Abstract
Debaryomyces hansenii is a hemiascomycetous yeast commonly found in natural substrates and in various types of cheese. Pichia guilliermondii is widely distributed in nature and is a common constituent of the normal human microflora. Both species have been described in human infections but are extremely difficult to differentiate phenotypically. Thus, frequent errors in identification occur. The 62 clinical and environmental isolates sent between 2000 and 2007 to the French National Reference Center for Mycoses and Antifungals as D. hansenii or P. guilliermondii were analyzed by using the carbon assimilation pattern, the presence of pseudohyphae, and sequencing of the ITS and D1/D2 regions of the rRNA gene. The objective of this study was to assess using nucleotide sequences whether phenotypic identification was accurate and whether phenotypic characteristics could be used to differentiate the two species when sequencing was not available. We found that 58% of the isolates were misidentified and belong to seven different species: P. guilliermondii, P. caribbica, P. jadinii, D. hansenii, Candida palmioleophila, C. haemulonii type II, and Clavispora lusitaniae. In conclusion, D. hansenii may not be as common a human pathogen as previously thought. Sequencing of either ITS or D1/D2 regions is a good tool for differentiating the species more frequently confused with D. hansenii, keeping in mind that reliable databases should be used.
Debaryomyces hansenii is a hemiascomycetous yeast commonly found in natural substrates and in various types of cheese (3, 26). It has been described in human infections (11, 31, 35). However, its incidence during candidemia is low based on data surveillance implemented by the Centers for Disease Control and Prevention (9), whereas one review on worldwide-collected isolates states that D. hansenii accounts for 0.08 to 0.5% of isolates recovered during invasive candidiasis (22). Pichia guilliermondii is widely distributed in nature (routinely isolated from insects, soil, plants, atmosphere, seawater, the exudates of various trees, and processed foods) and is a common constituent of the normal human microflora (19). Globally, this species and its anamorphic state Candida guilliermondii accounts for 1 to 2% of all candidemia (14, 23). However, the species D. hansenii (Candida famata) and P. guilliermondii are extremely difficult to differentiate phenotypically (17, 18). Thus, frequent errors in identification occur. We undertook a retrospective analysis of all isolates, mostly recovered from clinical specimens sent to the French National Reference Center for Mycoses and Antifungals (NRCMA) as D. hansenii or P. guilliermondii. The objective of the present study was to assess using nucleotide sequences whether phenotypic identification was correct and whether phenotypic characteristics could be used to differentiate the two species when sequencing was not available.
MATERIALS AND METHODS
Strains.
All of the epidemiologically unrelated clinical or environmental isolates (n = 41) sent between September 2000 and April 2007 as D. hansenii or P. guilliermondii to the NRCMA for confirmation of the identification were selected. In addition, 21 epidemiologically unrelated clinical or environmental isolates were collected for this specific study (Table 1) . Type strains for the anamorph of each species were obtained from the Centraalbureau voor Schimmelcultures (CBS; Utrecht, The Netherlands) (see Table S1 in the supplemental material). All isolates were stored frozen in 40% glycerol at −80°C.
TABLE 1.
Molecular identification, ID32C code, and production of pseudohyphae for the 62 clinical and environmental isolates sent to the NRCMA between September 2000 and April 2007 as D. hansenii or P. guilliermondii
| Strain | Site of isolation | Yr of isolation | City, country | First identification | Identification sequence | ID32C code | Presence (+) or absence (-) of pseudohyphae |
|---|---|---|---|---|---|---|---|
| 200500815 | Blood | 2001 | Hambourg, Germany | D. hansenii | D. hansenii | 5777375317 E+ | - |
| 200600362 | Sputum | 2006 | Paris, France | D. hansenii | D. hansenii | 5777174137 E+ | - |
| 200600935 | Table surface | 2006 | Fort de France, Martinique | D. hansenii | D. hansenii | 5777751137 E- | - |
| 200501145 | Sputum | 2005 | Paris, France | D. hansenii | P. caribbica | 7577752117 E+ | + |
| 200501146 | Mouth | 2005 | Paris, France | D. hansenii | P. caribbica | 7577352117 E+ | + |
| 200600033 | Blood | 2006 | Reims, France | D. hansenii | P. caribbica | 5577370117 E+ | + |
| 200700035 | BALa | 2006 | Paris, France | D. hansenii | P. caribbica | 7577770117 E+ | + |
| 200700175 | Blood | 2006 | Paris, France | D. hansenii | P. caribbica | 7577350117 E+ | + |
| 200700593 | Blood | 2007 | Paris, France | D. hansenii | P. caribbica | 7577350117 E+ | + |
| 200500086 | Blood | 2005 | Paris, France | D. hansenii | P. guilliermondii | 7577350117 E+ | + |
| 200500816 | Environment | 2001 | Saint Malo, France | D. hansenii | P. guilliermondii | 7577352117 E+ | - |
| 200500821 | Environment | 2002 | Dreux, France | D. hansenii | P. guilliermondii | 7577350117 E+ | - |
| 200501141 | Stomach | 2001 | Paris, France | D. hansenii | P. guilliermondii | 7577352117 E+ | + |
| 200501142 | Urine | 2001 | Paris, France | D. hansenii | P. guilliermondii | 7577350117 E+ | + |
| 200501144 | BAL | 2005 | Paris, France | D. hansenii | P. guilliermondii | 7577352117 E+ | + |
| 200501305 | Blood | 2005 | Pontoise, France | D. hansenii | P. guilliermondii | 7577350117 E+ | + |
| 200700040 | Blood | 2000 | Reims, France | D. hansenii | P. guilliermondii | 7577352117 E+ | + |
| 200700041 | Blood | 2000 | Reims, France | D. hansenii | P. guilliermondii | 7577352117 E+ | + |
| 200700261 | Blood | 1999 | Reims, France | D. hansenii | P. guilliermondii | 7577352117 E+ | + |
| 200400824 | Sputum | 2004 | Fort de France, Martinique | D. hansenii | C. haemulonii type II | 7167370317 E+ | - |
| 200600079 | Catheter | 2001 | Paris, France | D. hansenii | C. haemulonii type II | 7167370317 E- | + |
| 200500823 | Blood | 2002 | Paris, France | D. hansenii | C. lusitaniae | 5357350117 E+ | - |
| 200501223 | Tongue | 2002 | Paris, France | D. hansenii | C. lusitaniae | 7357370117 E+ | + |
| 200500813 | Catheter | 2001 | Bicêtre, France | D. hansenii | C. palmioleophila | 5367352315 E- | - |
| 200500825 | Mouth | 2002 | Reims, France | D. hansenii | C. palmioleophila | 5367352315 E- | - |
| 200500840 | Kidney | 2003 | Paris, France | D. hansenii | C. palmioleophila | 5367352315 E+ | - |
| 200500808 | Nose | 2000 | Paris, France | P. guilliermondii | P. caribbica | 7577352117 E- | + |
| 200500812 | Blood | 2003 | Paris, France | P. guilliermondii | P. caribbica | 7577750117 E- | + |
| 200500862 | Blood | 2003 | Paris, France | P. guilliermondii | P. caribbica | 7577352117 E+ | + |
| 200500949 | Blood | 2005 | Paris, France | P. guilliermondii | P. caribbica | 7577350117 E+ | + |
| 200501000 | Blood | 2005 | Paris, France | P. guilliermondii | P. caribbica | 7577370117 E+ | + |
| 200501201 | Blood | 2005 | Paris, France | P. guilliermondii | P. caribbica | 7577750117 E+ | + |
| 200501316 | BAL | 2005 | Paris, France | P. guilliermondii | P. caribbica | 7577350117 E+ | + |
| 200501317 | Sputum | 2002 | Paris, France | P. guilliermondii | P. caribbica | 7577350117 E+ | + |
| 200600196 | Sputum | 2006 | Paris, France | P. guilliermondii | P. caribbica | 7577350117 E+ | + |
| 200600368 | BAL | 2006 | Paris, France | P. guilliermondii | P. caribbica | 7577370117 E+ | - |
| 200601082 | Sputum | 2006 | Paris, France | P. guilliermondii | P. caribbica | 7577750117 E+ | + |
| 200601194 | Blood | 2006 | Paris, France | P. guilliermondii | P. caribbica | 7577750117 E+ | + |
| 200500030 | Blood | 2004 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352115 E+ | + |
| 200500719 | Blood | 2005 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200500809 | Selles | 2003 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200500861 | Blood | 2003 | Paris, France | P. guilliermondii | P. guilliermondii | 7577372137 E+ | + |
| 200500863 | Blood | 2004 | Paris, France | P. guilliermondii | P. guilliermondii | 5577352117 E+ | - |
| 200500864 | Spleen | 2004 | Marseille, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200500950 | Blood | 2005 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352115 E+ | - |
| 200501202 | Blood | 2005 | Paris, France | P. guilliermondii | P. guilliermondii | 7577350117 E+ | - |
| 200501314 | Blood | 2002 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200501319 | Throat | 2002 | Paris, France | P. guilliermondii | P. guilliermondii | 7577350117 E+ | + |
| 200501320 | Sputum | 2002 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200501343 | Urine | 2002 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200600032 | Blood | 2005 | Angers, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200600130 | Blood | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577350117 E+ | + |
| 200600335 | Blood | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577350117 E+ | + |
| 200600561 | BAL | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200600803 | Blood | 2006 | Bondy, France | P. guilliermondii | P. guilliermondii | 7577772117 E+ | + |
| 200600938 | Blood | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E- | - |
| 200601010 | BAL | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200601023 | Blood | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200601042 | Blood | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E+ | + |
| 200601191 | BAL | 2006 | Paris, France | P. guilliermondii | P. guilliermondii | 7577352117 E- | + |
| 200700329 | Blood | 2007 | Chambéry, France | P. guilliermondii | P. guilliermondii | 7577372117 E+ | + |
| 200500811 | Blood | 2003 | Paris, France | P. guilliermondii | P. jadinii | 4374350117 E+ | + |
BAL, bronchoalveolar lavage.
Phenotypic characterization of isolates.
Carbon assimilation patterns were obtained with the commercialized strips (ID32C; bioMérieux, Marcy l'Etoile, France). The presence of pseudohyphae was determined after growth for 24 h and 5 days of incubation at 30°C in potato-carrot-ox bile medium (used routinely in France instead of cornmeal agar or rice agar) (5). The MICs of amphotericin B, flucytosine, fluconazole, voriconazole, and caspofungin were determined by using the EUCAST microdilution method (7). The concentrations corresponding to the MICs that inhibited 50% (MIC50) and 90% (MIC90) of the isolates were determined.
Molecular characterization.
After 24 h of incubation at 27°C on Sabouraud dextrose agar plates, single colonies were transferred to 1 ml of distilled water in a microcentrifuge tube, and DNA extraction was performed by using a High-Pure PCR template preparation kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. Universal fungal primers were used for the amplification of the ITS1-5.8S-ITS2 (primers V9D [8] and LS266 [15]) and 26S (primers NL1 and NL4) (20) ribosomal DNA regions. Reaction volumes of 50 μl contained 3 μl of genomic DNA, 2.5 U of AmpliTaq Gold, 5 μl of PCR buffer 10×, 5 μl of MgCl2 25 mM, 5 μl of 2.5 mM deoxynucleoside triphosphate (Roche), and 1.25 μl of 20 μM primers. The PCR products were amplified by using an iCycler thermocycler (Bio-Rad, Marnes-La-Coquette, France) set up with a first cycle of denaturation for 10 min at 95°C, followed by 30 cycles of denaturation at 94°C for 30 s and 30 s at 58°C and elongation at 72°C for 30 s, with a final extension step of 10 min at 72°C. Both strands of purified amplified fragments were sequenced at the Genopole of the Pasteur Institute, on an ABI Prism 3700 DNA analyzer (Applied Biosystems, Courtaboeuf, France), with the same primers that were used in the PCR step. Sequences were edited with Chromas Pro version 1.33 (Technelysium Pty., Ltd., Australia).
The sequences of the ITS1-5.8S-ITS2 regions were delimited by the sequences of the primers ITS1 and ITS4 (TCCGTAGGTGAACCTGCGG/GCATATCAATAAGCGGAGGA) (36). In the same way, the sequences of the D1/D2 region of the 26S subunit were delimited by the sequence TTGRAATC (R = A or G) and by the sequence of the primer U1 (GTGAAATTGTTGAAAGGGAA) (29). The sequences were compared to those of the type strains to confirm species identification. The D1/D2 and ITS bounded sequences, including the sequences of the type strains, were aligned by using CLUSTAL W software (32). Phylogenetic analyses were performed by using the Bayesian Markov chain Monte Carlo method based on MrBayes software (28) and were based on a concatenated alignment of the ITS and D1/D2 datasets.
RESULTS
Among the 36 isolates first identified as P. guilliermondii, species identification was confirmed for 23 isolates, 12 were identified as P. caribbica based on the comparison of their ITS and 26S sequences with the type strain sequences (CBS 2022), and one was identified as P. jadinii based on both carbon assimilation profile and nucleotide sequences. Of the 26 isolates initially identified as D. hansenii, only 3 were confirmed as such, 10 were identified as P. guilliermondii, 6 were identified as P. caribbica, 3 were identified as Candida palmioleophila, and 2 belong to the currently recognized species Candida haemulonii type II and 2 to Clavispora lusitaniae (Table 1).
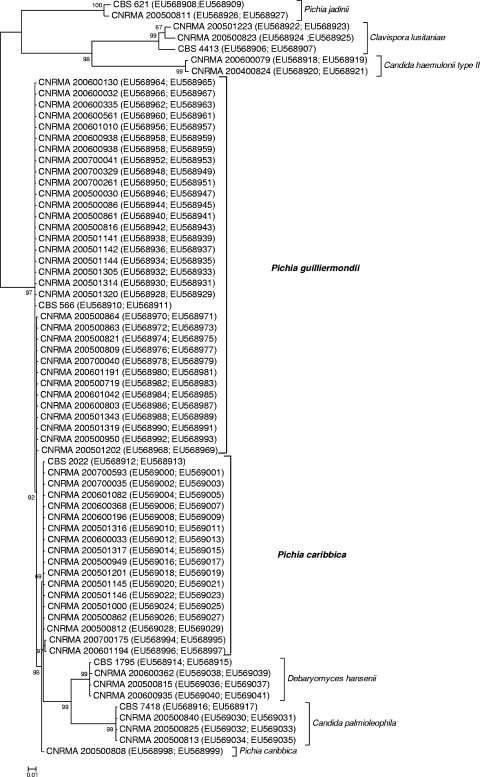
A Bayesian tree was constructed by using the bounded sequences of the ITS and 26S regions of all isolates, including the type strains (Fig. 1). The isolates of P. guilliermondii are divided into two closely related groups: one group included the type strain (CBS 566) and 20 of 33 isolates, and the second group contained 13 isolates. P. caribbica isolates were clustered into three groups: one group included the type strain (CBS 2022) and 15 of 18 isolates, and the second and the third groups included, respectively, 2 isolates and 1 isolate. Both species were closely related. The isolates of D. hansenii, including the type strain CBS 1795, were closely related to those of C. palmioleophila. Isolates of P. jadinii, C. lusitaniae, and C. haemulonii type II are well separated from these species and would have been using only one of the nucleotide sequences.
FIG. 1.
Bayesian phylogenetic majority rule consensus tree of the combined D1/D2 and ITS datasets for the 62 clinical and environmental isolates and the type strains of D. hansenii, P. guilliermondii, P. caribbica, C. palmioleophila, C. lusitaniae, and P. jadinii. CBS numbers correspond to type strains. GenBank accession numbers D1/D2 and ITS datasets are indicated in parentheses. The scale bar represents the distance between strains (10%).
Analysis of carbon assimilation patterns by each species showed that all three isolates of D. hansenii assimilated dl-lactate, whereas none of the P. guilliermondii and P. caribbica isolates did so (Table 2). None of the D. hansenii isolates was resistant to 0.01% cycloheximide, whereas >90% of P. guilliermondii and P. caribbica isolates were. For C. palmioleophila, the carbon assimilation patterns were ambiguous with D. hansenii profiles. For C. haemulonii type II, C. lusitaniae, and P. jadinii the majority of the codes were lacking in the ID32C database or show a low discrimination with D. hansenii. None of the D. hansenii isolates was able to produce pseudohyphae, whereas 27 of 33 and 17 of 18 isolates of P. guilliermondii and P. caribbica, respectively, produce them. For isolates eventually identified as P. guilliermondii (n = 33) or P. caribbica (n = 18), different carbon assimilation profiles were obtained (eight and nine, respectively), some of which were shared by both species, one being a frequent code for both (see Table S2 in the supplemental material). Each isolate of D. hansenii had a unique profile that was not found for any of the P. guilliermondii and P. caribbica isolates. An unweighted pair-group method for arithmetic averages tree was constructed by using the significant carbon sources of the ID32C assimilation pattern for the 62 clinical and environmental isolates (see Fig. S1 in the supplemental material).
TABLE 2.
Percentage of the clinical and environmental isolates of P. guilliermondii, P. caribbica, and D. hansenii forming pseudohyphae and exhibiting growth in the presence of discriminatory carbon sourcesa
| Carbon source | % Species
|
||
|---|---|---|---|
| P. guilliermondii (n = 33) | P. caribbica (n = 18) | D. hansenii (n = 3) | |
| Cycloheximide (0.01%) | 97 | 94 | 0 |
| dl-Lactate | 0 | 0 | 100 |
| d-Xylose | 100 | 100 | 66 |
| Ribose | 6 | 33 | 33 |
| Ramnose | 9 | 22 | 66 |
| Erythritol | 0 | 0 | 66 |
| Melibiose | 74 | 22 | 0 |
| Glucuronate | 0 | 0 | 66 |
| Gluconate | 0 | 0 | 33 |
| Lactose | 3 | 0 | 66 |
| Sorbose | 94 | 100 | 100 |
| Esculine | 91 | 89 | 66 |
| Pseudohyphae | 83 | 94 | 0 |
n, Number of isolates.
MICs of amphotericin B and flucytosine were similar for all P. guilliermondii, P. caribbica, and D. hansenii isolates (data not shown). The MICs of fluconazole (range, 0.124 to 2 μg/ml), and to a lesser degree those of voriconazole (<0.015 μg/ml) and caspofungin (<0.015 to 0.03 μg/ml), were lower for D. hansenii than those determined for P. guilliermondii (MIC50/MIC90 [range] = 8/64 μg/ml [2 to ≥64] for fluconazole, 0.06/0.5 μg/ml [0.03 to 2] for voriconazole, and 0.125/1 μg/ml [0.014 to 2] for caspofungin) and for P. caribbica (MIC50/MIC90 [range] = 4/64 μg/ml [1 to ≥64] for fluconazole, 0.125/0.25 μg/ml [0.06 to ≥8] for voriconazole, and 0.125/0.25 μg/ml [0.03 to 0.5] for caspofungin) isolates.
DISCUSSION
D. hansenii (teleomorph of C. famata) has been repeatedly associated with catheter-related bloodstream infection and rarely with other infections (4, 12, 24, 25, 27, 31). However, in most of the reported cases, the species was identified by using phenotypic parameters. Based on our results and on previous reports, one may wonder whether some of these cases could not have been misidentified. In our study, 36 of 62 (58%) of the isolates classified as D. hansenii or P. guilliermondii were misidentified. Isolates initially identified as D. hansenii belong to six different species (D. hansenii, P. guilliermondii, P. caribbica, C. palmioleophila, C. haemulonii type II, and C. lusitaniae). Among the 36 isolates initially identified as P. guilliermondii, 12 belong to the very closely related species P. caribbica and one was P. jadinii (Table 1). D. hansenii may thus be a frequent species of the gut microflora in individuals who like cheese. It does not seem to represent a frequent cause of fungemia. Whether nucleotide sequencing was used for the identification of the isolates to the species level in the ARTEMIS study reporting up to 0.5 and 1% of C. famata and C. guilliermondii among isolates responsible for invasive candidiasis is unknown. In the current active surveillance program on yeast fungemia in the Paris area, no cases were due to D. hansenii, whereas P. guilliermondii and P. caribbica were recovered in, respectively, 1 and 0.5% of the 1975 cases recorded thus far over the 5-year survey period (YEASTS program, unpublished data).
D. hansenii is an uncommon yeast characterized by its cryo- and halotolerance (6), but these criteria, as well as the production of ascospores, are not used in routine yeast identification. Analysis of carbon assimilation patterns showed that, in contrast to what is found for the majority of P. guilliermondii and P. caribbica isolates, all isolates of D. hansenii assimilated dl-lactate but were unable to produce pseudohyphae and were susceptible to 0.01% cycloheximide (Table 2) (3). No single D. hansenii carbon assimilation profile was shared with the other species studied here. Furthermore, there was a trend toward lower azole and caspofungin MICs than for P. guilliermondii, a finding not reported for echinocandins by Pfaller et al. (21).
P. guilliermondii is a genetically heterogeneous complex comprising several phenotypically indistinguishable taxa that have been brought into synonymy (3, 13, 34) or assigned new names (30). One of these synonymous species is Candida fermentati, described by Bai in 1996 (1). Changes in nomenclature could also explain additional mistakes such as the P. jadinii synonym of C. guilliermondii var. nitratophila. The teleomorphic state of C. fermentati has been recently described by Vaughan-Martini in 2005 based on molecular data and is named Pichia caribbica (34). Since P. caribbica has been described, many analyses using molecular tools (RAPD [random(ly) amplified polymorphic DNA], electrophoretic karyotyping, DNA composition, chromosomal DNA banding profiles, and D1/D2 sequences) reclassified clinical and environmental isolates phenotypically identified as P. guilliermondii into several species, including P. guilliermondii but also P. caribbica (2, 14, 30, 34). In our analyses, some carbon assimilation profiles were shared by the two species, one of the codes being a frequent code for both (see Table S2 in the supplemental material), and the similarity of the ITS and 26S sequences was greater than 98%.
In 1997, Nishikawa et al. designed specific primers in the large subunit region to differentiate D. hansenii and P. guilliermondii (17). Lan and Xu in 2006 proposed the sequencing of the RIBO gene to differentiate P. caribbica and P. guilliermondii, but the sequences seem too divergent within the same species (14). Recently, Tsui et al. (33) demonstrated that a polygenic sequencing method is best able to differentiate closely related species such as P. guilliermondii and P. caribbica. Here, sequencing of the ITS and 26S regions was sufficient, but using both enhances the chances of differentiation.
We also want to underline another pitfall related to these species' identification. Sequences recorded before 2004 and the complete sequencing of D. hansenii genome by Dujon et al. (10) in the GenBank database are sometimes erroneous. For example, the ITS sequence of strain CBS8417 (accession no. AF209874) deposited in 2001 as D. hansenii var. fabryii was reidentified in the CBS database (http://www.cbs.knaw.nl/yeast/BioloMICS.aspx) as P. guilliermondii in 2006. In the same way, the ITS sequence of strain BPY-01 recorded in GenBank as D. hansenii var. fabryii in 2002 (accession no. AY125962) is homologous with the ITS sequence of P. guilliermondii type strain CBS566. This underlines the importance of using databases such as that of the CBS, where taxonomy is more reliable than in public repositories as pointed out by Nilsson et al. (16).
In conclusion, D. hansenii may not be as common a human pathogen as previously thought. Sequencing of either the ITS or the D1/D2 region is a good tool for differentiating the species more frequently confused with D. hansenii, keeping in mind that reliable databases should be used.
Supplementary Material
Acknowledgments
We thank the Institut de Veille Sanitaire for its financial support. The technical help of the sequencing facilities and specifically that of Isabelle Iteman, Anne-Sophie Delannoy, Rachel Lavenir, and Laure Diancourt (PF-8) is gratefully acknowledged.
Footnotes
Published ahead of print on 13 August 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bai, F. Y. 1996. Separation of Candida fermentati comb. nov. from Candida guilliermondii by DNA base composition and electrophoretic karyotyping. Syst. Appl. Microbiol. 19178-181. [Google Scholar]
- 2.Bai, F. Y., H. Y. Liang, and J. H. Jia. 2000. Taxonomic relationships among the taxa in the Candida guilliermondii complex, as revealed by comparative electrophoretic karyotyping. Int. J. Syst. Evol. Microbiol. 50(Pt. 1)417-422. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, J. A., R. W. Payne, and D. Yarrow. 2000. Yeasts: characteristics and identification, 3rd ed. Cambridge, University Press, Cambridge.
- 4.Carrega, G., G. Riccio, L. Santoriello, M. Pasqualini, and R. Pellicci. 1997. Candida famata fungemia in a surgical patient successfully treated with fluconazole. Eur. J. Clin. Microbiol. Infect. Dis. 16698-699. [DOI] [PubMed] [Google Scholar]
- 5.Casal, M., and M. J. Linares. 1981. The comparison of six media for chlamydospore production by Candida albicans. Mycopathologia 76125-128. [DOI] [PubMed] [Google Scholar]
- 6.Corredor, M., A. M. Davila, S. Casaregola, and C. Gaillardin. 2003. Chromosomal polymorphism in the yeast species Debaryomyces hansenii. Antonie van Leeuwenhoek 8481-88. [DOI] [PubMed] [Google Scholar]
- 7.Cuenca-Estrella, M., C. B. Moore, F. Barchiesi, J. Bille, E. Chryssanthou, D. W. Denning, J. P. Donnelly, F. Dromer, B. Dupont, J. H. Rex, M. D. Richardson, B. Sancak, P. E. Verweij, J. Rodríguez-Tudela, et al. 2003. Multicenter evaluation of the reproducibility of the proposed antifungal susceptibility testing method for fermentative yeasts of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (AFST-EUCAST). Clin. Microbiol. Infect. 9467-474. [DOI] [PubMed] [Google Scholar]
- 8.de Hoog, G. S., and G. A. H. van den Ende. 1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41183-189. [DOI] [PubMed] [Google Scholar]
- 9.Diekema, D. J., S. A. Messer, A. B. Brueggemann, S. L. Coffman, G. V. Doern, L. A. Herwaldt, and M. A. Pfaller. 2002. Epidemiology of candidemia: 3-year results from the emerging infections and the epidemiology of Iowa organisms study. J. Clin. Microbiol. 401298-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 43035-44.15229592 [Google Scholar]
- 11.Gupta, A., H. Mi, C. Wroe, B. Jaques, and D. Talbot. 2006. Fatal Candida famata peritonitis complicating sclerosing peritonitis in a peritoneal dialysis patient. Nephrol. Dial. Transplant. 212036-2037. [DOI] [PubMed] [Google Scholar]
- 12.Krcmery, V., and A. Kunova. 2000. Candida famata fungemia in a cancer patient: case report. J. Chemother. 12189-190. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman, C. P., and J. W. Fell. 1998. The yeasts: a taxonomic study. Elsevier Science B.V., Amsterdam, The Netherlands.
- 14.Lan, L., and J. Xu. 2006. Multiple gene genealogical analyses suggest divergence and recent clonal dispersal in the opportunistic human pathogen Candida guilliermondii. Microbiology 1521539-1549. [DOI] [PubMed] [Google Scholar]
- 15.Masclaux, F., E. Gueho, G. S. de Hoog, and R. Christen. 1995. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J. Med. Vet. Mycol. 33327-338. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson, R. H., M. Ryberg, E. Kristiansson, K. Abarenkov, K. H. Larsson, and U. Köljalg. 2006. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLos ONE 1e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishikawa, A., T. Sugita, and T. Shinoda. 1997. Differentiation between Debaryomyces hansenii/Candida famata complex and Candida guilliermondii by polymerase chain reaction. FEMS Immunol. Med. Microbiol. 19125-129. [DOI] [PubMed] [Google Scholar]
- 18.Nishikawa, A., H. Tomomatsu, T. Sugita, R. Ikeda, and T. Shinoda. 1996. Taxonomic position of clinical isolates of Candida famata. J. Med. Vet. Mycol. 34411-419. [DOI] [PubMed] [Google Scholar]
- 19.Odds, F. C. 1988. Ecology of Candida and epidemiology of candidosis, p. 68-82. In Candida and candidosis: a review and bibliography, 2nd ed. Baillière Tindall, London, England.
- 20.O'Donnell, K. 1993. Fusarium and its near relatives, p. 225-233. In D. R. Reynolds and J. W. Taylor (ed.), The fungal holomorph: mitotic, meiotic, and pleomorphic speciation in fungal systematics. CAB International, Wallingford, United Kingdom.
- 21.Pfaller, M. A., L. Boyken, R. J. Hollis, J. Kroeger, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2008. In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance. J. Clin. Microbiol. 46150-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaller, M. A., D. J. Diekema, M. Mendez, C. Kibbler, P. Erzsebet, S. C. Chang, D. L. Gibbs, and V. A. Newell. 2006. Candida guilliermondii, an opportunistic fungal pathogen with decreased susceptibility to fluconazole: geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J. Clin. Microbiol. 443551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prinsloo, B., G. F. Weldhagen, and R. W. Blaine. 2003. Candida famata central nervous system infection. S. Afr. Med. J. 93601-602. [PubMed] [Google Scholar]
- 25.Quindos, G., F. Cabrera, M. C. Arilla, A. Burgos, R. Ortiz-Vigon, J. L. Canon, and J. Ponton. 1994. Fatal Candida famata peritonitis in a patient undergoing continuous ambulatory peritoneal dialysis who was treated with fluconazole. Clin. Infect. Dis. 18658-660. [DOI] [PubMed] [Google Scholar]
- 26.Quiros, M., P. Martorell, M. J. Valderrama, A. Querol, J. M. Peinado, and M. I. de Siloniz. 2006. PCR-RFLP analysis of the IGS region of rDNA: a useful tool for the practical discrimination between species of the genus Debaryomyces. Antonie van Leeuwenhoek 90211-219. [DOI] [PubMed] [Google Scholar]
- 27.Rao, N. A., A. V. Nerenberg, and D. J. Forster. 1991. Torulopsis candida (Candida famata) endophthalmitis simulating Propionibacterium acnes syndrome. Arch. Ophthalmol. 1091718-1721. [DOI] [PubMed] [Google Scholar]
- 28.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 191572-1574. [DOI] [PubMed] [Google Scholar]
- 29.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 332913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.San Millan, R. M., L. C. Wu, I. F. Salkin, and P. F. Lehmann. 1997. Clinical isolates of Candida guilliermondii include Candida fermentati. Int. J. Syst. Bacteriol. 47385-393. [DOI] [PubMed] [Google Scholar]
- 31.St-Germain, G., and M. Laverdiere. 1986. Torulopsis candida, a new opportunistic pathogen. J. Clin. Microbiol. 24884-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsui, C. K., H. M. Daniel, V. Robert, and W. Meyer. 2008. Re-examining the phylogeny of clinically relevant Candida species and allied genera based on multigene analyses. FEMS Yeast Res. 8651-659. [DOI] [PubMed] [Google Scholar]
- 34.Vaughan-Martini, A., C. P. Kurtzman, S. A. Meyer, and E. B. O'Neill. 2005. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 5463-469. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, D., A. Sander, H. Bertz, J. Finke, and W. V. Kern. 2005. Breakthrough invasive infection due to Debaryomyces hansenii (teleomorph Candida famata) and Scopulariopsis brevicaulis in a stem cell transplant patient receiving liposomal amphotericin B and caspofungin for suspected aspergillosis. Infection 33397-400. [DOI] [PubMed] [Google Scholar]
- 36.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p. 315-322. In M.A Innis et al. (ed.), PCR protocols. Academic Press, Inc., San Diego, CA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.