from specimens which were kit positive and MRSA culture negative were also tested by the kit, but growths from all of these non-MRSA cultures yielded negative results with the kit.
Statistics.
The sensitivity, specificity, and positive and negative predictive values of the Xpert MRSA kit compared with those obtained using direct culture, enrichment culture, and amended results are shown in Table 3. With nasal swabs, when the amended results were considered, the sensitivity of the kit was 95%. In comparison, its sensitivity for specimens from the throat was only 75%. When the Xpert MRSA kit results from specimens from all sites were compared with amended results, the overall sensitivity, specificity, and positive and negative predictive values were 90%, 97%, 86%, and 98%, respectively (Table 3). When this analysis was restricted to specimens from patients who were previously recorded as being MRSA positive, the sensitivity of the kit for nasal specimens was 93%, whereas with specimens from patients with no record of being previously positive (i.e., new MRSA patients), the sensitivity for nasal specimens was 100%.
TABLE 3.
Sensitivity, specificity, and positive and negative predictive values of the Xpert MRSA kit compared with those from direct and enrichment cultures from nose, throat, and groin/perineum specimens
| Reference method and analysis group | Sited | Value (%) of Xpert MRSA kit
|
|||
|---|---|---|---|---|---|
| Sensitivity | Specificity | PPVe | NPVf | ||
| Direct culture | All | 88 | 92 | 53 | 99 |
| Enrichment culture | All | 79 | 94 | 71 | 96 |
| Nose | 84 | 92 | 67 | 97 | |
| Throat | 63 | 98 | 83 | 93 | |
| Groin/perineum | 92 | 94 | 69 | 99 | |
| Amended resultsa | |||||
| All patients | All | 90 | 97 | 86 | 98 |
| Nose | 95 | 98 | 90 | 99 | |
| Throat | 75 | 98 | 88 | 96 | |
| Groin/perineum | 97 | 96 | 80 | 99 | |
| Previously positive patientsb | All | 87 | 100 | 100 | 87 |
| Nose | 93 | 100 | 100 | 88 | |
| Throat | 73 | 100 | 100 | 77 | |
| Groin/perineum | 95 | 100 | 100 | 96 | |
| Patients who were not previously positivec | All | 96 | 97 | 62 | 100 |
| Nose | 100 | 97 | 67 | 100 | |
| Throat | 83 | 98 | 63 | 99 | |
| Groin/perineum | 100 | 95 | 59 | 100 | |
For further explanation of amended results, see Table 2, footnote c.
Analysis restricted to specimens from patients who were previously MRSA positive. With these specimens, kit-positive culture-negative specimens were also included as true positives because the patients were known to have been previously positive for MRSA.
Analysis restricted to specimens from patients with no record of being previously MRSA positive. With these specimens, kit-positive culture-negative specimens were also included as true positives if the patient was positive for MRSA at another site.
All, nose, throat, and groin/perineum.
PPV, positive predictive value.
NPV, negative predictive value.
DISCUSSION
Ireland has a serious problem with MRSA, and the proportion of MRSA among S. aureus blood culture isolates is among the highest in Europe (11). It has been reported that in areas where MRSA is endemic, the introduction of a full “search-and-destroy” approach, including screening of all patient contacts of all index cases and the institution of precautionary isolation for all such contacts, could reduce the prevalence of MRSA to <1% within 6 years (1). It has also been reported that rapid detection of carriage has an important role to play in such a “search-and-destroy” strategy (1). A study investigating the value of rapid diagnostic tests (RDTs) for MRSA when used for admission screening to a critical care area reported a reduction in the incidence of transmission of MRSA from 13.89/1,000 patient days to 4/1,000 patient days (5).
While RDTs may play a major role in reducing the prevalence of MRSA, the chosen method must be sensitive and specific and have good positive and negative predictive values. Failure to recognize a particular strain of MRSA can have very serious consequences, as was recently experienced in The Netherlands, where phenotypic tests failed to detect a mecA-positive strain with a low oxacillin MIC in one hospital. Over a 2-year period, this strain spread extensively not only within that hospital but in other hospitals also and it took over a year to control the resulting outbreaks (8).
As more molecular RDTs for MRSA become available, the choice of an appropriate assay becomes more difficult. Evaluating new molecular assays is expensive and time-consuming, and it is hard to exclude confounding variables, even when assays are evaluated at the same time. For example, the present study required duplicate specimens to allow comparison between the Xpert MRSA kit and enrichment culture. Because sampling using the routine Transwab method had to be undertaken prior to sampling with Copan swabs to ensure that the routine diagnostic culture protocol was not compromised by the clinical trial, it is possible that less material was available on the Copan swabs for detection by the Xpert MRSA assay. In contrast, in an earlier study evaluating the IDI-MRSA kit on the SmartCycler platform, swabs were cultured by salt enrichment after they had been used with the kit (16). In that evaluation, however, the LoD of salt enrichment culture (by use of Transwabs) was 240 CFU/ml (55 CFU/swab), whereas the LoD of salt enrichment culture (by use of Transwabs) in the present study was 9 CFU/swab, indicating that the Xpert MRSA kit was subjected to a more rigorous evaluation. A possible explanation for the lower LoD of salt enrichment culture in the present study is that the volume of medium used for salt enrichment was 4 ml, whereas the protocol in the IDI-MRSA kit evaluation used 1-ml volumes and enrichment culture was performed after material on the swab was removed for testing with the IDI-MRSA kit. Interestingly, the inclusion of enrichment culture increased the number of culture-positive specimens by 23% in the earlier study and by 51% (30/59) in the present study (16).
The Xpert MRSA assay demonstrated an average LoD (58 CFU/swab) threefold lower than that for direct culture (171 CFU/swab) but sixfold higher than that for enrichment culture (9 CFU/swab). In comparison, the earlier IDI-MRSA kit evaluation reported an average LoD value of 2,000 CFU/ml (equivalent to 190 CFU/swab), which was similar to the LoD of direct culture but 10-fold higher than the LoD of enrichment culture (16). The LoDs of direct culture with Transwabs were comparable in both studies: 171 CFU/swab in the present study and 800 CFU/ml (182 CFU/swab) in the earlier study (16).
The Xpert MRSA assay detected all MRSA isolates investigated, including control isolates exhibiting SCCmec V, VT, and VI (2, 4, 13). Although not investigated here, both Xpert MRSA assay and IDI-MRSA are reported to lack sensitivity when detecting the pulsed-field gel electrophoresis-nontypeable MRSA strain associated with pigs in The Netherlands (15, 19).
In the clinical trial, the Xpert MRSA assay exhibited sensitivities of 95% and 97% for detecting MRSA from nasal and groin/perineum sites but had difficulty detecting MRSA DNA from throat specimens (sensitivity, 75%). Interestingly although the previous evaluation of the IDI-MRSA kit reported lower sensitivities for nose and groin/perineum specimens (90% and 88%, respectively), specimens from the throat yielded a higher sensitivity (89%) (16).
A major difficulty evaluating molecular assays for the detection of MRSA from clinical specimens is defining true-positive and -negative specimens. In the present study, data were analyzed by direct comparison with culture, but to overcome the problem of considering all kit-positive culture-negative specimens as false-positive kit results, kit performance was analyzed using amended results where kit-positive culture-negative results from patients who were previously positive for MRSA were regarded as true-positive results. By use of these amended results for comparison, the sensitivities and specificities of direct culture, enrichment culture, and the Xpert MRSA assay were 60.6% and 99.6%, 84.2% and 98.3%, 89.5% and 97.3%, respectively. However, this comparison is not completely valid and may overestimate the sensitivity of the assay and underestimate that of culture because culture-negative results from patients who have undergone successful treatment for MRSA should not be considered false-negative culture results.
Despite the encouraging findings of the present study, the overall performance of the Xpert MRSA assay might be improved if result interpretation took account of all available assay data. The manufacturer's interpretation is based on the application of a CT cutoff value of 36 cycles, with specimens being recorded as MRSA positive if the CT is ≤36 and MRSA negative if the CT is >36, regardless of the presence or absence of evidence of amplification (the former being demonstrated by an amplification curve indicating an exponential rise in fluorescence emission). Data obtained in the present study suggested that this CT cutoff value might have introduced false-negative results both in the LoD investigations and during the clinical trial, where some tests (n = 14) demonstrated clear evidence of MRSA amplification but CT values exceeded 36 cycles (Fig. 1A, B, and F). Conversely, false-positive results may have also occurred (with the MSSA component of the mixed-culture cocktail, e.g.) where CT values of ≤36 were obtained and reported as positive but amplification curves failed to demonstrate an exponential rise. The inclusion of an equivocal interpretative category for tests where the amplification data of the MRSA target DNA fail to support the CT cutoff result warrants investigation.
Aside from the interpretative difficulties associated with the use of the CT cutoff value, findings from the present study also indicate that false-positive results may have occurred, due perhaps to a lack of specificity of the assay's proprietary target sequence for MRSA at the SCCmec-orfX junction. There are increasing numbers of reports of SCC elements that do not contain mecA, for example, SCCcap1 in MSSA, which integrates into the SCCmec chromosomal attachment site attB at the end of orfX (9, 10, 12). Although the Xpert assay recorded negative results for the MSSA control, S. aureus ATCC 25923, on some occasions the accompanying amplification curves demonstrated evidence of amplification of target DNA (Fig. 1D), suggesting that like the IDI-MRSA assay, the Xpert MRSA kit may have the problem of detecting SCC in MSSA strains such as S. aureus ATCC 25923, which may be why the manufacturer recommends the use of S. epidermidis ATCC 12228 as the negative control (6). This problem requires further investigation.
While the Xpert MRSA assay requires more interpretation than currently suggested by the manufacturer, it was found to perform well with nasal and groin/perineum specimens. It is rapid and easy to use and has the advantage of random access (i.e., an urgent specimen can be accommodated because specimens do not need to be tested in batches). Relative to culture, it is expensive, and results need to be confirmed by culture. A more extensive prospective study is warranted to determine its role in clinical practice in Ireland. A question that such a prospective clinical study might investigate is the duration of kit-positive results from specimens from patients who have been successfully treated for MRSA and whether there is a period during which previously culture-positive patients should not be screened by molecular methods. These data could inform how kit-positive results from patients who are known to have been previously positive for MRSA should be interpreted.
Acknowledgments
We thank the participant patients, the infection control staff (Lisa Fetherstone and Catherine O'Reilly), and the clinical staff at SJH for duplicate specimens. Thanks to Mary Kelleher, surveillance scientist, Microbiology Dept., SJH, for data on diagnostic specimens. We thank Biofact Ireland and Cepheid for providing the GeneXpert machines for the duration of the evaluation and for providing the Xpert MRSA kits at an evaluation price. We thank Teruyo Ito (Juntendo University, Tokyo, Japan), Robert Daum (University of Chicago, Chicago, IL), and Herminia de Lancastre (Instituto de Technologia Quimica e Biologica, Oeiras, Portugal) for the gift of control strains carrying SCCmec IV subtypes, SCCmec V, and SCCmec VI.
There are no conflicting interests to declare.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Bootsma, M. C., O. Diekmann, and M. J. Bonten. 2006. Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing. Proc. Natl. Acad. Sci. USA 1035620-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle-Vavra, S., B. Ereshefsky, C. C. Wang, and R. S. Daum. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 434719-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, D. F., D. I. Edwards, P. M. Hawkey, D. Morrison, G. L. Ridgway, K. J. Towner, and M. W. Wren. 2005. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J. Antimicrob. Chemother. 561000-1018. [DOI] [PubMed] [Google Scholar]
- 4.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 501001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham, R., P. Jenks, J. Northwood, M. Wallis, S. Ferguson, and S. Hunt. 2007. Effect on MRSA transmission of rapid PCR testing of patients admitted to critical care. J. Hosp. Infect. 6524-28. [DOI] [PubMed] [Google Scholar]
- 6.Desjardins, M., C. Guibord, B. Lalonde, B. Toye, and K. Ramotar. 2006. Evaluation of the IDI-MRSA assay for detection of methicillin-resistant Staphylococcus aureus from nasal and rectal specimens pooled in a selective broth. J. Clin. Microbiol. 441219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 421875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kluytmans, J. 2007. Control of meticillin-resistant Staphylococcus aureus (MRSA) and the value of rapid tests. J. Hosp. Infect. 65(Suppl. 2)100-104. [DOI] [PubMed] [Google Scholar]
- 9.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 1843623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mongkolrattanothai, K., S. Boyle, T. V. Murphy, and R. S. Daum. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 481823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy, O. M., S. Murchan, D. Whyte, H. Humphreys, A. Rossney, P. Clarke, R. Cunney, C. Keane, L. E. Fenelon, and D. O'Flanagan. 2005. Impact of the European Antimicrobial Resistance Surveillance System on the development of a national programme to monitor resistance in Staphylococcus aureus and Streptococcus pneumoniae in Ireland, 1999-2003. Eur. J. Clin. Microbiol. Infect. Dis. 24480-483. [DOI] [PubMed] [Google Scholar]
- 12.Noto, M. J., B. N. Kreiswirth, A. B. Monk, and G. L. Archer. 2008. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J. Bacteriol. 1901276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira, D. C., C. Milheirico, and H. de Lencastre. 2006. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 503457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2180-189. [DOI] [PubMed] [Google Scholar]
- 15.Roosendaal, R., J. A. J. W. Kluytmans, J. H. C. Woudenberg, X. Huijsdens, and C. M. J. E. Vandenbroucke-Grauls. 2007. Methicillin-resistant Staphylococcus aureus strains from animal origin are recognized by IDI-MRSA PCR. Clin. Microbiol. Infect. 13S234.
- 16.Rossney, A. S., C. M. Herra, M. M. Fitzgibbon, P. M. Morgan, M. J. Lawrence, and B. O'Connell. 2007. Evaluation of the IDI-MRSA assay on the SmartCycler real-time PCR platform for rapid detection of MRSA from screening specimens. Eur. J. Clin. Microbiol. Infect. Dis. 26459-466. [DOI] [PubMed] [Google Scholar]
- 17.Rossney, A. S., M. J. Lawrence, P. M. Morgan, M. M. Fitzgibbon, A. Shore, D. C. Coleman, C. T. Keane, and B. O'Connell. 2006. Epidemiological typing of MRSA isolates from blood cultures taken in Irish hospitals participating in the European Antimicrobial Resistance Surveillance System (1999-2003). Eur. J. Clin. Microbiol. Infect. Dis. 2579-89. [DOI] [PubMed] [Google Scholar]
- 18.Shore, A., A. S. Rossney, C. T. Keane, M. C. Enright, and D. C. Coleman. 2005. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 492070-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wassenberg, M. W. M., S. Nijssen, X. W. Huijsdens, A. M. C. Bergmans, C. M. J. E. Vandenbroucke-Grauls, M. J. M. Bonten, and J. A. J. W. Kluytmans. 2007. Performance of the GeneXpert system for detection of methicillin-resistant Staphylococcus aureus including the non-typeable clone from animal origin. Clin. Microbiol. Infect. 13S233-S234. [Google Scholar]