Abstract
Based on analysis of 16,392 bp encompassing the complete open reading frames (ORFs) 1, 5, 31, 36, 37, 47, 60, 62, 67, and 68 of the genome of genotype M1 varicella-zoster virus (VZV) was found in swab samples originating from eight Tanzanian zoster patients. Moreover, sequence analysis suggests recombination events between different VZV genotypes within ORFs 1, 31, 60, and 67.
Varicella-zoster virus (VZV; also known as Human herpesvirus 3), belonging to the subfamily Alphaherpesvirinae of the family Herpesviridae, is the cause of chickenpox (varicella) and may reactivate from latency, causing shingles (zoster). Until the beginning of the 21st century, the double-stranded DNA genome of VZV was believed to be highly conserved. Therefore, efforts to analyze entire genome sequences of VZV strains were very rare, and only the entire genome of the VZV prototype strain Dumas was known for decades (2). The VZV genotypes demonstrate a specific geographical distribution. Genotypes E1 and E2 have been detected mainly in Europe and the Americas, whereas genotype J strains are dominant in Japan. Genotype M2 strains probably arose through recombination of genotype E1 and E2 strains with genotype J strains and currently are distributed in countries where the European/American genotypes may have recombined with the Japanese genotype (12). In contrast, genotype M1 strains seem to be associated with nonwhite patients (3, 4, 8). The genotype M1 prototype strain CA123 was isolated in 1990 in California (4).
Based upon the results of partial and full-genome sequence analyses of several VZV strains, an out-of-Africa model for VZV evolution, suggesting that VZV coevolves with humankind and diversified from ancestral VZV genotypes into Japanese (J) and European/American (E1 and E2) genotypes (7, 12), was proposed. Another model suggests that VZV evolution is driven by climatic factors and that VZV strain distribution is associated with temperate and tropical climate conditions (1, 3, 8). Recombination analysis suggests that genotype M2 strains and genotype M1 strains are recombinant strains that originated from strains of the European/American (E1/E2) and Japanese (J) genotypes, respectively (6).
Several VZV genotyping schemes based on partial VZV genome sequences were proposed for genotype classification (3, 5, 12). Full-genome sequence analysis allowed the development of two classification schemes, resulting in different VZV genotype nomenclatures (4, 6, 7). Recently, we described a simple and reliable VZV genotyping scheme based on analysis of a 1,990-bp region originating from open reading frames (ORFs) 51 to 58. This procedure allows the typing of VZV wild-type strains by high-throughput procedures directly from clinical samples without intermediate virus propagation. Genotyping by this novel procedure and that based on full-genome phylogenetic analysis resulted in the same classification of all strains analyzed (9, 10).
As there is limited sequence information available for African VZV wild-type strains, our objectives were (i) to identify the genotype of circulating VZV wild-type strains in Tanzania and (ii) to reconstruct their evolutionary history.
Vesicle fluid samples were obtained in 2007 from four male and four female nonwhite adults who were zoster patients that were referred to the Ikonda hospital, Makete district, Iringa region, Tanzania. DNA was isolated from the samples by using the RTP DNA/RNA virus mini kit (Invitek, Berlin, Germany) according to the manufacturer's instructions. DNA amplification reactions were carried out with 25-μl volumes with 5 μl of extract or control DNA, 12.5 μl Taq PCR master mix (Qiagen, Hilden, Germany), corresponding to 0.6 U Taq polymerase, 1 μl of each forward and reverse primer (for detailed information, see Table 1), corresponding to 0.5 μM, and 5.5 μl water. Thermal cycling comprised an initial hot start at 95°C for 3 min, followed by 45 cycles of denaturation at 95°C for 30 s, annealing at primer-pair-specific temperatures (Table 1) for 30 s, and polymerization at 72°C for 2 min. Finally, an extension step at 72°C for 10 min was carried out. The PCR products were visualized with a UV transilluminator following separation on 1.5% agarose gels containing ethidium bromide. Purification and sequencing of PCR products, as well as VZV sequence and phylogenetic analyses, were performed as described earlier (9, 10).
TABLE 1.
Primers used to amplify and sequence genes in VZV strains
| Primer | Sequence (5′-3′) | Nucleotide positionsa | ORF no.b | Amplicon size (bp)b | Tm (°C)c |
|---|---|---|---|---|---|
| ORF1F | ATATTTTTGGGATCCGCA | 933-916 | 1 | 362 | 51.5 |
| ORF1R | TCCGGAAGGGGAAGGTA | 571-587 | 54.7 | ||
| ORF5F | TGCCATCTTCCACGGGTC | 5292-5275 | 5 | 1,058 | 58.7 |
| ORF5R | GCGTGTTTAGTCACATGA | 4234-4251 | 47.2 | ||
| ORF31F | GCGTTTTCATAACCTCCGTTACG | 56952-56974 | 31 | 965 | 60.6 |
| ORF31R | CCGTTCGTTTTGGCTTCCAG | 57917-57895 | 59.4 | ||
| ORF31F1 | GAACCTGCAGCGCGGAAC | 57845-57862 | 31 | 875 | 60.5 |
| ORF31R1 | CTCGTTATCTGTTCCAAGCTGGC | 58720-58698 | 62.4 | ||
| ORF31F2 | CTACGCGTTGTTATAGCCGTCC | 58632-58653 | 31 | 1,038 | 62.1 |
| ORF31R2 | CCTGTGATGCGTAATGGAGACAC | 59692-59670 | 62.4 | ||
| ORF36F | CGTTTGTCTACAATAAAC | 64789-64806 | 36 | 1,064 | 40.4 |
| ORF36R | ATACGTAAATACTAGGTATA | 65853-65834 | 35.8 | ||
| ORF37F | GCGGTGATATTGTAGCGCAAG | 66042-66062 | 37 | 1,042 | 59.8 |
| ORF37R | GCGGTAATCCAAACTCTCTTCGG | 67084-67062 | 62.4 | ||
| ORF37F1 | GTTTATTTACTTGGACGTGGGTTGG | 66992-67016 | 37 | 1,084 | 61.3 |
| ORF37R1 | CTGTGTCCCTGGACAGCTGG | 68076-68057 | 63.5 | ||
| ORF37F2 | ACTCCACACCCATGTACCAGAAG | 67981-68003 | 37 | 740 | 62.4 |
| ORF37R2 | TTTGGTAGAGTGCACCAAACACC | 68721-68699 | 60.6 | ||
| ORF47F | GACGAAGCGTTACTTACACA | 83148-83167 | 47 | 1,570 | 51.0 |
| ORF47R | TTTTGGCTGGCTGGGGGC | 84718-84701 | 65.3 | ||
| ORF60F | AGGGAAAACACAAGCGTC | 101667-101650 | 60 | 518 | 53.5 |
| ORF60R | TTTGAATCCGATAGTTTCA | 101149-101167 | 46.5 | ||
| ORF62F | GGCGCTCACGAGAAAAGGAG | 105172-105191 | 62 | 1,204 | 61.4 |
| ORF62R | CTGTCGACCCGAGACCTGG | 106376-106358 | 63.1 | ||
| ORF62F1 | CAAAGCGGGTCCATCCCTG | 106245-106263 | 62 | 1,249 | 61 |
| ORF62R1 | CCAAGCTGACCGGTGTCAACTC | 107494-107473 | 64 | ||
| ORF62F2 | CGGAACGGGAGACGCTACG | 107388-107406 | 62 | 1,299 | 63.1 |
| ORF62R3 | GGAAACGGGCAGAGGTACGAC | 108687-108667 | 63.7 | ||
| ORF62F4 | ACCGCTGGTCTTCCCGTTG | 108578-108596 | 62 | 622 | 61 |
| ORF62R4 | TCGCAATCCTTTGAAGGCTG | 109200-109181 | 57.3 | ||
| ORF67F | GCGCCTCATTTAATCGCG | 114478-114496 | 67 | 1,103 | 58.2 |
| ORF67R | TAAAATCCGGGATAATTAG | 115581-115563 | 45.1 | ||
| ORF68F | ATTCCGAGGGTCGCCTGTAA | 115787-115806 | 68 | 1,910 | 60.1 |
| ORF68R | GTTGCCCCGGTTCGGTGA | 117697-117680 | 63.9 |
The nucleotide positions are given according to the numbering in the Dumas reference strain (accession number NC001348).
The values refer to both the forward and reverse primers.
Tm, melting temperature.
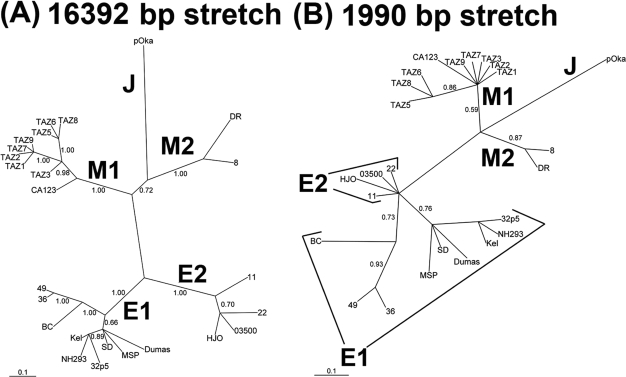
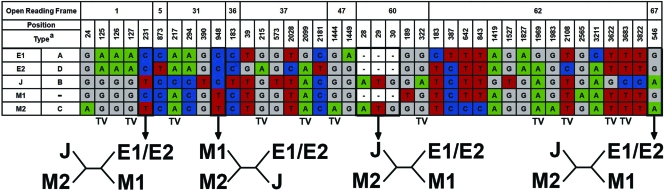
Phylogenetic analysis based on stretches of 16,392 and 1,990 bp of the genotype M1 strains from Tanzanian zoster patients demonstrated a close relationship to the recently described genotype M1 prototype strain CA123 (Fig. 1A and B). Analysis of the genotype M1 strains from Tanzanian patients revealed genotype M1-specific single-nucleotide polymorphisms in ORF 60 (G189T) and ORF 62 (G2565A) (Fig. 2) and within the ORF 51 genotyping area (G1947A) (data not shown). Moreover, the amplification and sequence analysis of more than 16 kb of the VZV genome revealed putative sites within ORFs 1, 31, 60, and 67, which show that viral recombination events may have occurred between genotype M1 or M2 and E1 or E2 (Fig. 2). In comparison to a previous study (6), two out of four sites (ORF 31 and ORF 67, nucleotides [nt] 948 and 546, respectively) were confirmed in this study, whereas the remaining sites (ORF1 and ORF60, nt 231 and 28 to 30, respectively) were found exclusively in this study (Fig. 2).
FIG. 1.
Phylogenetic trees of the novel Tanzanian VZV strains and the previously described genotype E1, E2, M1, M2, and J VZV strains. Trees based on the 16,392-bp stretch (A) and based on the 1,990-bp stretch (B) that are used in the genotyping scheme described recently (9). Posterior probabilities are shown on each branch.
FIG. 2.
Schematic representation of a recombination analysis of a 16,392-bp stretch of VZV, including only phylogenetically informative sites (n = 40) that are found in all strains of the respective genotypes. Genotype E1 includes nine strains: Dumas (NC001348), from The Netherlands; BC (AY548171), from British Columbia, Canada; 36 (DQ479958), from New Brunswick, Canada; 49 (DQ479959), from New Brunswick, Canada; MSP (AY548170), from Minnesota; 32p5 (DQ479961), from Texas; Kel (DQ479954), from Iowa; SD (DQ479953), from South Dakota; and NH293 (DQ674250), from the United States. Genotype E2 includes four strains: 11 (DQ47995), from New Brunswick, Canada; 22 (DQ479956), from New Brunswick, Canada; 03-500 (DQ479957), from Alberta, Canada; and HJO (AJ871403), from Germany. Genotype J includes one strain, pOka (AB097933), from Japan. Genotype M1 includes the prototype CA123 strain (DQ457052), from California, and the eight novel Tanzanian strains. Genotype M2 includes two strains: 8 (DQ479960), from New Brunswick, Canada; and DR (DQ452050), from the United States. The four putative sites that are informative for recombination events are indicated in with arrows and bold text, and the possible tree topologies of the four sites are shown below the table. TV, transversions. The genotype designations are based on those used in references 4 and 12. Dashes indicate nucleotide deletions.
Interestingly, two of the most conserved herpesvirus genes (ORF 31 and ORF 67), coding for the envelope glycoproteins B and I (12), were obviously included in the potential recombination event. Furthermore, the in-frame translation initiation ATG insertion within the envelope glycoprotein L gene (ORF 60) linked the African genotype M1 strains to the European/American genotype E1/E2 strains (Fig. 2) and may again suggest recombination between these genotypes. Moreover a unique 9-nt deletion (3733 to 3742) resulting in an amino acid deletion of two glycines and one asparagine within the ORF 62 protein in TAZ5 was observed (data not shown). As ORF 62 codes for the VZV immediate-early regulatory gene IE62, polymorphisms have been considered to be a potential reason for attenuation and linked to transversions at nt 1882, 2872, and 3778 (11). Interestingly, the deletion observed in the ORF 62 of TAZ5 is located closely to position 3778.
In conclusion, genotype M1 VZV wild-type strains circulating in Tanzania seem to be recombinant strains and associated with African origins. There is an urgent need to analyze more VZV wild-type strains from different regions of Africa to explain why the recombinant-like genotype M1 is common in a region from which Homo sapiens dispersed out of Africa.
Acknowledgments
The work was supported by the Hospital of the Johann Wolfgang Goethe University and Paul und Ursula Klein Stiftung.
Footnotes
Published ahead of print on 13 August 2008.
REFERENCES
- 1.Barrett-Muir, W., F. T. Scott, P. Aaby, J. John, P. Matondo, Q. L. Chaudhry, M. Siqueira, A. Poulsen, K. Yaminishi, and J. Breuer. 2003. Genetic variation of varicella-zoster virus: evidence for geographical separation of strains. J. Med. Virol. 70(Suppl. 1)S42-S47. [DOI] [PubMed] [Google Scholar]
- 2.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 671759-1816. [DOI] [PubMed] [Google Scholar]
- 3.Loparev, V. N., A. Gonzalez, M. Deleon-Carnes, G. Tipples, H. Fickenscher, E. G. Torfason, and D. S. Schmid. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 788349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loparev, V. N., E. N. Rubtcova, V. Bostik, D. Govil, C. J. Birch, J. D. Druce, D. S. Schmid, and M. C. Croxson. 2007. Identification of five major and two minor genotypes of varicella-zoster virus strains: a practical two-amplicon approach used to genotype clinical isolates in Australia and New Zealand. J. Virol. 8112758-12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muir, W. B., R. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 761971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norberg, P., J. A. Liljeqvist, T. Bergstrom, S. Sammons, D. S. Schmid, and V. N. Loparev. 2006. Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J. Virol. 809569-9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters, G. A., S. D. Tyler, C. Grose, A. Severini, M. J. Gray, C. Upton, and G. A. Tipples. 2006. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J. Virol. 809850-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlivan, M., K. Hawrani, W. Barrett-Muir, P. Aaby, A. Arvin, V. T. Chow, T. J. John, P. Matondo, M. Peiris, A. Poulsen, M. Siqueira, M. Takahashi, Y. Talukder, K. Yamanishi, M. Leedham-Green, F. T. Scott, S. I. Thomas, and J. Breuer. 2002. The molecular epidemiology of varicella-zoster virus: evidence for geographic segregation. J. Infect. Dis. 186888-894. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt-Chanasit, J., M. Stürmer, A. Hahn, S. G. Schäd, G. Gross, R. G. Ulrich, G. Heckel, and H. W. Doerr. 2007. Novel genotyping approach for varicella-zoster virus strains from Germany. J. Clin. Microbiol. 453540-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt-Chanasit, J., K. Bleymehl, S. G. Schäd, G. Gross, R. G. Ulrich, and H. W. Doerr. 2008. Novel varicella-zoster virus glycoprotein E gene mutations associated with genotypes A and D. J. Clin. Microbiol. 46325-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyler, S. D., G. A. Peters, C. Grose, A. Severini, M. J. Gray, C. Upton, and G. A. Tipples. 2007. Genomic cartography of varicella-zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology 359447-458. [DOI] [PubMed] [Google Scholar]
- 12.Wagenaar, T. R., V. T. Chow, C. Buranathai, P. Thawatsupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 71072-1081. [DOI] [PubMed] [Google Scholar]