Abstract
Dientamoeba fragilis is a parasite that has been recognized to be a causative agent of gastrointestinal symptoms. Because in most studies only some infected persons experience symptoms, it is possible that D. fragilis is a heterogeneous species with variants that display similar morphologies but different pathogenicities. The search for genetic variation in D. fragilis was based on the small-subunit rRNA gene, which was not found to be useful for molecular epidemiology. In this report, we describe the isolation and characterization of additional rRNA gene cluster sequences, the internal transcribed spacer 1 (ITS-1)-5.8S rRNA gene-ITS-2 region. For comparative purposes, we also isolated the ITS-1-5.8S rRNA gene-ITS-2 region of Histomonas meleagridis, a protozoan parasite of birds and a close relative of D. fragilis. This region was found to be highly variable, and 11 different alleles of the ITS-1 sequence could be identified. Variation in the ITS-1 region was found to be intragenomic, with up to four different alleles in a single isolate. So-called C profiles were produced from the ITS-1 repertoire of single isolates,. Analysis of the C profiles of isolates from nonrelated patients identified several clearly distinguishable strains of D. fragilis. Within families, it was shown that members can be infected with the same or different strains of D. fragilis. In conclusion, the ITS-1 region can serve as a molecular epidemiological tool for the subtyping of D. fragilis directly from feces. This may serve as a means of studying the transmission, geographical distribution, and relationships between strains and the pathogenicity of this parasite.
The protozoan Dientamoeba fragilis is found in the mucosal crypts of the large intestines of humans. For a long time, this parasite was ignored by most clinicians; but recently, evidence has been accumulating that D. fragilis is an important enteric pathogen associated with abdominal pain, bloating, diarrhea, and irritable bowel syndrome (4). Despite its global distribution and its apparent causative role in gastrointestinal complaints, very little is known about its pathogenicity, route of transmission, epidemiology, or genetics. This neglect of D. fragilis is partly due to the difficulties associated with the diagnosis of D. fragilis infection. The organism degenerates rapidly in passed stool and lacks a cyst stage, making microscopic detection possible only with freshly passed stool or by the use of fixatives and permanent stains. Recently, PCR-based methods that detect D. fragilis in a clinical sample without the need for prior culturing of the organism have been developed (9, 12, 16). These methods make use of the multicopy, small-subunit (ssu) rRNA gene, the sequence of D. fragilis that has been the best studied to date. The ssu rRNA gene is useful for the sensitive and specific detection of D. fragilis by diagnostic PCR analysis, but due to a lack of variability in both symptomatic and asymptomatic individuals, this target is not suitable for epidemiological studies. Although heterogeneity in the D. fragilis ssu rRNA gene sequence, which results in the differentiation of genotypes 1 and 2, was reported (9), the latter genotype was found in only two cases and not in subsequent studies of larger groups of patients in The Netherlands (9), England (17), and Australia (13). This indicated that the ssu rRNA gene of D. fragilis displays insufficient variability to be used as a molecular epidemiological marker.
In an effort to detect genetic variations in D. fragilis that could be used as molecular epidemiological markers, we isolated and characterized part of the ribosomal gene cluster region containing internal transcribed spacer 1 (ITS-1), the 5.8S rRNA gene (5.8S gene), and ITS-2 of D. fragilis and the closely related avian parasite Histomonas meleagridis. The ITS-1-5.8S gene-ITS-2 region has been extensively used in phylogenetic studies and as a molecular epidemiological marker for other parasites. Concurrently, intragenomic variation in the ITS region of D. fragilis was observed by Windsor et al. (18), who dismissed this target as a marker for strain subtyping, as sequence traces derived from direct amplification would be too complex. However, in the closely related avian parasite Histomonas meleagridis, the complexity of ITS sequencing electropherograms was successfully reduced by a method called “C profiling” (14).
The present paper describes the identification of 11 different variants of the ITS-1 region of D. fragilis, confirming and extending the results obtained by Windsor et al. (18). As observed with Histomonas, analysis of the C nucleotide residues in these complex chromatograms produced chromatograms that were reproducible and easy to interpret. This method could be applied to generate a molecular epidemiological marker of D. fragilis directly from stool specimens that can be used to study the transmission and geographical distribution of strains, the relationships between strains, and the pathogenicity of this parasite.
MATERIALS AND METHODS
Stool specimens and DNA extraction.
Intestinal parasites, including D. fragilis, were detected by the triple feces test (15). Stool specimens that were D. fragilis positive by microscopy were collected from patients presenting with gastrointestinal symptoms in routine clinical practice of the Academic Medical Center of Amsterdam. In addition, stool specimens microscopically positive for D. fragilis from asymptomatic carriers with no gastrointestinal complaints during the previous 2 months were included. Details of the processing of the fecal samples and subsequent DNA isolation were described earlier (9). A limiting dilution of D. fragilis trophozoites was obtained by 10-fold serial dilution of freshly passed stool samples from two patients in phosphate-buffered saline, followed by DNA extraction. D. fragilis genotype 2 DNA was isolated from strain Bi/pa (ATCC 30948). Trichomonas vaginalis was obtained from a vaginal swab specimen, followed by culturing in Trypticase-yeast extract-maltose. Histomonas meleagridis was cultured from the cecum of an infected turkey (14). DNA was isolated from the T. vaginalis and H. meleagridis cultures as described above for the D. fragilis samples.
PCR method.
The design of forward primer ssu2 (5′-GGAATCCCTTGTAAATGCGT-3′) was based on the sequences of the ssu rRNA genes of D. fragilis (GenBank accession number U37461), H. meleagridis (GenBank accession number AF293056), and T. vaginalis (GenBank accession number AY677680). Reverse primer lsu1 (5′-AGTTCAGCGGGTCTTCCTG-3′) was complementary to a conserved region in the 5′ end of the large-subunit (lsu) rRNA genes of Trichomonad species. The ssu2 and lsu1 primer combination was used for PCR amplification of the ITS-1-5.8S gene-ITS-2 region. Reverse primer 5.8s1 (5′-TGTGAGGAGCCAAGACATCC-3′) was complementary to the 5′ end of the 5.8S rRNA gene of D. fragilis. The ssu2 and 5.8s1 primer combination was used for PCR amplification of the ITS-1 region. For each series of samples, a negative control containing no input DNA was added. The PCR mixture (50 μl) contained 20 μl of DNA solution, 100 ng of each primer, 0.5 mM of a deoxynucleoside triphosphate mixture, 5 μl of 10× PCR buffer (Promega), 6 μl of 25 mM MgCl2, 5 μl of bovine serum albumin (5 mg/ml), 5 μl of α-casein (20 mg/ml) to relieve PCR inhibition by fecal substances, and 0.2 U Taq polymerase (Promega). After 3 min of initial denaturation at 94°C, 40 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1 min were performed, followed by a final extension at 72°C for 5 min. The amplification products were size fractionated on 1% agarose gels and visualized by ethidium bromide staining.
The sensitivity of the assay was assessed in several PCR experiments with a range of dilutions (in Tris-EDTA buffer with bacteriophage lambda DNA) of a control pGEMT vector containing a single copy of ITS-1. It could be established how many copies were minimally needed under our assay conditions to still obtain a PCR signal. A minimum of 10 copies (representing one individual parasite containing 11 copies) was needed.
Cloning and sequencing.
One microliter of the PCR product was ligated in the PCR cloning vector pGEMTeasy, as described by the manufacturer (Promega). After transformation into Escherichia coli DH5α, individual clones were tested for the presence of an insert by a colony PCR for 25 cycles with either the primer combination ssu2 and lsu1 (specific for the ITS-1-5.8S gene-ITS-2 region) or the primer combination ssu2 and 5.8s1 (specific for the ITS-1 region). Individual clones with an insert were subjected to sequencing with primer ssu2. For direct sequencing of the ITS-1 PCR products, an Applied Biosystems Prism 310 dye terminator fluorescence-based genetic analyzer was used. Homology searches were done by using the BLAST program with the default settings (available at http://www.ncbi.nlm.nih.gov/blast/). Multiple-sequence alignments, C profiles, and insertion/deletion (indel) analysis were performed by using CodonCode software (CodonCode Corporation, Dedham, MA).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this study for the D. fragilis ITS-1-5.8S gene-ITS-2 region appear in the European Molecular Biology Laboratory (EMBL) GenBank database under accession number DQ167586.
RESULTS
Isolation of ITS-1-5.8S gene-ITS-2 region of D. fragilis and related species.
To obtain sequence information on the ITS-1-5.8S gene-ITS-2 ribosomal gene region of D. fragilis, several PCR primer pairs were designed to amplify the sequences between the 3′ end of the ssu rRNA gene and the 5′ end of the lsu rRNA gene. The forward primers were complementary to the published ssu rRNA sequences of D. fragilis, H. meleagridis, and T. vaginalis (GenBank accession numbers U37461, AF293056, and AY677680 respectively) and contained several nucleotides at their 3′ ends that did not match the ssu rRNA gene sequences of other parasites prevalent in the human gut (data not shown). We therefore designed reverse primers in a conserved region at the utmost 5′ end of the lsu rRNA gene, based on published lsu rRNA gene sequences of trichomonads (data not shown). Only the primer combination ssu2 and lsu1 amplified a product of approximately 500 bp from DNA extracted from human feces containing D. fragilis, cultured T. vaginalis parasites, and cultured H. meleagridis parasites. These PCR products were excised from the gels, cloned, and sequenced. Sequence analysis showed that we indeed successfully cloned the ITS-1-5.8S gene-ITS-2 region of all three species. Both the ITS-1 region and the ITS-2 region of D. fragilis and H. meleagridis were found to be extremely AT rich (∼94%), while the 5.8S rRNA genes had an AT content of approximately 60%. The putative 5′ and 3′ ends of the ITS-1, 5.8S rRNA gene, and ITS-2 were based on homology with 5.8S rRNA genes of other trichomonads (data not shown) and abrupt changes in the AT content of the sequence. The 5.8S rRNA genes of D. fragilis and H. meleagridis appeared to be truncated at the 3′ end and were approximately 45 nucleotides shorter than those of the other trichomonads (data not shown).
The ITS-1 sequence of D. fragilis is variable.
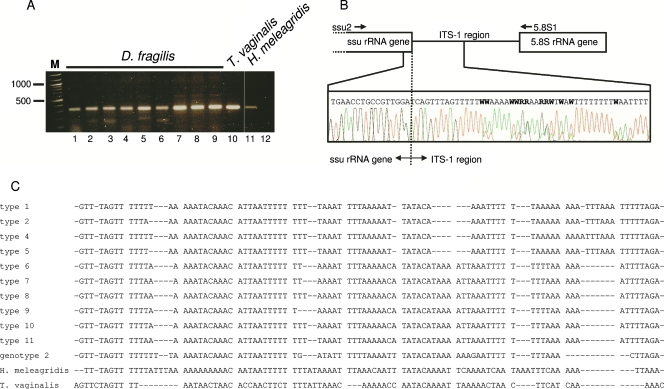
The PCR for amplification of the complete ITS-1-5.8S gene-ITS-2 region appeared to be too inefficient for the study of the genetic variability of D. fragilis parasites directly from human feces. We therefore focused on the ITS-1 region and developed a PCR using ssu rRNA-specific primer ssu2 and 5.8S rRNA gene-specific primer 5.8s1. This PCR was able to efficiently amplify the 3′ end of the ssu rRNA and the complete ITS-1 region of D. fragilis directly from fecal DNA and from cultured T. vaginalis and H. meleagridis parasites (Fig. 1A). Analysis of the sequences of the PCR products from D. fragilis showed a full match with the published ssu rRNA gene sequence and an aberrant ITS-1 sequence, with many double base calls detected in the sequencing chromatograms (Fig. 1B). This suggested that the ITS-1 sequence of D. fragilis is heterogeneous and that several ITS-1 variants were present in a single fecal specimen. An aberrant ITS-1 sequence with many double base calls was also observed in H. meleagridis but not in two clinical isolates of T. vaginalis (data not shown). The PCR products of D. fragilis from fecal samples from eight nonrelated individuals were cloned to further delineate the sequence heterogeneity within the ITS-1 region. Full-length sequencing of individual clones showed that among these eight fecal samples, six samples each contained three ITS-1 alleles, while two and four alleles were detected in one fecal sample each. A total of 11 different ITS-1 alleles could be identified. On the basis of the divergence in the 3′ part of the sequence, it was possible to separate these 11 ITS-1 alleles into two groups: alleles 1 to 5 and alleles 6 to 11 (Fig. 1c). Within these two groups of ITS-1 alleles, sequence variation was limited to single A/T indels. In addition to the sequences of D. fragilis genotype 1, we also analyzed the ITS-1 region of D. fragilis genotype 2 using the same ssu2 and 5.8s1 primer combination. Similar to genotype 1, the ITS-1 region of genotype 2 displayed sequence ambiguity, indicating heterogeneous ITS-1 sequences. The cloned ITS-1 region of D. fragilis genotype 2 also showed a high AT content, but its sequence was different from the ITS-1 sequences of D. fragilis genotype 1 isolates (Fig. 1C, genotype 2).
FIG. 1.
(A) PCR amplification of the ITS-1 region of D. fragilis directly from human feces (lanes 1 to 9) and from cultured T. vaginalis (lane 10) and H. meleagridis (lane 11) parasites. Lane 12, results of PCR without template. The size marker (lane M) is a 100-bp ladder. (B) Schematic representation of the region amplified by primers ssu2 and 5.8s1. The partial sequencing chromatogram of the 3′ end of the ssu RNA gene and the 5′ end of the ITS-1 region of D. fragilis illustrates the ambiguities and double base calls in the ITS-1 sequence. (C) Sequence alignment of the 11 different cloned ITS-1 regions of D. fragilis from fecal samples from eight patients and cloned ITS-1 regions of D. fragilis genotype 2, T. vaginalis, and H. meleagridis. The D. fragilis ITS-1 regions can be divided into two groups (types 1 to 5 and types 6 to 11, respectively) on the basis of the presence of the C residue at position 52 and sequence divergence in the 3′ part of the ITS-1 region.
The variability of the ITS-1 sequence can be used as a molecular epidemiological marker for D. fragilis.
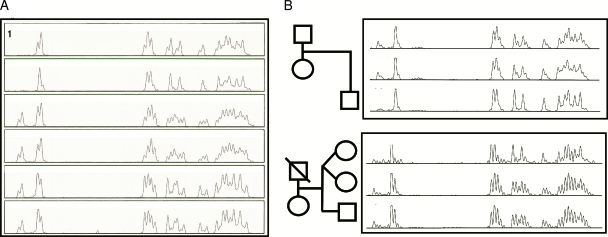
The presence of multiple ITS-1 alleles of D. fragilis in a single human fecal sample resulted in a complex and aberrant chromatogram during sequence analysis (Fig. 1B). A reduction of this complex chromatogram by deleting the peaks for the A, T, and G nucleotides gave a pattern of C nucleotides only that was relatively easy to interpret (Fig. 2). In this so-called C profile, each C residue is represented by multiple adjoining peaks. The multiple peaks for each C residue are caused by A/T nucleotide indels between the individual ITS-1 alleles. The relative heights of the individual peaks of the C profile depend on the relative abundance of the different ITS-1 alleles within a fecal sample. For example, the first two adjacent peaks in the C profiles shown in Fig. 2A correspond to the C residue at position 52 of ITS-1 types 6 to 11 (Fig. 1C). In the C profile of patient samples 1 and 2, these first two peaks were missing. This indicates that in these samples, only ITS-1 sequences missing the C residue at position 52 are present (Fig. 1C, alleles [types] 1 to 5). The reproducibility of the C profile was evaluated by repeating the DNA extraction and sequence analysis of three fecal samples containing D. fragilis. This demonstrated that the C profile was indeed reproducible for a particular sample (data not shown).
FIG. 2.
(A) C profiling of the D. fragilis ITS-1 regions directly from fecal specimens of nonrelated patients. Rows 1, 2, 3, and 5, the four different C profiles identified; row 4, C profile from a patient similar to the C profile in row 3; row 6, C profile from a patient similar to the C profile in row 5. (B) C profiling of the D. fragilis ITS-1 region in families. C profiles from parents asymptomatically infected with D. fragilis and their symptomatically infected son are shown in the top panel. The C profiles of twin sisters and their brother are shown in the bottom panel. All three children were asymptomatically infected with D. fragilis.
Analysis of fecal samples containing D. fragilis from samples from 10 nonrelated patients showed four different C profiles, which were present in one, two, two, and five samples, respectively (Fig. 2A). In addition, we tested fecal samples containing D. fragilis from the members of two families. For the family whose results are shown in the top panel of Fig. 2B, both the father and the mother were infected with D. fragilis but showed no clinical signs. In contrast, their infected son (age, 6 years) did have gastrointestinal complaints. The samples from both parents displayed very similar C profiles, while the sample from their son had a different C profile. Interestingly, the C profile of the sample from the son (C profile 9) was very similar to the C profile of the sample from another patient (Fig. 2A, C profile 2). In the family whose results are shown in the bottom panel of Fig. 2B, the twin sisters (ages, 4 years) and their brother (age, 7 years) were asymptomatically infected with D. fragilis. The twin sisters were infected with different D. fragilis variants (compare C profiles 7 and 8, respectively), while their brother was infected with the same variant as one of his sisters (compare C profiles 8 and 9).
The ITS-1 variation in D. fragilis is intragenomic.
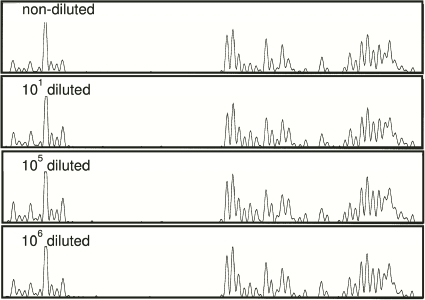
The results presented above show that a fecal sample from an infected individual contains different D. fragilis ITS-1 alleles. This can be explained either by the presence of multiple D. fragilis strains within a single fecal sample or by intragenomic variation of the ITS-1 alleles within a single D. fragilis trophozoite, or by a combination of both possibilities. To discriminate between these possibilities, a limiting dilution of freshly passed stool samples from two patients containing D. fragilis trophozoites was made. If different strains with different genotypes were present with different relative abundances, then the profile would change as the parasite populations were diluted out. DNA was extracted from each dilution step, and a C profile was produced from the nondiluted sample and from the 10-, 105-, and 106-fold dilutions (Fig. 3). In the 106-fold dilution, which corresponded to the detection limit, no significant change in the C profile compared to that for the nondiluted sample could be observed. Similar results were obtained with the sample from the second patient (data not shown). It is highly unlikely that more than one subtype of D. fragilis was present in both stool samples with equal abundances and that at the 106-fold dilution the complete array of ITS-1 types would still be present. The weight of the evidence therefore indicates that ITS-1 sequence variation is of intragenomic origin only.
FIG. 3.
Limiting dilution of a fecal sample containing D. fragilis trophozoites. The C profiles for a nondiluted sample and samples diluted 101, 105, and 106 times are shown. No significant differences between nondiluted and diluted samples were observed.
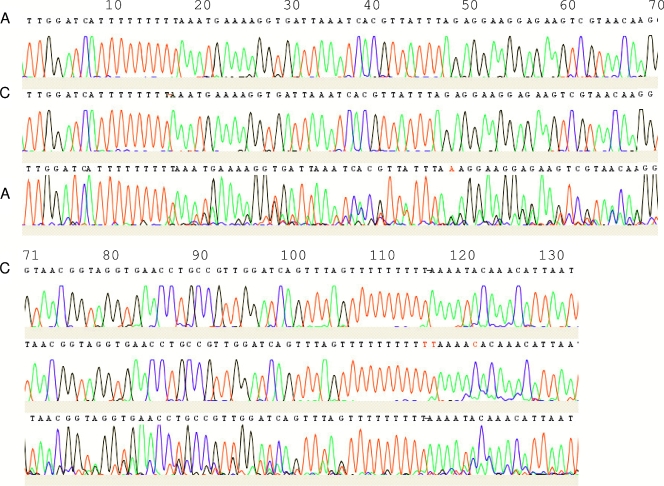
Similarly, variation in the ITS of strain Bi/pa (genotype 2) was observed. Since this culturable strain represents a clonal population, variation in this isolate must be intragenomic. Direct sequencing of a PCR product obtained from this strain yielded a complex chromatogram (Fig. 4), for which the CodonCode program predicted heterozygous indels. The sequencing of individual clones derived from the PCR product itself confirmed that the observed chromatogram was the result of sequence heterogeneity, including indel events in mononucleotide stretches in the sequence (Fig. 4).
FIG. 4.
Sequence chromatograms of two individual clones (clones a and b; top two rows) derived from the PCR product of strain Bi/pa (genotype 2) and the PCR product itself (c; bottom row). CodonCode predicts an indel at position 17, which corresponds to a difference in the stretch of T residues at about position 10 between the two clones (clone a, eight T residues; clone b, nine T residues). From this point onward, each peak is preceded by a smaller identical peak. Variation in the stretch of T residues at about position 110 (clone a, 9 T residues; clone b, 11 T residues) and the T-C transition at position 120 result in even more complexity after position 110.
DISCUSSION
The search for genetic variation in D. fragilis that can be used for molecular epidemiology has been the topic of several recent papers by research groups from the United Kingdom, The Netherlands, and Australia (5, 9, 12, 13, 17). These studies focused on the detection of sequence heterogeneity in the ssu rRNA gene of D. fragilis, which, to date, is the only sequence information available for this organism. Surprisingly, all but 1 of the 154 D. fragilis ssu rRNA sequences obtained from clinical samples showed no heterogeneity, and all sequences were assigned to genotype 1. The genotype 2 ssu rRNA gene was found in only one clinical sample, and its sequence was identical to that of D. fragilis strain Bi/pa (ATCC 30948). From these observations, it was concluded that D. fragilis appears to be a clonal species and that the genotype 1/2 variation could not be used for epidemiological studies. In the current study we isolated the ITS-1-5.8S gene-ITS-2 region of D. fragilis and examined it for genetic variation. For reasons of comparison. the ITS-1-5.8S gene-ITS-2 region of the closely related bird parasite H. meleagridis was also isolated. The ITS regions of the ribosomal gene cluster have no function in the mature ribosome and are thus likely to sustain much more sequence variation than coding regions. ITS regions are among the markers that are the most frequently used to study intra- and interspecific evolution in all kinds of organisms, including human parasites (2, 6, 8). Analysis of the ITS-1-5.8S gene-ITS-2 region of D. fragilis showed that the 5.8S rRNA genes of D. fragilis and H. meleagridis are surprisingly short (111 bp) compared to the lengths of the 5.8S rRNA genes of other trichomonads (∼150 bp). The high degree of homology between the 5.8S rRNA genes of D. fragilis and H. meleagridis further substantiates the observation that these organisms are closely related (4, 11). The ITS-1 and ITS-2 sequences were extremely AT rich and showed extensive variation in D. fragilis, both within and between clinical isolates. Cloning and sequencing of individual ITS-1 regions from D. fragilis revealed 11 different ITS-1 alleles which could be separated into two different groups, confirming and extending the observations of Windsor et al. (18).
Up to four different D. fragilis ITS-1 sequences could be observed in a single fecal specimen. Although this could indicate polyparasitism, in which the host is infected with multiple parasite variants (8), limiting-dilution experiments with intact trophozoites demonstrated that the variation in the ITS-1 region is intragenomic. H. meleagridis also shows this intragenomic heterogeneity of the ITS-1 region (14), while the two clinical isolates of T. vaginalis were devoid of ITS-1 sequence variation. The intragenomic heterogeneity of the ITS regions has already been observed in many different organisms. The extent varies greatly between taxa and is thought to arise by mechanisms such as slipped-strand mispairing events (19).
With the C-profiling technique, we found four different types of D. fragilis parasites among 10 randomly chosen, nonrelated patients. This suggests that the variability that exists in D. fragilis is sufficient for the use of this simple technique for the generation of a molecular epidemiological marker. This was substantiated by the analysis of two families infected with D. fragilis. In the first family, both parents were asymptomatically infected with the same type, while their 6-year-old son had gastrointestinal complaints and was infected with a different, possibly more virulent strain also detected in another symptomatic subject. Twin sisters of the second family were infected with different types, even though these girls spent most of their time together and visited the same day care center. Interestingly, their brother was infected with the same type as one of his sisters. Clearly, more data from large groups of asymptomatic individuals and patients with different symptomatologies are needed to establish a possible link between infection with a particular D. fragilis type and the clinical outcome of the infection. Moreover, more types may exist in different geographical locations.
The specificity of the ITS-1 PCR was not investigated in detail in this paper. However, the 204 bp of the ssu rRNA gene sequence that is part of the ∼300-bp ITS-1 PCR amplicon was always found to be identical to the sequence of the 3′ end of the D. fragilis genotype 1 ssu rRNA. DNA was not amplified from other (gut) protozoa, including Entamoeba dispar, Entamoeba coli, Entamoeba hartmanni, Entamoeba histolytica, Blastocystis hominis, Saccharomyces cerevisiae, and Encephalitozoon hellem, or from smear-negative controls (data not shown).
Our study clearly shows that the intragenomic variation of the ITS-1 region of D. fragilis can be used as a molecular epidemiological marker. Variation in this region could be directly analyzed from human feces by PCR amplification and sequencing without the need for prior culturing of the parasite. Techniques such as restriction fragment polymorphism analysis and single-strand conformation polymorphism analysis have also been successfully used for the detection of ITS sequence variations in human parasites (1, 3, 6, 7, 10). However, due to the extreme AT content and intragenomic variation of the D. fragilis ITS sequences, these techniques did not give satisfying results in our hands (data not shown). By analyzing only the peaks for C nucleotides from a sequencing chromatogram of the ITS-1 region of D. fragilis amplified directly from human feces, a profile that is relatively easy to interpret is obtained. Most research laboratories now have access to fluorescence-based sequence analyzers and should be able to create such C profiles. The ITS-1 C profile can be considered a fingerprint of D. fragilis which is found to be highly reproducible. This technique can now be used to discriminate between persistent infection and reinfection after drug treatment and for molecular epidemiological studies that are aimed at investigating the geographical distribution of D. fragilis strains, their transmission, and, most importantly, the possible correlation between various parasite strains and different degrees of virulence.
Acknowledgments
We thank Ron Peek for initiating this study and Youssef Hsaini for expert technical assistance.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Chalmers, R. M., C. Ferguson, S. Caccio, et al. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35397-410. [DOI] [PubMed] [Google Scholar]
- 2.Chilton, N. B., and R. B. Gasser. 1999. Sequence differences in the internal transcribed spacers of DNA among four species of hookworm (Ancylostomatoidea: Ancylostoma). Int. J. Parasitol. 291971-1977. [DOI] [PubMed] [Google Scholar]
- 3.Cupolillo, E., L. R. Brahim, C. B. Toaldo, et al. 2003. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J. Clin. Microbiol. 413126-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson, E. H., J. J. Windsor, and C. G. Clark. 2004. Emerging from obscurity: biological, clinical, and diagnostic aspects of Dientamoeba fragilis. Clin. Microbiol. Rev. 17553-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson, J. A., and C. G. Clark. 2000. Cryptic genetic diversity in Dientamoeba fragilis. J. Clin. Microbiol. 384653-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar, M., and P. K. Shukla. 2005. Use of PCR targeting of internal transcribed spacer regions and single-stranded conformation polymorphism analysis of sequence variation in different regions of rRNA genes in fungi for rapid diagnosis of mycotic keratitis. J. Clin. Microbiol. 43662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nejsum, P., E. D. Parker, Jr., J. Frydenberg, et al. 2005. Ascariasis is a zoonosis in Denmark. J. Clin. Microbiol. 431142-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivier, C., S. van de Pas, P. W. Lepp, et al. 2001. Sequence variability in the first internal transcribed spacer region within and among Cyclospora species is consistent with polyparasitism. Int. J. Parasitol. 311475-1487. [DOI] [PubMed] [Google Scholar]
- 9.Peek, R., F. R. Reedeker, and T. van Gool. 2004. Direct amplification and genotyping of Dientamoeba fragilis from human stool specimens. J. Clin. Microbiol. 42631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schonian, G., L. Schnur, M. el Fari, et al. 2001. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans. R. Soc. Trop. Med. Hyg. 95217-224. [DOI] [PubMed] [Google Scholar]
- 11.Silberman, J. D., C. G. Clark, and M. L. Sogin. 1996. Dientamoeba fragilis shares a recent common evolutionary history with the trichomonads. Mol. Biochem. Parasitol. 76311-314. [DOI] [PubMed] [Google Scholar]
- 12.Stark, D., N. Beebe, D. Marriott, et al. 2005. Detection of Dientamoeba fragilis in fresh stool specimens using PCR. Int. J. Parasitol. 3557-62. [DOI] [PubMed] [Google Scholar]
- 13.Stark, D., N. Beebe, D. Marriott, et al. 2005. Prospective study of the prevalence, genotyping, and clinical relevance of Dientamoeba fragilis infections in an Australian population. J. Clin. Microbiol. 432718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Heijden, H. M., W. J. Landman, S. Greve, et al. 2006. Genotyping of Histomonas meleagridis isolates based on internal transcribed spacer-1 sequences. Avian Pathol. 35330-334. [DOI] [PubMed] [Google Scholar]
- 15.van Gool, T., R. Weijts, E. Lommerse, et al. 2003. Triple faeces test: an effective tool for detection of intestinal parasites in routine clinical practice. Eur. J. Clin. Microbiol. Infect. Dis. 22284-290. [DOI] [PubMed] [Google Scholar]
- 16.Verweij, J. J., B. Mulder, B. Poell, et al. 2007. Real-time PCR for the detection of Dientamoeba fragilis in fecal samples. Mol. Cell. Probes 21400-404. [DOI] [PubMed] [Google Scholar]
- 17.Windsor, J. J., C. G. Clark, and L. Macfarlane. 2004. Molecular typing of Dientamoeba fragilis. Br. J. Biomed. Sci. 61153-155. [DOI] [PubMed] [Google Scholar]
- 18.Windsor, J. J., L. Macfarlane, and C. G. Clark. 2006. Internal transcribed spacer dimorphism and diversity in Dientamoeba fragilis. J. Eukaryot. Microbiol. 53188-192. [DOI] [PubMed] [Google Scholar]
- 19.Worheide, G., S. A. Nichols, and J. Goldberg. 2004. Intragenomic variation of the rDNA internal transcribed spacers in sponges (phylum Porifera): implications for phylogenetic studies. Mol. Phylogenet. Evol. 33816-830. [DOI] [PubMed] [Google Scholar]