Abstract
The performance of a plasma real-time PCR (cytomegalovirus [CMV] PCR kit; Abbott Diagnostics) was compared with that of the antigenemia assay for the surveillance of active CMV infection in 42 allogeneic hematopoietic stem cell transplantation (Allo-SCT) recipients. A total of 1,156 samples were analyzed by the two assays. Concordance between the two assays was 82.2%. Plasma DNA levels correlated with the number of pp65-positive cells, particularly prior to the initiation of preemptive therapy. Fifty-seven episodes of active CMV infection were detected in 37 patients: 18 were defined solely by the PCR assay and four were defined on the basis of the antigenemia assay. Either a cutoff of 288 CMV DNA copies/ml or a 2.42-log10 increase of DNAemia levels between two consecutive PCR positive samples was an optimal value to discriminate between patients requiring preemptive therapy and those not requiring therapy on the basis of the antigenemia results. The real-time PCR assay allowed an earlier diagnosis of active CMV infection and was a more reliable marker of successful clearance of CMV from the blood. Analysis of the kinetics of DNAemia levels at a median of 7 days posttreatment allowed the prediction of the response to CMV therapy. Two patients developed CMV colitis. The PCR assay tested positive both before the onset of symptoms and during the disease period. The plasma real-time PCR from Abbott is more suitable than the antigenemia assay for monitoring active CMV infection in Allo-SCT recipients and may be used for guiding preemptive therapy in this clinical setting.
Preemptive antiviral therapy based on the detection of cytomegalovirus (CMV) in blood has been shown to significantly reduce the incidence of CMV disease in the early posttransplant period following allogeneic hematopoietic stem cell transplantation (Allo-SCT) (2, 7). The pp65 antigenemia assay has been adopted by many transplant centers as the “gold standard” for monitoring active CMV infection and guiding preemptive therapy in this clinical setting. The antigenemia assay, however, requires rapid processing of specimens, is labor-intensive, cannot be automated, is subjective in reading, and is unfeasible during periods of severe neutropenia. In addition, it does not reflect the viral load in the blood compartment accurately (12) and sometimes displays negative results in the presence of CMV disease (2, 31, 32, 36).
Quantification of CMV DNA in blood by PCR is emerging as an alternative to the antigenemia assay and may soon become the standard for the surveillance of CMV infection in Allo-SCT recipients. Viral load quantification in blood allows early detection of active CMV infection, close monitoring of the response to antivirals, prediction of the risk of viremic relapse, emergence of resistant strains, and eventual development of CMV disease (8, 31). In addition, it may be safely used for guiding preemptive therapy (11, 16, 23, 24, 32, 37). Nevertheless, there is some controversy as to which is the optimal clinical specimen (plasma, whole blood, or leukocytes) for these purposes. In fact, discrepant results have been published on this subject. (1, 4, 6, 9, 10, 19, 21, 27, 31, 35, 38, 39, 40). Recent data on the performance of several highly sensitive real-time PCRs, both laboratory-developed and commercial assays (5, 20, 22), however, demonstrate that both plasma and whole blood are equally suitable for monitoring active CMV infection in Allo-SCT recipients.
Quantitative assays based on real-time PCR technology offer some advantages over competitive PCRs, including a higher dynamic range, precision, accuracy, and reproducibility and a shorter turnaround time. A number of laboratory-developed real-time PCR assays have been evaluated for monitoring CMV infection in Allo-SCT recipients (3, 14, 17, 18, 20, 22, 23, 25, 27-30, 32, 33, 35, 36, 41, 43). These assays, however, are not well standardized and use different target sequences, primer sets, and extraction and detection methods, which result in different analytical performances. This makes it difficult to compare studies conducted at different centers. A few commercial real-time PCRs are available (5, 13, 15, 42), and these have not been extensively evaluated with Allo-SCT recipients. In the present study a commercially available real-time plasma PCR assay (CMV PCR kit; Abbott Diagnostics, Des Plaines, IL) coupled with fully automated DNA extraction was evaluated for CMV DNA detection in plasma and compared with the antigenemia assay for monitoring active CMV infection in Allo-SCT recipients. We also addressed whether a strategy based on the quantification of CMV DNA in plasma could be established for triggering the initiation of CMV preemptive therapy in this clinical setting.
MATERIALS AND METHODS
Patients.
A total of 42 consecutive patients undergoing Allo-SCT at the bone marrow transplantation unit of the Hematology and Oncology Service of the University Clinic Hospital of Valencia, Spain, between January 2006 and November 2007 were included in the study. All patients gave their informed consent to participate in the study. Table 1 summarizes the clinical characteristics of the patients. From the time of hospital admission onwards, patients were given acyclovir (800 mg/three times daily orally) until day +30. CMV-seropositive (R+) or CMV-seronegative (R−) recipients receiving a graft from an unrelated HLA-mismatched CMV seropositive (D+) donor were treated with acyclovir at 500 mg/m2/8 h orally or intravenously (i.v.). No CMV-specific prophylaxis was given to D+/R− Allo-SCT recipients.
TABLE 1.
Characteristics of patients
| Parameter | Value |
|---|---|
| Total no. of patients | 42 |
| Median age, yrs (range) | 46 (18-70) |
| Sex, no. of male patients/no. of female patients | 22/20 |
| Diagnosis, no. (%) of patients | |
| Acute myeloid leukemia | 14 (33.3) |
| Acute lymphoblastic leukemia | 5 (11.9) |
| Chronic myeloid leukemia | 2 (4.8) |
| Primary myelofibrosis | 2 (4.8) |
| Myelodysplastic syndrome | 1 (2.4) |
| Non-Hodgkin's lymphoma | 6 (14.3) |
| Hodgkin's lymphoma | 1 (2.4) |
| Multiple myeloma | 4 (9.5) |
| Chronic lymphocytic leukemia | 3 (7.1) |
| Other | 4 (9.5) |
| CMV serology, no. (%) of patients | |
| D+/R+ | 20 (47.6) |
| D−/R+ | 16 (38.1) |
| D+/R− | 2 (4.8) |
| D−/R− | 4 (9.5) |
| Donor type, no. (%) of patients | |
| HLA-identical sibling | 18 (43.9) |
| Mismatched related donor | 1 (2.4) |
| Matched unrelated donor | 10 (24.4) |
| Mismatched unrelated donor | 12 (29.3) |
| Conditioning regimen, no. (%) of patients | |
| Nonmyeloablative | 27 (64.3) |
| Myeloablative | 15 (35.7) |
| Stem cell source, no. (%) of patients | |
| Peripheral blood | 31 (73,8) |
| Umbilical cord blood | 10 (23.8) |
| Bone marrow | 1 (2.4) |
| GvHDa prophylaxis, no. (%) of patients | |
| Cyclosporine plus methotrexate | 19 (45.2) |
| Cyclosporine plus mycophenolate mofetil | 11 (26.2) |
| Cyclosporine plus prednisone | 10 (23.8) |
| Other | 2 (4.8) |
| Acute GvHD incidence, no. (%) of patients | |
| Grades 0 to I | 24 (57.1) |
| Grades II to IV | 18 (42.9) |
GvHD, graft-versus-host disease.
Monitoring and management of active CMV infection.
The surveillance for active CMV infection was routinely performed with the pp65 antigenemia assay and the real-time PCR assay. Patients were monitored once a week during the first 100 days after transplant and every other week thereafter until day +180. During episodes of active CMV infections, patients were monitored twice or even three times a week when possible. An episode of active CMV infection was defined either by a single positive PCR, by antigenemia, or by both. Two consecutive negative results on both tests defined the end of a given episode. Preemptive therapy with oral valganciclovir (900 mg/12 h), i.v. ganciclovir (5 mg/kg of body weight/12 h), or i.v. foscarnet (60 mg/kg/12 h) was initiated upon a positive antigenemia result (≥1 pp65-positive cells/200,000 cells) as previously indicated (34). Preemptive therapy was discontinued after two consecutive negative antigenemia results after a minimum of 2 weeks of treatment. Persistence of positive antigenemia (≥3 weeks after initiation of therapy) was considered a treatment failure and prompted a change in the antiviral therapeutic schedule (either from an oral to an i.v. route, from valganciclovir to ganciclovir or foscarnet, or from ganciclovir to foscarnet or vice versa or a dose increment of the antiviral). The results of the PCR assay were not used for guiding preemptive therapy. CMV colitis was diagnosed on the basis of the clinical condition and the histological demonstration of CMV inclusions and the immunohistochemical detection of CMV proteins in tissue samples obtained at biopsy (26). CMV disease was treated with i.v. ganciclovir (5 mg/kg/12 h) for 21 days, followed by 5 mg/kg/day (5 days a week) until either day +90 or the resolution of the immunosuppressive condition, or i.v. foscarnet (60 mg/kg/8 h), following a similar schedule, as previously reported (34).
Virological assays.
Blood samples were obtained in EDTA-treated tubes and were processed within 2 h. Polymorphonuclear leukocytes (PMNLs) and plasma were separated by the standard dextran sedimentation method. The pp65 antigenemia assay was carried out by a standard immunofluorescence procedure as previously described (34). Results were reported as the number of pp65-positive cells/200,000 PMNLs. Real-time PCR with the Abbott CMV PCR kit (produced by Qiagen GmbH, Hilden, Germany, for Abbott Diagnostics, Des Plaines, IL; available in the United States as CMV ASR) was performed using the ABI PRISM 7000 system (Applied Biosystems Inc., Foster City, CA), or the m2000RT (Abbott Molecular) following the manufacturer's instructions. DNA extractions were performed using the Abbott mSample preparation system DNA kit on either the m1000 SP or the m2000 SP instrument (Abbott Molecular). DNA was extracted from 200 μl of plasma specimens. The analytical performance of the assay is similar regardless of the instrument used for either fluorescence quantification or DNA extraction. The CMV PCR kit contains reagents and enzymes for the specific amplification of a 105-bp region of the CMV major immediate-early gene. External positive controls (QS1, 6.54 log10 copies/μl; QS2, 5.54 log10 copies/μl; QS3, 4.54 log10 copies/μl; QS4, 3.54 log10 copies/μl) and an internal control (CMV IC) are supplied, which allow users to determine the viral load, control the efficiency of DNA isolation, and check for PCR inhibition. A 1/10 dilution of QS4 (QS5, 2.54 log10 copies/μl) was included in each run. According to the manufacturer, the limit of detection of the assay is approximately 25 CMV DNA copies/ml of plasma (95% probability that 25 copies/ml will be detected), and it shows a good linearity (R2 = 0.995), from 2.0 to 6.0 log10 CMV DNA copies/ml. The coefficient of variation of the assay at the lowest CMV DNA concentration (QS5) is approximately 10%.
Data analysis.
The Pearson test was used to assess the correlation coefficients between different variables. The Wilcoxon test was used for comparing paired values over time. The Mann-Whitney test and the Kruskal-Wallis test were used for comparing unpaired data from two groups and three or more groups, respectively. A P value of <0.05 was considered statistically significant. Receiver operating characteristic (ROC) plot analysis was performed to determine a threshold value of CMV DNA load in plasma for initiating preemptive treatment on the basis of the antigenemia assay (≥1 pp65-positive cells/200,000 PMNLs). The statistical analysis was performed with the aid of the statistical package SPSS (version 10.0). ROC curves were performed with the program Analyze-17 for Microsoft Excel (version 2.09).
RESULTS
Patients and samples.
Forty-two patients were included in the study. The patients were followe for a median of 192 days (range, 41 to 570 days). A median of 29 samples per patient (range, 9 to 60 samples) were available for analysis. Thirty-seven out of 42 patients (88.0%) developed one or more episodes of active CMV infection (19 D+/R+, 16 D−/R+, and 2 D+/R−). Twenty-nine of these patients received one or more courses of preemptive therapy upon a positive antigenemia result. Two patients (4.7%) developed CMV end-organ disease (colitis in both cases).
Performance of the plasma real-time PCR assay and the pp65 antigenemia assay for detection of active CMV infection.
A total of 1,220 samples were analyzed, both assays being performed on 1,156 samples. Sixty-four samples could not be analyzed by the antigenemia assay due to low neutrophil counts. CMV was detected by both assays in 176 samples (15.2%), and results were negative for 775 samples (67.0%). In addition, 201 samples (17.3%) tested positive in the PCR assay but negative in the antigenemia assay. To rule out the possibility of false-positive PCR results, different aliquots of a number of these samples were assayed. These specimens repeatedly tested positive. Four samples (0.3%) were positive in the antigenemia assay but negative in the PCR assay. These specimens were reanalyzed by PCR using different aliquots and found to actually be negative (the internal control was amplified, ruling out the possibility of PCR inhibition or inefficient DNA extraction). The concordance between the two assays was 82.2%. The data indicated that the plasma real-time PCR assay is more sensitive than the antigenemia assay (98.9% versus 47.2%) for detection of CMV in blood (if the specificities of both assays are considered to be 100%).
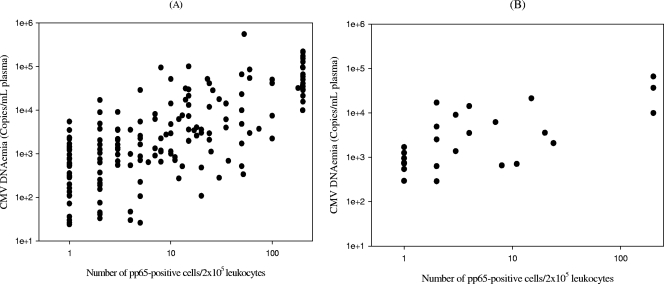
For samples testing positive in the PCR assay, plasma CMV DNA copy numbers increased significantly (P < 0.001 by the Kruskal-Wallis test) with the antigenemia values. As shown in Fig. 1A, overall CMV DNAemia levels correlated significantly with the number of pp65-positive cells (r = 0.501, P < 0.001, by the Pearson test). A higher correlation (r = 0.789, P < 0.001, by the Pearson test) was observed when only samples obtained before the initiation of preemptive therapy were considered for analysis (Fig. 1B).
FIG. 1.
Scatter diagrams (log10 scale) show the correlation between plasma CMV DNA levels and pp65 antigenemia values (number of pp65-positive cells/200,000 leukocytes examined) among all samples testing positive by the real-time PCR assay and the antigenemia assay during the study period (r = 0.501, P < 0.001, by the Pearson test) (A) and among samples drawn prior to the initiation of preemptive therapy testing positive by both methods (r = 0.789, P < 0.001, by the Pearson test) (B).
Detection of active CMV infection episodes by plasma real-time PCR and by the pp65 antigenemia assay.
Fifty-seven episodes of active CMV infection were detected in 37 patients. Thirteen out of the 37 patients experienced recurrent episodes (nine patients had two episodes, two patients had three episodes, and two patients experienced four episodes). A positive PCR was the only marker of active CMV infection in 18 episodes (in 16 patients). In 16 of these episodes, two or more consecutive samples tested positive. The remaining two episodes were defined by a single positive PCR result. Eight of the 16 patients displaying self-resolving PCR-positive/antigenemia-negative episodes did not develop recurrent episodes of CMV active infection. One patient had a second episode. PCR-positive/antigenemia-negative episodes in the remaining seven patients occurred following a prior PCR-positive/antigenemia-positive episode. No patients experiencing a PCR-positive/antigenemia-negative episode developed CMV end-organ disease.
Both assays eventually turned positive in 35 episodes. In 25 of these episodes, the first PCR-positive result preceded that of the antigenemia assay by a median of 13 days (range, 3 to 28 days). In the remaining 10 episodes both assays turned positive simultaneously. Four episodes in four different patients were diagnosed solely on the basis of the antigenemia assay. These episodes were defined by a single positive result (1 pp65-positive cell/200,000 PMNLs). The duration of the episodes diagnosed exclusively by PCR (median, 10 days; range, 6 to 60 days) was significantly (P < 0.0001, by the Mann-Whitney test) shorter than that of the episodes in which both assays turned positive (median, 59.1 days; range, 4 to 260 days, excluding the five episodes still active at the end of the study period). The antigenemia assay was intermittently negative during several of the latter episodes once preemptive therapy was initiated, whereas the PCR assay was consistently positive.
Guiding preemptive therapy on the basis of plasma CMV DNA quantification.
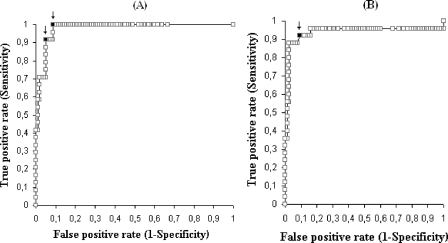
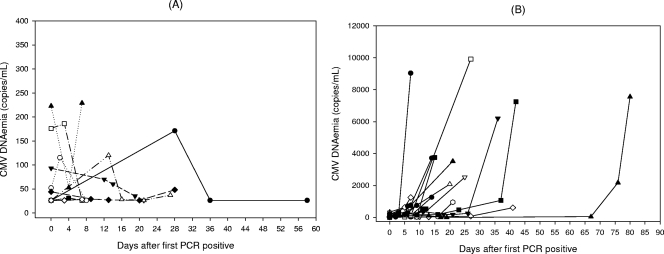
Occurrence of a positive antigenemia result triggers the initiation of preemptive anti-CMV therapy in our center. This strategy was shown to result in low incidence of CMV disease (34). In order to establish a corresponding CMV DNA copy number threshold value, we compared plasma DNA levels present at the moment of initiation of preemptive therapy with DNAemia levels found in patients with self-clearing infections (PCR positive/antigenemia negative) and those measured in PCR-positive/antigenemia-positive episodes prior to positive conversion of the antigenemia assay. The optimal DNAemia level cutoff was calculated using ROC curves for an antigenemia value of ≥1 pp65-positive cells/200,000 PMNLs (Fig. 2A). The threshold of 288 copies/ml was optimal to discriminate between patients who required preemptive therapy and those who did not on the basis of the antigenemia assay (sensitivity, 100%; specificity, 91.2%; negative predictive value [NPV], 100%; positive predictive value [PPV], 72.7%). Raising the cutoff value to 550 copies/ml increased the specificity and the PPV (95.1% and 81.4%, respectively) but decreased the sensitivity and the NPV (91.7% and 97.9%, respectively). As a complementary approach, we compared the kinetics of plasma DNA levels in PCR-positive/antigenemia-negative episodes of active CMV infection (n = 16, with two or more positive PCR results) with the kinetics of those in PCR-positive/antigenemia-positive episodes prior to initiation of preemptive therapy (n = 25, PCR-positive results preceding that of the antigenemia assay). Plasma DNA levels either decreased or remained relatively stable until clearance in the PCR-positive/antigenemia-negative episodes (Fig. 3A). Differences in DNAemia levels measured in consecutive samples within these episodes were not statistically significant (P > 0.5, by the Wilcoxon test). Contrarily, a significant (P < 0.001, by the Wilcoxon test) DNAemia level increase was observed in the PCR-positive/antigenemia-positive episodes concomitantly with the positive conversion of the antigenemia assay (Fig. 3B shows representative examples). ROC curve analysis data indicated that a DNAemia level increase of 2.42 log10 (266 copies/ml) between two consecutive positive PCR samples drawn a median of 7 days apart (range, 1 to 21 days) was the optimal value (sensitivity, 92%; specificity, 90.1%, PPV, 79.3%; NPV, 96.4%) for discriminating between patients who required preemptive therapy and those who did not (Fig. 2B).
FIG. 2.
ROC curves for an antigenemia value of ≥1 pp65-positive cells/200,000 PMNLs for establishing the optimal DNAemia level cutoff (A) and DNAemia level increase threshold between two consecutive PCR-positive samples obtained a median of 1 week apart (B) for triggering the initiation of anti-CMV preemptive therapy. Arrows in panel A point to the proposed DNAemia level thresholds (the left arrow corresponds to a value of 550 CMV DNA copies/ml, and the right arrow corresponds to a value of 288 CMV DNA copies/ml). The arrow in panel B points to a value of 2.42 log10 CMV DNA copies/ml.
FIG. 3.
(A) Kinetics of plasma CMV DNA load in self-clearing (PCR-positive/antigenemia-negative) episodes of active CMV infection defined by two or more PCR positive results (n = 16). (B) Kinetics of plasma CMV DNA load in 18 representative PCR-positive/antigenemia-positive episodes in which PCR-positive results preceded those of the antigenemia assay, until positive conversion of the antigenemia assay.
Monitoring of the response to CMV preemptive therapy by plasma real-time PCR and by the pp65 antigenemia assay.
Negative conversion of the antigenemia assay after initiation of preemptive therapy preceded that of the PCR assay in 20 out of 35 episodes of active CMV infection. Both assays turned negative at the same time point in 10 episodes. The remaining five episodes were still active at the end of the study period. Thus, no patient achieved a negative PCR result earlier than a negative antigenemia result. The median time intervals to obtain a negative PCR result and a negative antigenemia result after initiation of preemptive therapy were 28 days (range, 4 to 163 days) and 15.5 days (range, 2 to 123 days), respectively (P < 0.001, by the Wilcoxon test). We also investigated whether quantification of DNAemia levels early after initiation of preemptive therapy may predict the response to anti-CMV treatment. Data are shown in Table 2. A significant decrease (P < 0.001, by the Wilcoxon test) in the number of plasma CMV DNA copies was observed at a median of 7 days (range, 3 to 10 days) after initiation of therapy in 23 episodes. A significant increase (P = 0.007, by the Wilcoxon test) in CMV DNAemia levels was observed at the same time point (median, 7 days; range, 3 to 20 days) in the remaining 12 episodes. The duration of the latter episodes was significantly longer than that of the former episodes (P = 0.04, by the Mann-Whitney test, if defined by PCR, and P = 0.0005, by the Mann-Whitney test, if determined by the antigenemia assay) and required a change in the treatment more often (83% versus 19% of patients). The magnitude of the increase in CMV DNAemia load was significantly correlated (r = 0.634; P = 0.004, by the Pearson test) with the duration of the episode, as defined by the PCR assay. Contrarily, the magnitude of the decrease in the CMV DNAemia load was not significantly correlated with the duration of the episode (r = 0.1, P > 0.5, by the Pearson test). A reasonable correlation (r = 0.684, P < 0.001, by the Pearson test) between plasma CMV DNA levels at the moment of initiation of preemptive therapy and the duration of the episode as determined by PCR results was observed.
TABLE 2.
Plasma CMV DNA levels early after initiation of preemptive therapy and duration of episode of active CMV infection
| Episode of active CMV infection | DNAemia, median (range) no. of copies/ml, at time:
|
Episode duration, median (range) no. of days, determined by assay:
|
||
|---|---|---|---|---|
| Initiation of therapy | 1 wk (median) after therapy | PCR | Antigenemia | |
| Decreasing DNAemia (n = 23)a | 2,041 (33-99,898) | 111 (<25-3,078) | 21 (4-112) | 8 (2-100) |
| Increasing DNAemia (n = 12)b | 4,232 (288-169,487) | 11,727 (654-212,330) | 47 (17-570) | 35 (17-570) |
Decreasing CMV DNAemia levels at a median of 7 days (range, 3 to 10 days) after initiation of preemptive therapy.
Increasing CMV DNAemia levels at a median of 7 days (range, 3 to 20 days) after initiation of preemptive therapy. Dates of the last available positive sample were considered for the analysis of ongoing episodes (n = 5) at the end of the study period.
Performance of the plasma real-time PCR assay and the pp65 antigenemia assay in patients developing CMV disease.
Two patients developed CMV colitis during the study period despite initiation of preemptive therapy. The PCR assay tested consistently positive from 128 days and 69 days, respectively, prior to onset of disease. The antigenemia assay tested intermittently positive from 43 days prior to onset of symptoms in one patient and negative in the other patient (the assay result became positive during the disease period). Peak CMV DNAemia levels (5.2 log10 and 5.7 log10 copies/ml, respectively, corresponding to antigenemias of 200 pp65-positive cells/200,000 PMNLs) were reached within the symptomatic phase of the disease. Peak CMV DNAemia load in patients with CMV disease (median, 5.56 log10 copies/ml) was significantly higher (P = 0.028, by the Mann-Whitney test) than that reached by patients not developing CMV disease (median, 3.78 log10 copies/ml; range, 2.6 log10 to 5.74 log10 copies/ml). Of interest, one patient did not develop CMV disease and yet reached a peak DNAemia level of 5.74 log10 copies/ml.
DISCUSSION
In this study a commercially available plasma real-time PCR assay (CMV real-time PCR kit; Abbott Diagnostics) coupled with a fully automated DNA extraction was evaluated for the surveillance of active CMV infection in a cohort of adult Allo-SCT recipients. The Abbott assay has been previously shown to be more consistent than the ultrasensitive Cobas Amplicor Monitor in detecting low DNA levels in plasma (42). An equivalent test (Qiagen RealArt CMV LightCycler PCR; Qiagen, Germantown, MD) has also been shown to compare favorably with the Roche CMV UL54 analyte-specific reagent (Roche Diagnostics, Indianapolis, IN) and the Digene hybrid capture system cytomegalovirus DNA (version 2.0; Digene Corporation, Gaithersburg, MD) in terms of sensitivity (15). In another study, identical performances for the Qiagen real-time PCR, the Abbott test, and the ultrasensitive Amplicor CMV monitor were reported (5). Nevertheless, to the best of our knowledge, the performance of the Abbott plasma real-time PCR assay in comparison with that of the antigenemia assay for monitoring active CMV infection in Allo-SCT recipients has not been assessed. In the present study, the PCR assay proved to be much more sensitive than the antigenemia assay for detecting CMV in blood (sensitivity, 98.8% versus 45.2%). The PCR assay detected more episodes of active CMV infection than did the antigenemia assay. In fact, 18 episodes were defined solely on the basis of positive results with the PCR assay, whereas only four PCR-negative/antigenemia-positive episodes were observed. The latter episodes were defined by a single positive antigenemia assay (one pp65-positive cell/200,000 PMNLs). Our data are in accordance with those reported in earlier studies (3, 20, 22, 28, 29, 35, 36, 41) evaluating the performance of several laboratory-developed plasma real-time PCR assays in comparison with that of the antigenemia assay. The concordance between the antigenemia assay and the Abbott real-time PCR assay results was 82%, a percentage slightly lower than that previously reported by our group (34) for a qualitative plasma PCR (90%). The real-time PCR used in the current study is more sensitive than the Amplicor CMV test, which may account for this minimal difference. In our study, the CMV DNAemia load increased proportionally with the antigenemia levels, confirming previously reported data (3, 20, 22, 28, 36, 41). Nevertheless, a modest correlation between plasma CMV DNA levels and the number of pp65-positive cells was found, and samples yielding highly discrepant results were not uncommon, particularly after the initiation of preemptive therapy. In this sense, we observed a more robust correlation between the two assays when only pretreatment specimens were considered for analysis. Differences in the kinetics of DNAemia and antigenemia clearance after anti-CMV therapy may account for this finding.
The antigenemia assay has been adopted by many transplant centers as the method of choice for guiding preemptive therapy in Allo-SCT recipients. In our center, preemptive therapy is initiated upon a positive antigenemia result (≥1 pp65-positive cells/200,000 PMNLs) with an incidence of CMV disease lower than 5% (34). The occurrence of two consecutive positive PCR tests performed on whole blood has also been shown to be a clinically safe approach for triggering the initiation of preemptive therapy (7, 21). Currently, however, there are no consensus criteria for the initiation of anti-CMV preemptive therapy on the basis of a quantitative PCR monitoring strategy in this clinical setting. Several DNAemia level thresholds, ranging from 1,000 to 10,000 copies/ml of whole blood, as determined by laboratory-developed real-time PCR assays, have been prospectively evaluated and found to be clinically safe (11, 16, 24, 32, 37). To date, no DNA level cutoffs for plasma specimens have been clinically validated. If the decision to initiate preemptive therapy in our cohort had been based upon either the first detection of CMV DNA in plasma or the occurrence of two consecutive positive PCR results, 18 and 16 episodes of self-resolving active CMV infections, respectively, would have been unnecessarily treated. Thus, in accordance with data previously published by our group (34), the initiation of preemptive therapy on the basis of qualitative PCR results would lead to overtreatment. In order to determine the optimal DNAemia level cutoff for triggering the initiation of preemptive therapy, a ROC curve for an antigenemia value of ≥1 pp65-positive cells/200,000 PMNLs was calculated. The analysis of data indicated that a CMV DNAemia level of 288 copies/ml was a convenient value for discriminating between self-clearing infections and those requiring preemptive therapy. With this strategy, none of the self-resolving (PCR-positive/antigenemia-negative) episodes would have been treated, and all episodes treated on the basis of the antigenemia results would also have been treated; however, six episodes would have been overtreated (initiation of therapy a median of 7 days earlier). Raising the DNAemia level threshold to 550 copies/ml would have resulted in the following: (i) all episodes treated on the basis of the antigenemia results would also have been treated (four of them, however, a median of 7 days earlier and four of them, on the basis of the next PCR positive result, a median of 10 days later); (ii) none of the self-resolving (PCR-positive/antigenemia-negative) episodes would have been treated. Thus, the latter DNAemia threshold value may prove clinically safe. Several plasma DNA cutoff values, ranging from 200 to 10,000 copies/ml, have been suggested for triggering the initiation of anti-CMV preemptive therapy in Allo-SCT recipients (20, 25, 28, 36); however, none of these have been clinically validated. As an alternative approach, we investigated whether analysis of the kinetics of plasma CMV DNAemia could also be used for guiding preemptive therapy. As expected on the basis of the aforementioned DNAemia cutoff, ROC curve analysis of data indicated that a DNAemia level increase of 2.42 log10 (266 copies/ml) between two consecutive positive PCR samples drawn a median of 1 week apart is the optimal value for discriminating between patients who require preemptive therapy and those who do not. On the basis of this criterion, none of the self-resolving (PCR-positive/antigenemia-negative) episodes would have been treated preemptively, and all episodes treated on the basis of the antigenemia results would also have been treated (six of them a median of 7 days earlier and two of them a median of 7 days later).
According to our data, the real-time PCR assay displayed several advantages over the antigenemia assay for monitoring active CMV infection in Allo-SCT recipients. Firstly, in agreement with previously published data (3, 22, 35, 36, 41), the PCR assay allowed an earlier diagnosis of active CMV infection. Secondly, a negative PCR result was a more reliable marker of successful clearance of CMV from blood, since negative conversion of the antigenemia assay preceded that of the PCR assay in most episodes. Similar data have been previously published (2, 14, 22), suggesting that discontinuation of therapy upon a negative PCR result is probably safer than doing so on the basis of a negative antigenemia result. In support of this view, we observed intermittently negative antigenemias within several ongoing episodes of active CMV infection. Thirdly, analysis of the kinetics of CMV DNAemia—but not that of the antigenemia—either at the time of initiation of preemptive therapy or early thereafter may allow the prediction of the response to CMV treatment. In this sense, plasma CMV DNA levels at the moment of initiation of preemptive therapy directly correlated with the duration of the episode, as defined by PCR results. In addition, the duration of the episode was significantly shorter in those in which a rapid decrease (7 days after initiation of treatment) in the CMV DNAemia load was observed than in those with increasing DNAemia levels at this time point. Most of the latter episodes (83.3%) required a change in the antiviral therapy schedule. Thus, early assessment of the kinetics of CMV DNAemia after initiation of preemptive therapy may prove useful for deciding optimal therapeutic schedules on an individual basis.
A known drawback of the antigenemia assay is the lack of sensitivity for detecting CMV in blood prior to, or even during, CMV gastrointestinal disease (2). A similar concern has been raised about several in-house-developed plasma real-time PCRs (32, 36). Two patients of our cohort developed CMV colitis despite preemptive therapy. In both cases, the PCR assay tested consistently positive from 2 to 4 months prior to the onset of disease. The antigenemia assay, however, was positive for one patient but negative for the other (it became positive during the disease period). Thus, in agreement with an earlier report (3), the plasma real-time PCR appears to be suitable for the surveillance of active CMV infection in patients eventually developing CMV gastrointestinal disease. Interestingly, peak DNAemia levels in patients with CMV disease were significantly higher than those reached in patients not developing CMV disease. Nevertheless, one patient did not develop CMV disease and yet reached a peak DNAemia level comparable to that found in patients with disease. Thus, it is uncertain whether the presence of an ongoing CMV disease may be inferred on the basis of the number of CMV DNA copies in plasma.
In conclusion, our data indicate that the Abbott plasma real-time PCR is more suitable than the antigenemia assay for detecting CMV in blood and monitoring the response to anti-CMV treatment. A strategy for triggering the initiation of preemptive therapy on the basis of plasma CMV DNA loads was proposed. A comparative randomized clinical trial is nevertheless required to prove its efficacy in preventing the development of CMV disease.
Acknowledgments
This research was supported in part by FIS (Fondo de Investigación Sanitaria) (grant 06/1738).
We thank Julia García and Matilde Pastor for technical assistance.
Footnotes
Published ahead of print on 27 August 2008.
REFERENCES
- 1.Boeckh, M., G. M. Gallez-Hawkins, D. Myerson, J. Zaia, and R. A. Bowden. 1997. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic bone marrow transplantation. Transplantation 64108-113. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh, M., T. A. Gooley, D. Myerson, T. Cunningham, G. Schoch, and R. A. Bowden. 1996. Cytomegalovirus pp65 antigenemia-guided early treatment with ganciclovir versus ganciclovir at engraftment after allogeneic marrow transplantation: a randomized double-blind study. Blood 884063-4071. [PubMed] [Google Scholar]
- 3.Boeckh, M., M. Huang, J. Ferrenberg, T. Stevens-Ayers, L. Stensland, W. G. Nichols, and L. Corey. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 421142-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boivin, G., R. Belanger, R. Delage, C. Beliveau, C. Demers, N. Goyette, and J. Roy. 2000. Quantitative analysis of cytomegalovirus (CMV) viremia using the pp65 antigenemia assay and the COBAS AMPLICOR CMV MONITOR PCR test after blood and marrow allogeneic transplantation. J. Clin. Microbiol. 384356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caliendo, A. M., J. Ingersoll, A. M. Fox-Canale, S. Pargman, T. Bythwood, M. K. Hayden, J. W. Bremer, and N. S. Lurain. 2007. Evaluation of real-time PCR laboratory-developed test using analyte-specific reagents for cytomegalovirus quantification. J. Clin. Microbiol. 451723-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caliendo, A. M., K. St. George, S. Y. Kao, J. Allega, B. H. Tan, R. LaFontaine, L. Bui, and C. R. Rinaldo. 2000. Comparison of quantitative cytomegalovirus (CMV) PCR in plasma and antigenemia assay: clinical utility of the prototype AMPLICOR CMV MONITOR test in transplant recipients. J. Clin. Microbiol. 382122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Einsele, H., G. Ehninger, H. Hebart, K. M. Whittkowski, U. Schuler, G. Jahn, P. Mackes, M. Herter, T. Klingebiel, J. Löffler, S. Wagner, and C. A. Müller. 1995. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood 862815-2820. [PubMed] [Google Scholar]
- 8.Emery, V. C., C. A. Sabin, A. V. Cope, D. Gor, A. F. Hassan-Walker, and P. D. Griffiths. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 3552032-2036. [DOI] [PubMed] [Google Scholar]
- 9.Flexman, J., I. Kay, R. Fonte, R. Herrmann, E. Gabay, and S. Palladino. 2001. Differences between the quantitative antigenemia assay and the Cobas Amplicor Monitor quantitative PCR assay for detecting CMV viraemia in bone marrow and solid organ transplant recipients. J. Med. Virol. 64275-282. [DOI] [PubMed] [Google Scholar]
- 10.Gentile, G., A. Picardi, A. Capobianchi, A. Spagnoli, L. Cudillo, T. Dentamaro, A. Tendas, L. Cupelli, M. Ciotti, A. Volpi, S. Amadori, P. Martino, and P. de Fabritiis. 2006. A prospective study comparing quantitative cytomegalovirus (CMV) polymerase chain reaction in plasma and pp65 antigenemia assay in monitoring patients after allogeneic stem cell transplantation. BMC Infect. Dis. 6167-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerna, G., D. Lilleri, D. Caldera, M. Furione, L. Zenone-Bragotti, and E. P. Alessandrino. 2008. Validation of a DNAemia cutoff for preemptive therapy of cytomegalovirus in adult hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 41873-879. [DOI] [PubMed] [Google Scholar]
- 12.Gerna, G., M. Zavattoni, F. Baldanti, A. Sarasini, L. Chezzi, P. Grossi, and M. G. Revello. 1998. Human cytomegalovirus (HCMV) leukoDNAemia correlates more closely with clinical symptoms than antigenemia and viremia in heart and heart-lung transplant recipients with primary HCMV infection. Transplantation 651378-1385. [DOI] [PubMed] [Google Scholar]
- 13.Gouarin, S., A. Vabret, C. Scieux, F. Agbalika, J. Cherot C. Mengelle C. Deback, J. Petitjean, J. Dina, and F. Freymuth. 2007. Multicentric evaluation of a new commercial cytomegalovirus real-time PCR quantification assay. J. Virol. Methods 146147-154. [DOI] [PubMed] [Google Scholar]
- 14.Griscelli, F., M. Barrois, S. Chauvin, S. Lastere, D. Bellet, and J. H. Bourhis. 2001. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J. Clin. Microbiol. 394362-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson, K. E., L. B. Reller, J. Kurtzberg, M. Horwitz, G. Long, and B. D. Alexander. 2007. Comparison of the Digene hybrid capture system cytomegalovirus (CMV) DNA (version 2.0), Roche CMV UL54 analyte-specific reagent, and Qiagen RealArt CMV LightCycler PCR reagent test using AcroMetrix OptiQuant CMV DNA quantification panels and specimens from allogeneic-stem-cell transplant recipients. J. Clin. Microbiol. 451972-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington, S. M., R. S. Buller, G. A. Storch, L. Li, S. H. Fisher, P. R. Murray, and J. C. Gea-Banacloche. 2007. The effect of quantification standards used in real-time CMV PCR assays on guidelines for initiation of therapy in allogeneic stem cell transplant patients. Bone Marrow Transplant. 39237-238. [DOI] [PubMed] [Google Scholar]
- 17.Herrman, B., V. C. Larsson, C. J. Rubin, F. Sund, B. M. Eriksson, J. Arvidson, Z. Yun, K. Bondeson, and J. Blomberg. 2004. Comparison of a duplex real-time PCR assay and the COBAS Amplicor CMV monitor test for detection of cytomegalovirus. J. Clin. Microbiol. 421909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong, K. M., H. Najjar, M. Hawley, and R. D. Press. 2004. Quantitative real-time PCR with automated sample preparation for diagnosis and monitoring of cytomegalovirus infection in bone marrow transplant patients. Clin. Chem. 50846-856. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser, L., L. Perrin, B. Chapuis, K. Hadaya, L. Kolarova, C. Deffernez, S. Huguet, C. Helg, and W. Wunderli. 2002. Improved monitoring of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation by an ultrasensitive plasma DNA PCR assay. J. Clin. Microbiol. 404251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalpoe, J. S., A. C. M. Kroes, M. D. de Jong, J. Shinkel, C. S. de Brouwer, M. F. C. Beersma, and E. C. J. Claas. 2004. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J. Clin. Microbiol. 421498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lengerke, C., T. Ljubicic, C. Meisner, J. Loeffler, C. Sinzger, H. Einsele, and H. Hebart. 2006. Evaluation of the COBAS Amplicor CMV Monitor for early detection and monitoring of human cytomegalovirus infection after allogeneic stem cell transplantation. Bone Marrow Transplant. 3853-60. [DOI] [PubMed] [Google Scholar]
- 22.Leruez-Ville, M., M. Ouachée, R. Delarue, A. S. Sauget, S. Blanche, A. Buzyn, and C. Rouzioux. 2003. Monitoring cytomegalovirus infection in adult and pediatric bone marrow transplant recipients by a real-time PCR assay performed with blood plasma. J. Clin. Microbiol. 412040-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lilleri, D., F. Baldanti, M. Gatti, F. Rovida, L. Dossena, S. De Grazia, M. Torsellini, and G. Gerna. 2004. Clinically-based determination of safe DNAemia cutoff levels for preemptive therapy of human cytomegalovirus infections in solid organ and hematopoietic stem cell transplant recipients. J. Med. Virol. 73412-418. [DOI] [PubMed] [Google Scholar]
- 24.Lilleri, D., G. Gerna, M. Furione, M. E. Bernardo, G. Giorgiani, S. Telli, F. Baldanti, and F. Locatelli. 2007. Use of a DNAemia cut-off for monitoring human cytomegalovirus infection reduces the number of preemptively treated children and young adults receiving hematopoietic stem-cell transplantation compared with qualitative antigenemia. Blood 1102757-2760. [DOI] [PubMed] [Google Scholar]
- 25.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183377-382. [DOI] [PubMed] [Google Scholar]
- 26.Ljungman, P., P. Griffiths, and C. Payá. 2002. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin. Infect. Dis. 341094-1097. [DOI] [PubMed] [Google Scholar]
- 27.Machida, U., M. Kami, T. Fukui, Y. Kazuyama, M. Kinoshita, Y. Tanaka, Y. Kanda, S. Ogawa, H. Honda, S. Chiba, K. Mitani, Y. Muto, K. Osumi, S. Kimura, and H. Hirai. 2000. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol. 382536-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori, T., S. Okamoto, R. Watanabe, T. Yajima, Y. Iwao, R. Yamazaki, T. Nakazato, N. Sato, T. Iguchi, H. Nagayama, T. Hibi, and Y. Ikeda. 2002. Dose-adjusted preemptive therapy for cytomegalovirus disease based on real-time polymerase chain reaction after allogeneic stem cell transplantation. Bone Marrow Transplant. 29777-782. [DOI] [PubMed] [Google Scholar]
- 29.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, H. Ellerbrok, G. Pauli, and W. Siegert. 2000. Detection of cytomegalovirus DNA by real-time quantitative PCR. J. Clin. Microbiol. 382734-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pumannova, M., K. Roubalova, A. Vitek, and J. Sajdova. 2006. Comparison of quantitative competitive polymerase chain reaction-enzyme-linked immunosorbent assay with LightCycler-based polymerase chain reaction for measuring cytomegalovirus DNA in patients after hematopoietic stem cell transplantation. Diagn. Microbiol. Infect. Dis. 54115-120. [DOI] [PubMed] [Google Scholar]
- 31.Razonable, R. R., C. Paya, and T. F. Smith. 2002. Role of the laboratory in diagnosis and management of cytomegalovirus infection in hematopoietic stem cell and solid organ transplant recipients. J. Clin. Microbiol. 40746-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruell, J., C. Barnes, K. Mutton, B. Foulkes, J. Chang, J. Cavet, M. Guiver, I. Menasce, M. Dougal, and R. Chopra. 2007. Active CMV disease does not always correlate with viral load detection. Bone Marrow Transplant. 4055-61. [DOI] [PubMed] [Google Scholar]
- 33.Schaade, L., P. Kockelkorn, K. Ritter, and M. Kleines. 2000. Detection of cytomegalovirus DNA in human specimens by LightCycler PCR. J. Clin. Microbiol. 384006-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solano, C., I. Muñoz, A. Gutiérrez, A. Farga, F. Prósper, J. García-Conde, D. Navarro, and C. Gimeno. 2001. Qualitative plasma assay (AMPLICOR CMV test) versus pp65 antigenemia assay for monitoring cytomegalovirus viremia and guiding preemptive ganciclovir therapy in allogeneic stem cell transplantation. J. Clin. Microbiol. 393938-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuge, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60455-462. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, Y., Y. Kanda, M. Kami, S. Mori, T. Hamaki, E. Kusumi, S. Miyakoshi, Y. Nannya, S. Chiba, Y. Arai, K. Mitani, H. Hirai, and Y. Mutuo. 2002. Monitoring cytomegalovirus infection by antigenemia assay and two distinct real-time PCR methods after hematopoietic stem cell transplantation. Bone Marrow Transplant. 30315-319. [DOI] [PubMed] [Google Scholar]
- 37.Verkruyse, L. A., G. A. Storch, S. M. Devine, J. F. DiPersio, and R. Vij. 2006. Once daily ganciclovir as initial pre-emptive therapy delayed until threshold CMV load ≥10000 copies/ml: a safe and effective strategy for allogeneic stem cell transplant patients. Bone Marrow Transplant. 3751-56. [DOI] [PubMed] [Google Scholar]
- 38.Von Müller, L., J. Hinz, M. Bommer, W. Hampl, S. Kluwick, M. Wiedmann, D. Bunjes, and T. Mertens. 2007. CMV monitoring using blood cells and plasma: a comparison of apples with oranges? Bone Marrow Transplant. 39353-357. [DOI] [PubMed] [Google Scholar]
- 39.Von Müller, L., W. Hampl, J. Hinz, H. Meisel, A. Reip, E. Engelmann, R. Heilbronn, B. Gärtner, O. Krämer, H. Einsele, H. Hebart, T. Ljubicic, J. Löffler, and T. Mertens. 2002. High variability between results of different in-house tests for cytomegalovirus (CMV) monitoring and standardized quantitative plasma CMV PCR assay. J. Clin. Microbiol. 402285-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinberg, A., D. Schissel, and R. Giller. 2002. Molecular methods for cytomegalovirus surveillance in bone marrow transplant recipients. J. Clin. Microbiol. 404203-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yakushiji, K., H. Gondo, K. Kamenazi, K. Shigematsu, S. Hayashi, M. Kuroiwa, S. Taniguchi, Y. Ohno, K. Takase, A. Numara, K. Aoki, K. Kato, K. Nagafuji, K. Shimoda, T. Okamura, N. Kinukawa, N. Kasuga, M. Sata, and M. Harada. 2002. Monitoring of cytomegalovirus reactivation after allogeneic stem cell transplantation: comparison of an antigenemia assay and quantitative real-time polymerase chain reaction. Bone Marrow Transplant. 29599-606. [DOI] [PubMed] [Google Scholar]
- 42.Yerly, S., L. Perrin, C. Van Delden, S. Schaffer, S. Thamm, W. Wunderli, and L. Kaiser. 2007. Cytomegalovirus quantification in plasma by an automated real-time PCR assay. J. Clin. Virol. 38298-303. [DOI] [PubMed] [Google Scholar]
- 43.Yun, Z., I. Lewensohn-Fuchs, P. Ljungman, L. Ringholm, J. Jonsson, and J. Albert. 2003. A real-time TaqMan PCR for routine quantification of cytomegalovirus DNA in crude leukocyte lysates from stem cell transplant patients. J. Virol. Methods 11073-79. [DOI] [PubMed] [Google Scholar]