Abstract
Coagulase-negative staphylococci (CoNS) are the main cause of catheter-related infections, especially among immunosuppressed and neutropenic patients, as well as a source of bacterial contamination in blood cultures. Using biochemical identification and pulsed-field gel electrophoresis (PFGE), we sought to identify possible clonal isolates of bacteremia in patients with central lines in an oncology ward (OW), with comparison to isolates that were recovered by venipuncture from an adult emergency room (ER). A total of 243 CoNS isolates were identified to species level from the OW (126) and ER (117), with Staphylococcus epidermidis isolates being the most common (OW, 79.4%; ER, 45.3%). PFGE demonstrated a predominant clone of S. epidermidis (major subtype A) which was 35.5 times more likely (odds ratio [OR] = 35.5; 95% confidence interval [CI] = 4.7 to 267.0; P < 0.00001) to be present in the OW versus the ER. These (CoNS or major subtype A) isolates were more frequently resistant to gentamicin (OR = 2.83; 95% CI = 1.23 to 6.53; P = 0.016) and less frequently resistant to trimethoprim-sulfamethoxazole (OR = 0.38; 95% CI = 0.18 to 0.80; P = 0.013). Subset analysis of S. epidermidis isolates 2 years after the study period showed the persistence of the clone of major subtype A within the OW. This study demonstrates the presence of a predominant clone among central line isolates from an OW that is not present in CoNS venipuncture isolates from an ER.
Coagulase-negative staphylococci (CoNS) are the main cause of catheter-related infections, especially among immunosuppressed and neutropenic patients (6, 43, 48, 50). Moreover, studies have used biochemical identification and genotypic analysis to illustrate clonal infections that occur in specific units. (2, 5, 6, 12, 17-19, 23, 24, 28-30, 33-35, 38, 43, 44, 48, 49, 51) Other studies have shown differences in the production of possible virulence factors such as lipase, esterase, elastase, and biofilm (polysaccharide intercellular adhesin [ica operon]) among strains seen in patients with catheter-related infections versus a nonseptic control groups (7, 11, 13, 29, 32, 43, 53). Production of biofilm may allow these isolates to preferentially colonize the catheter, catheter hub, and other indwelling medical devices while escaping the immune system and antibiotics This may lead to an increased frequency of bacteremia, false-positive blood cultures, and catheter-related septicemia due to hub colonization (7, 9, 13, 15, 21, 32, 52, 53). Still, other studies suggest that gut translocation with its resultant bacteremia leads to a significant number of cases of CoNS septicemia, particularly in cancer patients (8, 9, 17, 33).
Blood culture contamination by CoNS is also a significant cause of positive blood cultures, leading to a problem in the determination of true bacteremia (9, 34, 36, 37, 52). As a result, there is a marked increased in the financial burden, with an estimated cost of $5,000 to $10,000 per false-positive blood culture due to more extensive laboratory testing, pharmacy costs, radiological procedures (echocardiograms), and unnecessary increased lengths of stay (4, 36, 37). Blood culture contamination rates are considerably increased due to hub colonization if the blood is withdrawn from central venous lines as opposed to by venipuncture. (3, 16, 26, 40-42, 52) The purpose of the current retrospective case-control study was four fold. First, we desired to determine, over a period of 1 year, the rate of CoNS blood contamination and infection in a group of patients from a unit (adult oncology) with a high frequency of central lines and thus high contamination and infection rates. Second, we sought to identify to species level the study group CoNS isolates (those from central lines in the oncology ward [OW]) and isolates from a control group (samples obtained from peripheral venipuncture from the adult emergency room [ER]) and compare the colonization/infection rates between the two groups of patients by examination of the medical record and utilizing a clinical algorithm for classification of infection versus contamination (5). Third, genotyping (by pulsed-field gel electrophoresis [PFGE]) of selected isolates from each group was performed to determine if any clonal patterns existed within the study group and/or control group. Fourth, we wanted to determine if any of the possible clonal groups were associated with clinical findings indicative of infection, as proposed in a previously published algorithm (5).
MATERIALS AND METHODS
Institutional review board permission was obtained for all aspects of this study.
Specimen selection.
The laboratory computer database was analyzed for CoNS blood isolates from both the adult OW and adult ER for a period of 1 year. Initial evaluation of the site of infection among the OW isolates demonstrated that most of them were drawn from central lines. In fact, most of the OW patients had central lines for chemotherapy, etc. (including a small number of patients who had a CoNS-positive blood culture that was designated as being from “peripheral”). Not all of these “peripheral” isolates were Staphylococcus epidermidis (which could be used in the epidemiologic comparison). After discussion with clinical staff, it was further determined that “peripheral” usually meant “peripheral line” and not “peripheral venipuncture.” Patients with OW venipuncture isolates usually also had a central line with the same sort of inherent risks for exposure to the clone (nurse to patient). Because of these factors, no significant number of true venipuncture isolates from the OW could be used for adequate epidemiological comparison with central line isolates from the OW.
A total of 243 isolates were evaluated. One hundred twenty-six were from the OW, and 117 were from the ER. Culture result records were obtained, and data were extracted for the study. Medical charts were examined, and relevant clinical information was obtained, such as hematology indices, immunosuppressive treatments and/or conditions, clinical diagnosis, vital signs at the time of culture, and the presence of any other focal signs/symptoms of infection. Specimens from the OW were included in the study if they were identified as being drawn from a central venous line. For the control group, isolates were included in the study if they were obtained from a peripheral venipuncture in the ER. In the control group, isolates were excluded from the study if they were from patients who had some form of central venous line or another type of implanted or indwelling device.
All S. epidermidis isolates (153) were examined further for clonality using PFGE, as this was the most frequent species isolated. For the analysis of the association between the largest clone of S. epidermidis (PFGE subtype A) with central line culture in the OW compared to peripheral venipuncture in the ER, duplicate clonal isolates were excluded (random exclusion of duplicate clonal isolates from the same patient). Non-S. epidermidis CoNS were also added to the analysis to more accurately represent the strength of the association of major subtype A compared to all CoNS.
Isolate identification.
All CoNS isolates utilized in this study were obtained from frozen stocks at −70°C and subcultured. These isolates were identified by the API Staph (bioMerieux, Inc., Durham, NC) commercial biochemical identification system according to the manufacturer's instructions.
PFGE.
PFGE was performed on all isolates of S. epidermidis from both the OW and the ER as described previously (46). Briefly, chromosomal SmaI digests were prepared by incubating agarose plugs containing lysed, pure organism overnight at room temperature with a buffer containing the SmaI restriction enzyme. Plugs were loaded on an agarose gel and run for 18 h overnight equilibrated at 14°C, with a constant voltage of 6 V/cm and pulses ramped from 5.3 s to 34.9 s at an angle of 120 degrees. Gels were stained in 0.1% ethidium bromide and photographed. Along with the specimens, a DNA size ladder as well as the same Staphylococcus positive control were run on each gel. Each image was captured with the exact same camera and camera settings. The gel lanes were cut and pasted next to one another for comparison, using image software without modifying the image dimensions. These methods allowed definitive, consistent comparison between gels. The banding patterns of all isolates evaluated with PFGE were compared by visual inspection, and the isolates were grouped based on number of band differences from the dominant primary clone identified in the S. epidermidis isolates from the OW and the ER as suggested by Tenover et al. (47).
Evaluation of infection status.
To evaluate associations between particular PFGE subtypes and central line use, we compared these types utilizing a standard for true infection that has been published previously (5), with a slight modification. The algorithm for deciding if an isolate from a patient was due to true infection is as follows.
) Those isolates that were from patients who had ≥1 additional CoNS-positive blood culture in a 5-day period were considered consistent with infection as the source.
) Those isolates that were from patients who had no additional positive blood cultures in a 5-day period from the index blood culture were considered consistent with infection as the source if both of the following conditions applied: (a) a white blood cell [WBC] count of <2,000 cells/μl or >14,000 cells/μl and (b) either a temperature of <36°C or >38.5°C or a systolic blood pressure of <90 mm Hg.
) Those isolates that did not meet either criterion 1 or 2 were considered contaminants.
Data analysis.
All data were entered into a database, and all statistical analyses were performed with SPSS version 10.0.1 for Windows (SPSS Inc., Chicago, IL) and GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego CA). P values were calculated by the two-tailed Fisher exact test unless otherwise specified. The independent-sample t test was utilized to evaluate the statistical significance of the comparisons of means. Differences between means were considered statistically significant if the P value was <0.05.
RESULTS
Two-hundred forty-three isolates were included in the study (117 from the ER and 126 from the OW). Only one isolate out of all of those tested was repeatedly identified by the API Staph ID system as Staphylococcus aureus. This isolate was also repeatedly Staphaurex negative and repeatedly tube coagulase negative at 2 h, 4 h, and 18 h. Table 1 shows the distribution of coagulase-negative organisms identified using the API Staph commercial system. The OW had a more narrow spectrum of species (total of 6) than the ER (total of 14). As expected, S. epidermidis was the predominant species out of all isolates identified (66.4% of 277 isolates). S. epidermidis was almost twice as frequent among central line isolates from the OW than among those venipuncture isolates identified from the ER (81.9% versus 45.3%; P < 0.00001). Staphylococcus haemolyticus was three times more frequent among isolates from the OW than among those from the ER (7.9% versus 2.6%), but this was not statistically significant (P = 0.087). Two isolates from each ward (four total) were identified as a Micrococcus sp.
TABLE 1.
Identification of CoNS isolates recovered from ER venipunctures and OW central lines
| Species | No. (%) of isolates
|
||
|---|---|---|---|
| OW | ER | Total | |
| Unidentifiable | 5 (4.0) | 13 (11.1) | 18 (7.4) |
| S. epidermidis | 100 (79.4) | 53 (45.3) | 153 (63.0) |
| S. haemolyticus | 10 (7.9) | 3 (2.6) | 13 (5.3) |
| S. saprophyticus | 1 (0.8) | 3 (2.6) | 4 (1.6) |
| S. hominis | 3 (2.4) | 13 (11.1) | 16 (6.6) |
| S. lugdunensis | 5 (4.0) | 8 (6.8) | 13 (5.3) |
| S. xylosus | 0 (0.0) | 3 (2.6) | 3 (1.2) |
| S. capitis | 0 (0.0) | 10 (8.5) | 10 (4.1) |
| S. cohnii | 0 (0.0) | 2 (1.7) | 2 (0.8) |
| S. lentus | 0 (0.0) | 1 (0.9) | 1 (0.4) |
| S. simulans | 0 (0.0) | 1 (0.9) | 1 (0.4) |
| S. warneri | 0 (0.0) | 3 (2.6) | 3 (1.2) |
| S. chromogenes | 0 (0.0) | 1 (0.9) | 1 (0.4) |
| S. aureus | 0 (0.0) | 1 (0.9) | 1 (0.4) |
| Micrococcus sp. | 2 (1.6) | 2 (1.7) | 4 (1.6) |
| Total | 126 (100.0) | 117 (100.0) | 243 (100.0) |
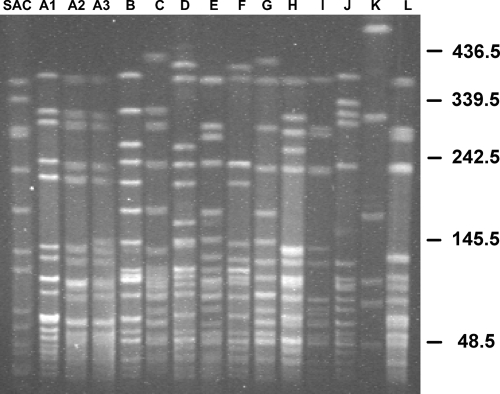
Figure 1 shows the PFGE band patterns for each of the major and minor subtypes observed among OW and ER S. epidermidis blood culture isolates. The predominant major subtype (A) had three minor subtypes (A1, A2, and A3), which corresponded to zero to three, four, and five to seven band differences, respectively, from the predominant A1 minor subtype band pattern seen in lane 2. Table 2 defines the distribution of the S. epidermidis subtypes exhibited in Fig. 1 between the OW and ER and contains all clonal isolates (including duplicates from the same patient). Eight different subtypes were seen in both the OW (A1, A2, A3, B, C, D, E, and F) and the ER (A1, B, G, H, I, J, K, and L). The two locations had in common two subtypes (A1 and B). Only 40% of OW S. epidermidis isolates were nonclonal, compared to 52.8% of ER S. epidermidis isolates. The most common S. epidermidis subtype was A1, at 21.6% of all isolates, and it comprised 32.0% and 1.9% of the OW and ER S. epidermidis isolates, respectively. The second most common major subtype (B) was present in equal proportions in the OW and ER. Six isolates in the ER exhibited a G major subtype, which was not present in the OW isolates.
FIG. 1.
S. epidermidis PFGE gel demonstrating all major and minor subtypes present among OW and ER blood culture isolates. SAC, Staphylococcus aureus control. DNA fragment size is measured in kilobases on the right. The A1 minor subtype showed a one- to three-band difference compared to the predominant A major subtype pattern. A2 showed a four-band difference from the predominant A major subtype pattern. A2 showed a five- to seven-band difference from the predominant A major subtype pattern.
TABLE 2.
S. epidermidis PFGE type distribution of the adult OW central line isolates and the adult ER isolates
| PFGE major (minor) subtypea | No. (%) of isolatesb
|
||
|---|---|---|---|
| OW | ER | Total | |
| A (1) | 32 (32.0) | 1 (1.9) | 33 (21.6) |
| A (2) | 4 (4.0) | 0 (0.0) | 4 (2.6) |
| A (3) | 6 (6.0) | 0 (0.0) | 6 (3.9) |
| B | 7 (7.0) | 4 (7.5) | 11 (7.2) |
| C | 2 (2.0) | 0 (0.0) | 2 (1.3) |
| D | 3 (3.0) | 0 (0.0) | 3 (2.0) |
| E | 3 (3.0) | 0 (0.0) | 3 (2.0) |
| F | 3 (3.0) | 0 (0.0) | 3 (2.0) |
| G | 0 (0.0) | 6 (11.3) | 6 (3.9) |
| H | 0 (0.0) | 3 (5.7) | 3 (2.0) |
| I | 0 (0.0) | 2 (3.8) | 2 (1.3) |
| J | 0 (0.0) | 4 (7.5) | 4 (2.6) |
| K | 0 (0.0) | 3 (5.7) | 3 (2.0) |
| L | 0 (0.0) | 2 (3.8) | 2 (1.3) |
| Nonclonal | 40 (40.0) | 28 (52.8) | 68 (44.4) |
| Total | 100 (100.0) | 53 (100.0) | 153 (100.0) |
The A1 subtype showed a one- to three-band difference from the predominant A major subtype pattern. A2 showed a four-band difference from the predominant A major subtype pattern. A2 showed a five- to seven-band difference from the predominant A major subtype pattern.
Includes duplicate clones from the same patient isolated on different days.
The association of S. epidermidis PFGE major subtype A with central line culture from the OW compared to peripheral venipunctures from the ER is demonstrated in Table 3. Duplicate clones from the same patient were excluded from the analysis to ensure that there was no artificial inflation of the strength of the association of the major subtype A and central line use among OW patients. Non-A includes those PFGE subtypes that were other than type A as well as those isolates that were Staphylococcus spp. other than S. epidermidis. Thus, all non-A samples were not clonally related to major subtype A. The total number of isolates included in the analysis was 221. This included 131 isolates from the data in Table 2 (153 minus 22 duplicates) and 90 non-S. epidermidis CoNS isolates.
TABLE 3.
Association of S. epidermidis PFGE major subtype A with central line culture from the OW compared to peripheral venipunctures from the ER
| Subtypea | No. (%) of isolatesb
|
OR (95% CI); P | ||
|---|---|---|---|---|
| OW | ER | Total | ||
| Non-A | 82 (75.9) | 112 (99.1) | 194 (87.8) | |
| A | 26 (24.1) | 1 (0.9) | 27 (12.2) | 35.5 (4.7-267.0); <0.00001 |
| Total | 108 (100.0) | 113 (100.0) | 221 (100.0) | |
Duplicate clones from the same patient have been excluded from the analysis.
Non-A includes those PFGE subtypes that were other than major type A as well as those isolates that were Staphylococcus spp. other than S. epidermidis. Thus, all non-A samples were not clonally related to major subtype A.
A total of 24.1% of OW isolates were the major subtype A versus non-subtype A, compared to 0.9% of ER isolates (odds ratio [OR] for major subtype A detected in OW = 35.5; 95% confidence interval [CI] = 4.7 to 267.0; P < 0.00001). Of the major subtype A, minor subtypes A1 and A3 had statistically significant associations with isolation from the OW central lines. Subtype A1 had an OR of 22.4 (95% CI = 2.9 to 171.0; P = 0.000014) for isolation from the OW central lines as opposed to the ER venipunctures. Subtype A3 was seen in 4.6% (95% CI = 2.0 to 10.4) of OW isolates, compared to 0% of ER isolates (P = 0.027 for difference). Among all other subtypes, no statistically significant association was found (P > 0.115) for those patients who did or did not have a central line.
As previously discussed, the same S. epidermidis subtype was seen in an individual patient on multiple blood cultures (n = 46). Patient 15 had seven blood culture isolates with the same exact minor subtype of A1, which was the most frequent occurrence among all patients. These seven isolates were obtained on four different days, three of which were 2 to 4 months apart. Thirty-nine of the total of 46 repeat positives (84.8%) were isolated from the OW. Out of all repeat positives, the major subtype A showed the largest number of repeat positives (24/46; 52.2%) Within the major subtype A, minor subtype A1 was the most frequent at 20 (43.5%), followed by minor subtypes A2 (4.3%) and A3 (4.3%). Another group of patients had repeat positive subtypes, and each of these subtypes was unique to one of these patients (not clonally related to more than one patient and thus not defined by a letter designation). This was the second largest group of repeat positive cultures from the same patient, with a total of 11 of 46 (23.9%) repeat positives.
Using a previously published algorithm for diagnosis of a true clinical infection, S. epidermidis major subtype A was evaluated for the possibility that it might be responsible for true infection as opposed to colonization and transient bacteremia among isolates from both the OW and ER (5). In the combined OW and ER patients with clonal isolates, there was no association between major subtype A and infection as defined by the algorithm (OR = 1.41; 95% CI = 0.58 to 3.42; P = 0.504; n = 85). Interestingly, major subtype G showed a statistically significant negative association with infection versus lack of infection (OR = 0.10; 95% CI = 0.01 to 0.88; P = 0.023) as defined by the algorithm. There were no other statistically significant associations between infection as assessed by the algorithm and the particular subtypes.
Table 4 shows the values among major subtype A (or a subtype other than A) for different laboratory and physical exam findings that are indicative of infection. Mean systolic blood pressure, mean diastolic blood pressure, and total WBC count were all lower in patients from whom major subtype A was isolated, compared to those with subtypes (P = 0.013, P = 0.022, and P = 0.002, respectively). Temperature was higher (P = 0.001) and there was a trend for higher respiratory rates (P = 0.072) among patients from whom major subtype A was isolated, compared to other subtypes.
TABLE 4.
Univariate analysis of values for clinical indicators of infection among the different subtypes
| Clinical parameter (unit) | Subtype | n | Mean ± SEM | P value for differencea |
|---|---|---|---|---|
| Temp (°C) | A | 50 | 38.5 ± 0.1 | 0.001 |
| Non-A | 43 | 37.7 ± 0.2 | ||
| Systolic blood pressure (mm Hg) | A | 50 | 122.6 ± 2.8 | 0.013 |
| Non-A | 43 | 134.2 ± 3.5 | ||
| Diastolic blood pressure (mm Hg) | A | 50 | 69.0 ± 2.3 | 0.022 |
| Non-A | 43 | 76.0 ± 2.0 | ||
| Heart rate (beats/min) | A | 50 | 104.0 ± 2.9 | 0.460 |
| Non-A | 43 | 101.1 ± 2.6 | ||
| Respiratory rate (breaths/min) | A | 50 | 21.5 ± 0.8 | 0.072 |
| Non-A | 43 | 19.7 ± 0.5 | ||
| Total WBC (cells/μl) | A | 50 | 2,791 ± 613 | 0.002 |
| Non-A | 43 | 6,858 ± 1,088 | ||
| Absolute neutrophil count (cells/μl) | A | 41 | 3,098 ± 915 | 0.220 |
| Non-A | 24 | 4,910 ± 982 |
By t test for independent samples.
Antibiotic susceptibility data for the central line isolates were available for vancomycin, penicillin, levofloxacin, clindamycin, minocycline, cefazolin, rifampin, methicillin, erythromycin, amoxicillin/clavulinate, gentamicin, and trimethoprim-sulfamethoxazole. Central line isolates that were major subtype A were more likely to be resistant versus sensitive/intermediate to gentamicin than central line isolates that were not major subtype A (OR = 2.83; 95% CI = 1.23 to 6.53; P = 0.016). These same central line isolates (major subtype A) were less likely to be resistant to trimethoprim-sulfamethoxazole (OR = 0.38; 95% CI = 0.18 to 0.80; P = 0.013). All central line isolates were sensitive to vancomycin and resistant to penicillin and minocycline. No other association of antibiotic susceptibility was observed when comparing central line isolates of major subtype A versus non-major subtype A. Antibiotic resistance was not associated with algorithm infection status (data not shown).
Sixty-nine percent of clonal OW central line isolates were cultured from patients with leukemia, and all of the OW patients with clonal isolates had some type of lymphoproliferative disorder. Acute myelogenous leukemia was the most common disorder in the OW (45.2%). A total of 57.1% of the ER venipuncture clonal isolates were cultured from patients with a neurological disorder who had impaired motor function.
Subset analysis of S. epidermidis isolates obtained 2 years after the initial study period demonstrated by PFGE that the major subtype A (minor subtype A1) continued to circulate within the OW central line isolates (data not shown).
DISCUSSION
Many of the previously mentioned studies in the literature have utilized controls from patients similar to, or the same group of patients as, those under study. We chose the adult ER so as to exclude those patients who would have possibly had an indwelling catheter or other medical device that might have been obtained at our hospital. This choice of a control group would also give a good comparison of the ecological diversity of CoNS in the community compared to the selective population of OW patients. From the data, we demonstrate that S. epidermidis is the predominant organism isolated from CoNS-positive blood cultures from ER venipunctures and OW central lines and that there is a more narrow spectrum of different species seen among OW line isolates than among ER venipuncture isolates. There is also a predominant clone of S. epidermidis that is circulating in our OW among patients who have central lines. Antibiotic usage in this unit has likely influenced the ecology and selected organisms with resistance to antibiotics, in particular, penicillins, minocycline, and gentamicin, which has been documented in other studies (1, 21, 22, 24, 27). Although we did not evaluate for the presence of biofilm production in these clonal isolates, evidence from other studies suggests that this helps establish and select organisms that are more resistant and invasive and evade the immune system (7, 21, 32). Our data appear to suggest that a predominant clone was consistently and frequently isolated from patients with central lines and that this clone (A) was isolated multiple times from the same patients (Table 4). This suggests a propensity of this isolate to colonize central lines, which is likely to involve extensive biofilm production.
We believe that a significant cause of the proliferation of clone A in our OW was patient-to-patient transfer by medical staff. In neonatal typing studies, clusters of CoNS have been shown to be distributed among both neonates and hospital staff, while CoNS isolates associated with sepsis may be more homogeneous (10, 24, 25, 35). In operating room environments and radiological suites, extensive shedding of CoNS into the air by medical staff is seen, even with contact barrier precautions and masks (31, 45). Frequent utilization of central lines for blood tests and intravenous administration of drugs promotes frequent distribution of the biofilm-producing clones that are more resistant and likely to produce central line infections. Further evidence that supports our hypothesis that patient-to-patient transfer of these organisms is occurring in our OW was the fact that the minor A1 subtype of the major subtype A was still present in central line isolates 2 years after the initial data were collected.
Another contributor to the clonal spread of sepsis-related S. epidermidis in our OW may have been colonization and gut mucosal translocation of these organisms. Several studies have implicated this pathogenic mechanism (1, 8, 9, 14, 17, 20, 39), especially in cancer patients. All of our OW patients with clonal isolates had some type of lymphoproliferative disorder (69% had leukemia). Many of these patients had recent transplants, gastrointestinal graft-versus-host disease, low WBC counts (clone A mean WBC = 2,791 cells/μl [Table 4]), and some form of chemotherapy which would make them particularly susceptible to mucosal translocation of colonizing S. epidermidis organisms. Another possibility is that mucosal translocation seeds the bloodstream with biofilm-producing isolates that then adhere to and colonize catheters, further promoting the spread of these organisms.
As previously defined, we utilized an algorithm of infection status to further characterize the possibility of clonal organisms causing catheter-related infections (5). There was no statistically significant increase in infections among those patients with major subtype A versus non-major subtype A, indicating that major subtype A isolates do not cause more infection than non-major subtype A isolates. In Table 4, the means of several clinical indicators were marginally but statistically significantly different between major subtype A and non-major subtype A isolates. These values were minimally from to the cutoffs that have been shown in the literature to be predictive of infection.(5) When these univariate indicators were incorporated into the algorithm, these effects did not predict infection. An explanation for the lower WBC count (not low enough to define infection) might be that this allows preferential colonization of a catheter by the clone, while being an indicator not of “infection” but of mere colonization.
The fact that major subtype A was not associated with our definition of infection suggests that these isolates may have been contaminants from extensively colonized central lines instead of true line infections or mucosally translocated organisms causing sepsis. Further, these major subtype A isolates may be no more pathogenic than non-major subtype A isolates, and this could be a possible explanation for the lack of an association with infection status. It is important to also note that the fact that many of the central line OW patients had leukemias/lymphoproliferative disorders may affect our definition of sepsis, because these patients do not always express every physical sign and symptom of infection.
In summary, we found a clonal group (group A) of isolates that was responsible for a significant number of positive blood cultures among OW patients, and only one major subtype group A isolate was present in the comparison group (ER patients). Central line usage for blood draws and administration of medications appears play a significant role in colonization, blood culture contamination, and bacteremia among OW patients. Blood cultures from only venipuncture sites should be the standard procedure for evaluating sepsis and clinically relevant bacteremia in cancer patients.
Acknowledgments
This work was supported by in part by a clinical and translational research award to K. L. Muldrew from the Department of Pathology, Vanderbilt University, Nashville, TN.
We thank Rosemary Verrall and Joni Williams from the Clinical Microbiology Laboratory at Vanderbilt University Medical Center for their excellent technical assistance.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Agvald-Ohman, C., B. Lund, and C. Edlund. 2004. Multiresistant coagulase-negative Staphylococci disseminate frequently between intubated patients in a multidisciplinary intensive care unit. Crit. Care. 8R42-R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agvald-Ohman, C., B. Lund, H. Hjelmqvist, G. Hedin, J. Struwe, and C. Edlund. 2006. ICU stay promotes enrichment and dissemination of multiresistant coagulase-negative Staphylococcal strains. Scand. J. Infect. Dis. 38441-447. [DOI] [PubMed] [Google Scholar]
- 3.Bach, A., H. Eberhardt, A. Frick, H. Schmidt, B. W. Bottiger, and E. Martin. 1999. Efficacy of silver-coating central venous catheters in reducing bacterial colonization. Crit. Care Med. 27515-521. [DOI] [PubMed] [Google Scholar]
- 4.Bates, D. W., L. Goldman, and T. H. Lee. 1991. Contaminant blood cultures and resource utilization. The true consequences of false-positive results. JAMA 265365-369. [PubMed] [Google Scholar]
- 5.Beekmann, S. E., D. J. Diekema, and G. V. Doern. 2005. Determining the clinical significance of coagulase-negative Staphylococci isolated from blood cultures. Infect. Control Hosp. Epidemiol. 26559-566. [DOI] [PubMed] [Google Scholar]
- 6.Bjorkqvist, M., B. Soderquist, E. Tornqvist, L. Sjoberg, H. Fredlund, I. Kuhn, P. Colque-Navarro, and J. Schollin. 2002. Phenotypic and genotypic characterisation of blood isolates of coagulase-negative Staphylococci in the newborn. APMIS 110332-339. [DOI] [PubMed] [Google Scholar]
- 7.Cafiso, V., T. Bertuccio, M. Santagati, F. Campanile, G. Amicosante, M. G. Perilli, L. Selan, M. Artini, G. Nicoletti, and S. Stefani. 2004. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin. Microbiol. Infect. 101081-1088. [DOI] [PubMed] [Google Scholar]
- 8.Costa, S. F., A. A. Barone, M. H. Miceli, I. M. van der Heijden, R. E. Soares, A. S. Levin, and E. J. Anaissie. 2006. Colonization and molecular epidemiology of coagulase-negative Staphylococcal bacteremia in cancer patients: a pilot study. Am. J. Infect. Control 3436-40. [DOI] [PubMed] [Google Scholar]
- 9.Costa, S. F., M. H. Miceli, and E. J. Anaissie. 2004. Mucosa or skin as source of coagulase-negative Staphylococcal bacteraemia? Lancet Infect. Dis. 4278-286. [DOI] [PubMed] [Google Scholar]
- 10.de Silva, G. D., A. Justice, A. R. Wilkinson, J. Buttery, M. Herbert, N. P. Day, and S. J. Peacock. 2001. Genetic population structure of coagulase-negative Staphylococci associated with carriage and disease in preterm infants. Clin. Infect. Dis. 331520-1528. [DOI] [PubMed] [Google Scholar]
- 11.de Silva, G. D., M. Kantzanou, A. Justice, R. C. Massey, A. R. Wilkinson, N. P. Day, and S. J. Peacock. 2002. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J. Clin. Microbiol. 40382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbins, B. M., P. Kite, A. Kindon, M. J. McMahon, and M. H. Wilcox. 2002. DNA fingerprinting analysis of coagulase negative Staphylococci implicated in catheter related bloodstream infections. J. Clin. Pathol. 55824-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foka, A., V. Chini, E. Petinaki, F. Kolonitsiou, E. D. Anastassiou, G. Dimitracopoulos, and I. Spiliopoulou. 2006. Clonality of slime-producing methicillin-resistant coagulase-negative Staphylococci disseminated in the neonatal intensive care unit of a university hospital. Clin. Microbiol. Infect. 121230-1233. [DOI] [PubMed] [Google Scholar]
- 14.Frebourg, N. B., B. Cauliez, and J. F. Lemeland. 1999. Evidence for nasal carriage of methicillin-resistant staphylococci colonizing intravascular devices. J. Clin. Microbiol. 371182-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fux, C. A., D. Uehlinger, T. Bodmer, S. Droz, C. Zellweger, and K. Muhlemann. 2005. Dynamics of hemodialysis catheter colonization by coagulase-negative Staphylococci. Infect. Control Hosp. Epidemiol. 26567-574. [DOI] [PubMed] [Google Scholar]
- 16.Gilsdorf, J. R., K. Wilson, and T. F. Beals. 1989. Bacterial colonization of intravenous catheter materials in vitro and in vivo. Surgery 10637-44. [PubMed] [Google Scholar]
- 17.Herwaldt, L. A., R. J. Hollis, L. D. Boyken, and M. A. Pfaller. 1992. Molecular epidemiology of coagulase-negative Staphylococci isolated from immunocompromised patients. Infect. Control Hosp. Epidemiol. 1386-92. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y. C., Y. H. Wang, Y. H. Chou, and R. I. Lien. 2006. Significance of coagulase-negative Staphylococci isolated from a single blood culture from neonates in intensive care. Ann. Trop. Paediatr. 26311-318. [DOI] [PubMed] [Google Scholar]
- 19.Huang, Y. C., Y. H. Wang, L. H. Su, Y. H. Chou, R. I. Lien, and T. Y. Lin. 2006. Determining the significance of coagulase-negative Staphylococci identified in cultures of paired blood specimens from neonates by species identification and strain clonality. Infect. Control Hosp. Epidemiol. 2770-73. [DOI] [PubMed] [Google Scholar]
- 20.Jones, J. W., R. J. Scott, J. Morgan, and J. V. Pether. 1992. A study of coagulase-negative Staphylococci with reference to slime production, adherence, antibiotic resistance patterns and clinical significance. J. Hosp. Infect. 22217-227. [DOI] [PubMed] [Google Scholar]
- 21.Klingenberg, C., E. Aarag, A. Ronnestad, J. E. Sollid, T. G. Abrahamsen, G. Kjeldsen, and T. Flaegstad. 2005. Coagulase-negative Staphylococcal sepsis in neonates: association between antibiotic resistance, biofilm formation and the host inflammatory response. Pediatr. Infect. Dis. J. 24817-822. [DOI] [PubMed] [Google Scholar]
- 22.Klingenberg, C., A. Sundsfjord, A. Ronnestad, J. Mikalsen, P. Gaustad, and T. Flaegstad. 2004. Phenotypic and genotypic aminoglycoside resistance in blood culture isolates of coagulase-negative Staphylococci from a single neonatal intensive care unit, 1989-2000. J. Antimicrob. Chemother. 54889-896. [DOI] [PubMed] [Google Scholar]
- 23.Krause, R., R. Haberl, A. Wolfler, F. Daxbock, H. W. Auner, G. J. Krejs, C. Wenisch, and E. C. Reisinger. 2003. Molecular typing of coagulase-negative Staphylococcal blood and skin culture isolates to differentiate between bacteremia and contamination. Eur. J. Clin. Microbiol. Infect. Dis. 22760-763. [DOI] [PubMed] [Google Scholar]
- 24.Krediet, T. G., E. M. Mascini, E. van Rooij, J. Vlooswijk, A. Paauw, L. J. Gerards, and A. Fleer. 2004. Molecular epidemiology of coagulase-negative staphylococci causing sepsis in a neonatal intensive care unit over an 11-year period. J. Clin. Microbiol. 42992-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milisavljevic, V., F. Wu, J. Cimmotti, J. Haas, P. Della-Latta, E. Larson, and L. Saiman. 2005. Genetic relatedness of Staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Identical clones found in two different hospital NICU's at 7 miles apart. Am. J. Infect. Control. 33341-347. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. J., B. Venus, and M. Mathru. 1984. Comparison of the sterility of long-term central venous catheterization using single lumen, triple lumen, and pulmonary artery catheters. Crit. Care Med. 12634-637. [DOI] [PubMed] [Google Scholar]
- 27.Monsen, T., C. Karlsson, and J. Wistrom. 2005. Spread of clones of multidrug-resistant, coagulase-negative Staphylococci within a university hospital. Infect. Control Hosp. Epidemiol. 2676-80. [DOI] [PubMed] [Google Scholar]
- 28.Muller-Premru, M., and P. Cernelc. 2004. Molecular epidemiology of catheter-related bloodstream infections caused by coagulase-negative Staphylococci in haematological patients with neutropenia. Epidemiol. Infect. 132921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ninin, E., N. Caroff, E. Espaze, J. Maraillac, D. Lepelletier, N. Milpied, and H. Richet. 2006. Assessment of ica operon carriage and biofilm production in Staphylococcus epidermidis isolates causing bacteraemia in bone marrow transplant recipients. Clin. Microbiol. Infect. 12446-452. [DOI] [PubMed] [Google Scholar]
- 30.Nouwen, J. L., A. van Belkum, S. de Marie, J. Sluijs, J. J. Wielenga, J. A. Kluytmans, and H. A. Verbrugh. 1998. Clonal expansion of Staphylococcus epidermidis strains causing Hickman catheter-related infections in a hemato-oncologic department. J. Clin. Microbiol. 362696-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nouwen, J. L., J. J. Wielenga, H. van Overhagen, J. S. Lameris, J. A. Kluytmans, M. D. Behrendt, W. C. Hop, H. A. Verbrugh, and S. de Marie. 1999. Hickman catheter-related infections in neutropenic patients: insertion in the operating theater versus insertion in the radiology suite. J. Clin. Oncol. 171304. [DOI] [PubMed] [Google Scholar]
- 32.Otto, M. 2004. Virulence factors of the coagulase-negative Staphylococci. Front. Biosci. 9841-863. [DOI] [PubMed] [Google Scholar]
- 33.Paulus, S. C., H. K. van Saene, S. Hemsworth, J. Hughes, A. Ng, and B. L. Pizer. 2005. A prospective study of septicaemia on a paediatric oncology unit: a three-year experience at The Royal Liverpool Children's Hospital, Alder Hey, UK. Eur. J. Cancer 412132-2140. [DOI] [PubMed] [Google Scholar]
- 34.Persson, L., H. Strid, U. Tidefelt, and B. Soderquist. 2006. Phenotypic and genotypic characterization of coagulase-negative Staphylococci isolated in blood cultures from patients with haematological malignancies. Eur. J. Clin. Microbiol. Infect. Dis. 25299-309. [DOI] [PubMed] [Google Scholar]
- 35.Raimundo, O., H. Heussler, J. B. Bruhn, S. Suntrarachun, N. Kelly, M. A. Deighton, and S. M. Garland. 2002. Molecular epidemiology of coagulase-negative Staphylococcal bacteraemia in a newborn intensive care unit. J. Hosp. Infect. 5133-42. [DOI] [PubMed] [Google Scholar]
- 36.Richter, S. S., S. E. Beekmann, J. L. Croco, D. J. Diekema, F. P. Koontz, M. A. Pfaller, and G. V. Doern. 2002. Minimizing the workup of blood culture contaminants: implementation and evaluation of a laboratory-based algorithm. J. Clin. Microbiol. 402437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruge, D. G., R. L. Sandin, S. A. Siegelski, J. N. Greene, and N. Johnson. 2002. Reduction in blood culture contamination rates by establishment of a policy for central intravenous catheters. Lab. Med. 10797-800. [Google Scholar]
- 38.Seo, S. K., L. Venkataraman, P. C. DeGirolami, and M. H. Samore. 2000. Molecular typing of coagulase-negative Staphylococci from blood cultures does not correlate with clinical criteria for true bacteremia. Am. J. Med. 109697-704. [DOI] [PubMed] [Google Scholar]
- 39.Silva, F. R., E. M. Mattos, M. V. Coimbra, B. T. Ferreira-Carvalho, and A. M. Figueiredo. 2001. Isolation and molecular characterization of methicillin-resistant coagulase-negative Staphylococci from nasal flora of healthy humans at three community institutions in Rio de Janeiro City. Epidemiol. Infect. 12757-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sitges-Serra, A., J. Linares, and J. Garau. 1985. Catheter sepsis: the clue is the hub. Surgery 97355-357. [PubMed] [Google Scholar]
- 41.Sitges-Serra, A., J. Linares, J. L. Perez, E. Jaurrieta, and L. Lorente. 1985. A randomized trial on the effect of tubing changes on hub contamination and catheter sepsis during parenteral nutrition. JPEN J. Parenter. Enteral. Nutr. 9322-325. [DOI] [PubMed] [Google Scholar]
- 42.Sitges-Serra, A., P. Puig, J. Linares, J. L. Perez, N. Farrero, E. Jaurrieta, and J. Garau. 1984. Hub colonization as the initial step in an outbreak of catheter-related sepsis due to coagulase negative Staphylococci during parenteral nutrition. J. Parenter. Enteral. Nutr. 8668-672. [DOI] [PubMed] [Google Scholar]
- 43.Spare, M. K., S. E. Tebbs, S. Lang, P. A. Lambert, T. Worthington, G. W. Lipkin, and T. S. Elliott. 2003. Genotypic and phenotypic properties of coagulase-negative Staphylococci causing dialysis catheter-related sepsis. J. Hosp. Infect. 54272-278. [DOI] [PubMed] [Google Scholar]
- 44.Spiliopoulou, I., I. Santos Sanches, C. Bartzavali, A. M. Ludovice, M. Aires de Sousa, G. Dimitracopoulos, and H. de Lencastre. 2003. Application of molecular typing methods to characterize nosocomial coagulase-negative Staphylococci collected in a Greek hospital during a three-year period (1998-2000). Microb. Drug Resist. 9273-282. [DOI] [PubMed] [Google Scholar]
- 45.Tammelin, A., P. Domicel, A. Hambraeus, and E. Stahle. 2000. Dispersal of methicillin-resistant Staphylococcus epidermidis by staff in an operating suite for thoracic and cardiovascular surgery: relation to skin carriage and clothing. J. Hosp. Infect. 44119-126. [DOI] [PubMed] [Google Scholar]
- 46.Tang, Y. W., J. Han, M. A. McCormac, H. J. Li, and C. W. Stratton. 2008. Staphylococcus pseudolugdunensis sp. nov, a pyrrolidonyl-arylamidase/ornithine decarboxylase-positive bacterium isolated from blood cultures. Diagn. Microbiol. Infect. Dis. 60351-359. [DOI] [PubMed] [Google Scholar]
- 47.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Pelt, C., J. Nouwen, E. Lugtenburg, C. van der Schee, S. de Marie, P. Schuijff, H. Verbrugh, B. Lowenberg, A. van Belkum, and M. Vos. 2003. Strict infection control measures do not prevent clonal spread of coagulase negative Staphylococci colonizing central venous catheters in neutropenic hemato-oncologic patients. FEMS Immunol. Med. Microbiol. 38153-158. [DOI] [PubMed] [Google Scholar]
- 49.Vermont, C. L., N. G. Hartwig, A. Fleer, P. de Man, H. Verbrugh, J. van den Anker, R. de Groot, and A. van Belkum. 1998. Persistence of clones of coagulase-negative staphylococci among premature neonates in neonatal intensive care units: two-center study of bacterial genotyping and patient risk factors. J. Clin. Microbiol. 362485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viale, P., and S. Stefani. 2006. Vascular catheter-associated infections: a microbiological and therapeutic update. J. Chemother. 18235-249. [DOI] [PubMed] [Google Scholar]
- 51.Villari, P., C. Sarnataro, and L. Iacuzio. 2000. Molecular epidemiology of Staphylococcus epidermidis in a neonatal intensive care unit over a three-year period. J. Clin. Microbiol. 381740-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative Staphylococci. Lancet Infect. Dis. 2677-685. [DOI] [PubMed] [Google Scholar]
- 53.Yao, Y., D. E. Sturdevant, and M. Otto. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191289-298. [DOI] [PubMed] [Google Scholar]