Epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) and EMRSA-16 are pandemic MRSA strains which have received global attention due to their worldwide spread and increased prevalence. Both are primarily hospital-associated pathogens responsible for a myriad of diseases, ranging from uncomplicated skin infections to life-threatening bacteremia and pneumonia.
EMRSA-15 and EMRSA-16 were first identified in the United Kingdom during the early 1990s and, within a few years, became the predominant health care-associated clones in that country (11, 12, 14). One or both strains have since been detected in the health care settings of many countries, including Denmark (4, 13), Sweden (3, 18), Belgium (13), Spain (10, 15), and Kuwait (20). However, despite the global distribution of these strains, only EMRSA-16 has been found within the United States (9). In this report, we describe the isolation and characterization of an EMRSA-15 variant in the central United States.
A 10-month-old male was admitted to a local children's hospital after developing a facial rash consisting of 2- to 3-mm-size pustular lesions with some shallow ulceration accompanied by fever. MRSA strain CRG1250 was cultured from the rash. As part of a hospital surveillance study, pulsed-field gel electrophoresis (PFGE) (1) was performed and indicated that CRG1250 was related to the classic EMRSA-15 type strain (Fig. 1) with a four-band difference. Although not available for direct comparison, both the EMRSA-15 type D1 strain from Kuwait (20) and EMRSA-15 variant B3 (14) appeared to be highly related to CRG1250 with estimated one- and two-band differences, respectively.
FIG. 1.
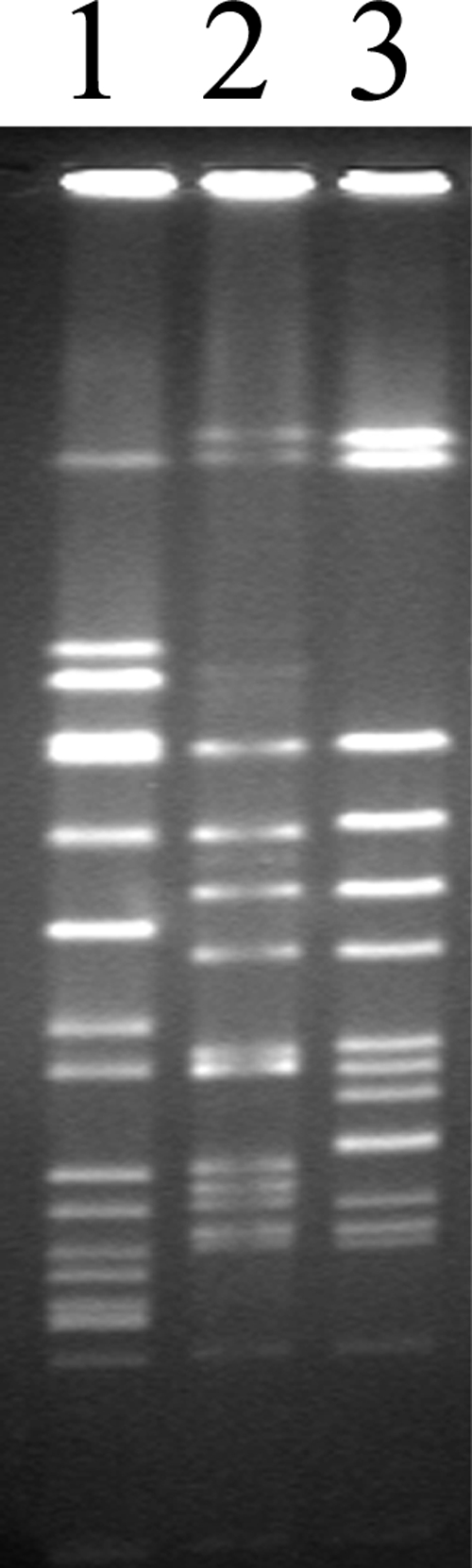
PFGE analysis of SmaI-digested chromosomal DNA from S. aureus isolates NCTC8325 (size standard; lane 1), classical EMRSA-15 (lane 2), and CRG1250 (lane 3). The resulting PFGE pattern was compared with known EMRSA strain types as described previously (19) using BioNumerics version 4.6 (Applied Maths, Sint-Martens-Latem, Belgium) with unweighted-pair group arithmetic averages and the Dice coefficient.
The molecular characterization of EMRSA-15 isolates from various geographical locations has demonstrated that these isolates are multilocus sequence type (MLST) 22 (ST22), accessory gene regulator (agr) type 1, and staphylococcal protein A (spa) type t022, t032, or t223 (3, 5, 16) and harbor the SCCmec type IV element (3). Phenotypically, EMRSA-15 isolates do not typically produce urease (17) and are commonly resistant to erythromycin and/or ciprofloxacin in addition to methicillin (14). Genotypic analysis (3, 6) indicated that CRG1250 was a single-locus variant of ST22 (a 1-bp change from glp-1 to glp-90), agr type 1, spa type t223, and SCCmec type IV. CRG1250 did not produce urease as determined by BBL urea agar slants (BD Diagnostic Systems, Sparks, MD) and was resistant to oxacillin, erythromycin, and ciprofloxacin by disk diffusion (2), all consistent with an EMRSA-15 designation.
Most EMRSA-15 strains produce enterotoxin C (SEC) but lack Panton-Valentine leukocidin (PVL) and toxic shock syndrome toxin 1 (TSST-1) (11, 17). However, some EMRSA-15 variants lack SEC or have acquired PVL (14, 20, 22). When screened for toxin-encoding genes by PCR (1, 21), CRG1250 was SEC negative in contrast to classical EMRSA-15 strains but similar to EMRSA-15 variants, such as strains B3 and Kuwait 2968 (14, 20). While classical EMRSA-15 strains and CRG1250 were PCR negative for PVL, CRG1250 additionally harbored the tst gene encoding TSST-1.
Taken together, the genotypic and phenotypic characteristics of CRG1250 strongly suggest that this strain is a SEC-negative, tst-positive variant of EMRSA-15. Interestingly, studies, thus far, have failed to identify tst in classical or variant EMRSA-15 strains (5, 14). However, the loss of SEC and acquisition of TSST-1 are not altogether surprising since the genes encoding these toxins reside on potentially mobile genetic elements (7, 8).
To our knowledge, this report represents the first observed incidence of EMRSA-15 in the United States. The patient and his immediate family reside in the central United States and had not recently traveled internationally. Thus, this isolate most likely does not represent a direct introduction of EMRSA-15 into the United States but rather a strain currently present in relatively small numbers, since it was the only such isolate we have encountered, which may or may not increase in incidence.
Footnotes
Published ahead of print on 30 July 2008.
REFERENCES
- 1.Bonnstetter, K. K., D. J. Wolter, F. C. Tenover, L. K. McDougal, and R. V. Goering. 2007. Rapid multiplex PCR assay for identification of USA300 community-associated methicillin-resistant Staphylococcus aureus isolates. J. Clin. Microbiol. 45141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 997687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 431836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghebremedhin, B., W. Konig, W. Witte, K. J. Hardy, P. M. Hawkey, and B. Konig. 2007. Subtyping of ST22-MRSA-IV (Barnim epidemic MRSA strain) at a university clinic in Germany from 2002 to 2005. J. Med. Microbiol. 56365-375. [DOI] [PubMed] [Google Scholar]
- 6.Larsen, A. R., S. Bocher, M. Stegger, R. Goering, L. V. Pallesen, and R. Skov. 2008. Epidemiology of European community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV strains isolated in Denmark from 1993 to 2004. J. Clin. Microbiol. 4662-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29527-543. [DOI] [PubMed] [Google Scholar]
- 8.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 5577-104. [DOI] [PubMed] [Google Scholar]
- 9.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montesinos, I., T. Delgado, D. Riverol, E. Salido, M. A. Miguel, A. Jimenez, and A. Sierra. 2006. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus associated with the emergence of EMRSA-16 at a university hospital. J. Hosp. Infect. 64257-263. [DOI] [PubMed] [Google Scholar]
- 11.Moore, P. C., and J. A. Lindsay. 2002. Molecular characterisation of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 51516-521. [DOI] [PubMed] [Google Scholar]
- 12.Murchan, S., H. M. Aucken, G. L. O'Neill, M. Ganner, and B. D. Cookson. 2004. Emergence, spread, and characterization of phage variants of epidemic methicillin-resistant Staphylococcus aureus 16 in England and Wales. J. Clin. Microbiol. 425154-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 411574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Neill, G. L., S. Murchan, A. Gil-Setas, and H. M. Aucken. 2001. Identification and characterization of phage variants of a strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-15). J. Clin. Microbiol. 391540-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Roth, E., F. Lorenzo-Diaz, N. Batista, A. Moreno, and S. Mendez-Alvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 424649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersdorf, S., K. Oberdorfer, and C. Wendt. 2006. Longitudinal study of the molecular epidemiology of methicillin-resistant Staphylococcus aureus at a university hospital. J. Clin. Microbiol. 444297-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson, J. F., and S. Reith. 1993. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 2545-52. [DOI] [PubMed] [Google Scholar]
- 18.Seeberg, S., L. Larsson, C. Welinder-Olsson, T. Sandberg, E. Skyman, B. Bresky, A. Lindqvist, and M. van Raalte. 2002. How an outbreak of MRSA in Gothenburg was eliminated: by strict hygienic routines and massive control-culture program. Lakartidningen 993198-3204. (In Swedish.) [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Udo, E. E., N. Al-Sweih, and B. Noronha. 2006. Characterisation of non-multiresistant methicillin-resistant Staphylococcus aureus (including EMRSA-15) in Kuwait hospitals. Clin. Microbiol. Infect. 12262-269. [DOI] [PubMed] [Google Scholar]
- 21.van der Mee-Marquet, N., G. Lina, R. Quentin, H. Yaouanc-Lapalle, C. Fièvre, N. Takahashi, and J. Etienne. 2003. Staphylococcal exanthematous disease in a newborn due to a virulent methicillin-resistant Staphylococcus aureus strain containing the TSST-1 gene in Europe: an alert for neonatologists. J. Clin. Microbiol. 414883-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wannet, W. J., M. E. Heck, G. N. Pluister, E. Spalburg, M. G. van Santen, X. W. Huijsdans, E. Tiemersma, and A. J. de Neeling. 2004. Panton-Valentine leukocidin positive MRSA in 2003: the Dutch situation. Euro Surveill. 928-29. [PubMed] [Google Scholar]


