Abstract
Motility in mycobacteria was described for the first time in 1999. It was reported that Mycobacterium smegmatis and Mycobacterium avium could spread on the surface of solid growth medium by a sliding mechanism and that the presence of cell wall glycopeptidolipids was essential for motility. We recently reported that Mycobacterium vaccae can also spread on growth medium surfaces; however, only smooth colonies presented this property. Smooth colonies of M. vaccae do not produce glycopeptidolipids but contain a saturated polyester that is absent in rough colonies. Here, we demonstrate that Mycobacterium chubuense, Mycobacterium gilvum, Mycobacterium obuense, and Mycobacterium parafortuitum, which are phylogenetically related to M. vaccae, are also motile. Such motility is restricted to smooth colonies, since natural rough mutants are nonmotile. Thin-layer chromatography analysis of the content of cell wall lipids confirmed the absence of glycopeptidolipids. However, compounds like the above-mentioned M. vaccae polyester were detected in all the strains but only in smooth colonies. Scanning electron microscopy showed great differences in the arrangement of the cells between smooth and rough colonies. The data obtained suggest that motility is a common property of environmental mycobacteria, and this capacity correlates with the smooth colonial morphotype. The species studied in this work do not contain glycopeptidolipids, so cell wall compounds or extracellular materials other than glycopeptidolipids are implicated in mycobacterial motility. Furthermore, both smooth motile and rough nonmotile variants formed biofilms on glass and polystyrene surfaces.
The genus Mycobacterium contains more than 100 species of nontuberculous mycobacteria (NTM) (28). Unlike the members of Mycobacterium tuberculosis complex and Mycobacterium leprae, NTM species are not obligated pathogens and are inhabitants of the environment. They can be found in natural water, water distribution systems, soil, protozoans, plants, and animals (24, 27). The emergence, in the last decade, of opportunistic NTM infections in humans has prompted the study of characteristics that allow mycobacteria to persist in their natural reservoirs. Motility has been related to the capacity of bacteria to colonize and persist in the environment. Mycobacteria were considered nonmotile microorganisms until 1999, when Martínez et al. (17) reported that Mycobacterium smegmatis and Mycobacterium avium spread on the surface of a growth medium by sliding motility. In these species, motility was related to colonial morphology and the presence of glycopeptidolipids (GPLs), a class of glycosylated peptidolipids located in the cell wall of some mycobacteria (for a recent review on GPLs, see the report of Chatterjee and Khoo [5]). Smooth colonies were motile and contained GPLs, but spontaneous rough colonies lacked GPLs and were nonmotile (17). Genetic evidence for the requirement of GPLs for sliding motility was later provided for M. smegmatis but not for M. avium (25). The latter species expresses a variety of colony morphotypes, the most prevalent of which are smooth opaque, smooth transparent, and rough. In the work of Martínez et al. (17), the smooth transparent variant produced the largest halos, and the smooth opaque variant also spread but formed halos with a different appearance. The rough strain, completely devoid of GPLs, did not spread, while another rough strain which was able to synthesize incomplete GPLs produced smaller spreading areas. In addition to these results, Cangelosi et al. have reported that when M. avium grows in agar media containing Congo red, new phenotypic variations can be detected since smooth strains give pink, red, and white colonies (4). These authors detected that red variants spread more aggressively than white ones, but no data about the content of GPLs or other compounds responsible for the different behaviors between the two morphotypes were provided.
Mycobacterium vaccae is a rapidly growing pigmented species that has been isolated from soil, watering ponds, wells, and lacteal glands and skin lesions in cattle (31). The M. vaccae ATCC 15483T strain displays smooth colonies, but natural rough variants can easily be obtained by subculturing on solid medium. We have recently reported that the smooth colonies of M. vaccae spread extensively on the surface of growth medium, whereas the natural rough variants were able to invade only a few millimeters of the growth surface (26). M. vaccae is phylogenetically distant from M. smegmatis and M. avium (28) and devoid of GPLs (5). Interestingly, we identified a new long-chain saturated fatty acid polyester (SP) produced by the smooth colonies of M. vaccae but not by the rough colonies (26). This polymer, not previously found in other mycobacteria, releases 1-tetradecanol (1-OH-14:0) by saponification. In addition to being present in M. vaccae, 1-tetradecanol has been described to occur in saponified lipidic extracts of Mycobacterium chubuense, Mycobacterium gilvum, Mycobacterium obuense, and Mycobacterium parafortuitum (29). 16S rRNA gene-based phylogenetic studies show a close relationship among these species, including M. vaccae (28). Consequently, with these data we decided to investigate the motility and presence of SP in these species. In this work, we studied the capacity of these species to spread on the surface of solid medium by comparing smooth and rough colonial morphotypes. We performed scanning electron microscopy (SEM) of smooth and rough colony variants. We analyzed the contents of cell wall surface lipids for all the strains and morphology variants while also looking for the presence of the SP which was previously described to occur in M. vaccae. Finally, as spreading motility has been correlated with biofilm formation in some mycobacteria (15, 25), we decided to investigate the capacity of each smooth and rough morphotype to form biofilms on glass (hydrophilic) and polystyrene (hydrophobic) surfaces.
MATERIALS AND METHODS
Strains and growth medium.
M. chubuense ATCC 27278T, M. gilvum DSM 43547, M. obuense ATCC 27023T, M. parafortuitum ATCC 19686T, and M. vaccae ATCC 15483T strains used in this study were grown on tryptone soy agar (TSA; Scharlau Chemie, Barcelona, Spain) medium for 2 weeks at 30°C. Cultures were observed with the naked eye, looking for rough colony variants.
SEM.
For SEM analysis, bacterial cells were grown on TSA medium at 30°C for 2 weeks. Then, the different colonies were collected with the surrounding agar by using a blade, deposited in cryotubes, and fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 2 hours at 4°C. After three washes at 5 min each in 0.2 M sodium cacodylate buffer, the cells were dehydrated in an ascending ethanol series (30, 50, 70, 80, and 90% for 10 min each and twice with 100% ethanol) and dried by critical-point drying with CO2. Samples were mounted on adhesive carbon films and then coated with gold. Bacilli were observed with an S-570 scanning electron microscope (Hitachi Ltd., Japan) at an accelerating voltage of 15 kV.
Motility.
Motility was evaluated using the surface spreading assay described by Martínez et al. (17). Briefly, the medium used was Middlebrook 7H9 broth (Difco Laboratories, Surrey, United Kingdom), not supplemented but solidified with 0.3% agarose, hereinafter referred to as motility medium. This medium was dispensed in petri dishes that were allowed to sit at room temperature overnight prior to inoculation. Single smooth and rough colonies were inoculated in the center of the plates containing the motility medium by poking the medium with a sterile bacteriologic loop. Plates were Parafilm sealed and incubated at 30°C for 6 days. Spreading ability was evaluated visually using a Leica MZ FLIII (Leica Microsystems, Wetzlar, Germany) binocular stereomicroscope.
Analyses of cell wall surface lipids.
Mycobacterial cells were scraped from the surface of TSA, and glycolipids, phospholipids, and apolar free lipids were extracted with methanol-chloroform (2:1, vol/vol) and then with chloroform-methanol (2:1, vol/vol) at room temperature with continuous stirring. The organic extracts were pooled, evaporated to dryness, and partitioned with chloroform-methanol-water (8:4:2, vol/vol/vol). The organic phase was recovered and evaporated to dryness.
For the analysis of glycolipids and phospholipids, dry extracts were resuspended in chloroform and examined by thin-layer chromatography (TLC) on silica gel-coated plates (G-60, 0.25-mm thickness; Merck, Darmstadt, Germany) developed with chloroform-methanol (85:15, vol/vol) and chloroform-methanol-water (90:10:1, 30:8:1, and 60:35:8, vol/vol/vol) (7). The glycolipids were visualized by spraying the plates with 1% (wt/vol) anthrone (Sigma, St. Louis, MO) in sulfuric acid, followed by heating at 120°C. Molybdenum blue spray reagent (Sigma) was used to reveal phosphorus-containing substances. From the same resuspended extracts, the presence of SP compounds was examined by TLC with methanol-chloroform (90:10, vol/vol) and visualized by spraying the TLC plates with 1% (wt/vol) anthrone in sulfuric acid (26).
For the analysis of apolar free lipids, dry extracts were redissolved in chloroform and analyzed by two-dimensional TLC with petroleum ether (bp, 60 to 80°C)-ethyl acetate (98:2, vol/vol) three times in the first direction and with petroleum ether (bp, 60 to 80°C)-acetone (98:2, vol/vol) once in the second direction. Apolar free lipids were visualized by spraying the plates with 10% (wt/vol) molybdophosphoric acid (Merck) in ethanol (20).
Mycolic acids were extracted and methylated by acid methanolysis as previously described (19). Methyl mycolates were analyzed by TLC using hexane-ether (85:15, vol/vol) three times. TLC plates were sprayed with 10% (wt/vol) molybdophosphoric acid in ethanol and heated at 120°C.
Detection of 1-tetradecanol.
The presence of 1-tetradecanol in smooth and rough variants was detected by gas chromatography-mass spectrometry (GC-MS). The chloroform-methanol extracts (approximately 1 mg) were saponified with 2 ml of 10% (wt/vol) KOH in methanolic solution at room temperature overnight. Then, 1 ml of water and 2 ml of n-hexane were added, vigorously mixed, and allowed to stand for 5 min. The n-hexane fraction containing the alcohols (called the neutral fraction) was removed into another tube, evaporated, and injected in an Agilent 6890 II GC coupled to a model 5973 MS (Agilent, CA) (26). The column used was a cross-linked methyl silicone column (HP-1; 30 m, 0.25-mm inside diameter, 0.25-μm film thickness; Agilent). The column was programmed to go from 60 to 300°C at 15°C/min. The injector temperature was 250°C. The identification of 1-tetradecanol was made by comparing the retention time and mass spectra with those of authentic 1-tetradecanol (Merck).
Biofilm formation assay.
The biofilm formation was assayed by determining the ability of cells to adhere to the surfaces of glass and polystyrene tubes. The protocol followed was similar to that described by Zamora et al. (32). Bacterial cells grown on TSA medium at 30°C for 2 weeks were suspended in phosphate-buffered saline (PBS) to a concentration equivalent to a McFarland standard of 0.5. Tubes containing 6 ml of tryptone soy broth (Scharlau Chemie, Barcelona, Spain) were inoculated with 15 μl of the PBS suspensions and incubated at 30°C with continuous stirring on an orbital shaker. Glass and polystyrene tubes with 6 ml of tryptone soy broth and without mycobacterial suspensions were used as negative controls.
The cellular adhesion was measured at 1, 2, 3, 4, and 7 days of incubation, and each sample was assayed in triplicate. After incubation, culture supernatants were discarded, and then nonadherent cells were removed by washing the tubes three times with 6 ml of sterile PBS. To remove adherent cells, 6 ml of PBS and five sterile glass beads were added to the tubes, which were briefly vortexed and sonicated. The attached bacteria were quantified by preparing serial dilutions from the sonicate and streaking 100 μl of each dilution on TSA plates. The cellular adhesion level was expressed as the number of CFU adhered per cm2.
Statistical analysis.
Sigma Stat (SPSS software) was used to compare differences in cellular adhesion between smooth and rough variants using the Student t test. Differences were significant when P was <0.05.
RESULTS
Spontaneous rough colony variants were obtained from all the smooth strains.
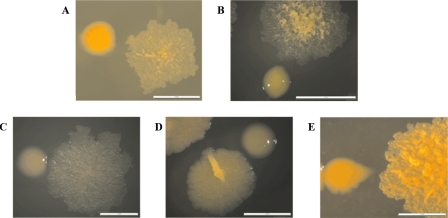
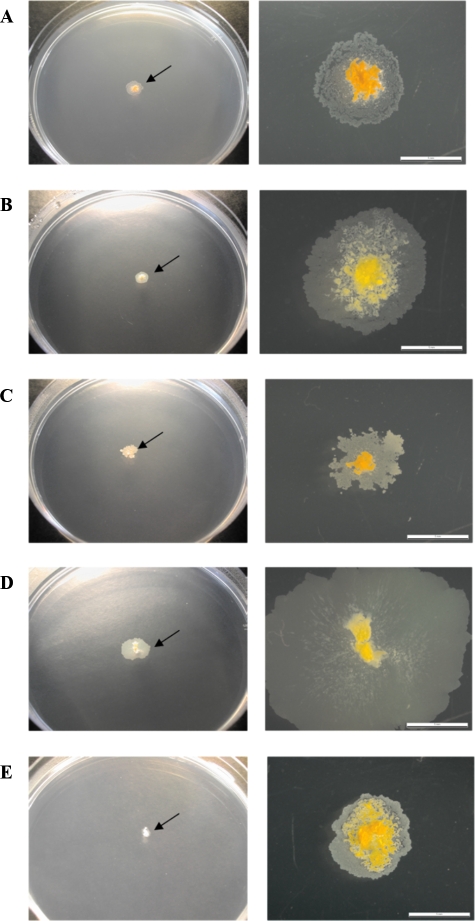
After the repeated culturing (two or three passages) of smooth colonies of M. chubuense, M. gilvum, M. obuense, and M. parafortuitum on TSA at 30°C, mainly smooth colonies were obtained, but a few spontaneous rough colonies were also detected for these strains (Fig. 1). For M. vaccae, we worked with the original smooth colonies and with rough natural mutants obtained in a previous work (26). Rough variants were characterized by an irregular surface with many wrinkles and crests (Fig. 1). The surfaces of rough colonies were significantly drier than the surfaces of smooth ones. A bright and moist texture was exhibited only by smooth colonies (Fig. 1). In subsequent cultures performed for 3 years on TSA, the rough variants did not switch to the smooth type.
FIG. 1.
Smooth and rough colonies of M. chubuense (A), M. gilvum (B), M. obuense (C), M. parafortuitum (D), and M. vaccae (E). The images are representative of hundreds of colonies obtained through the study.
SEM showed large differences between smooth and rough colonies in the arrangement of the cells in the colonies.
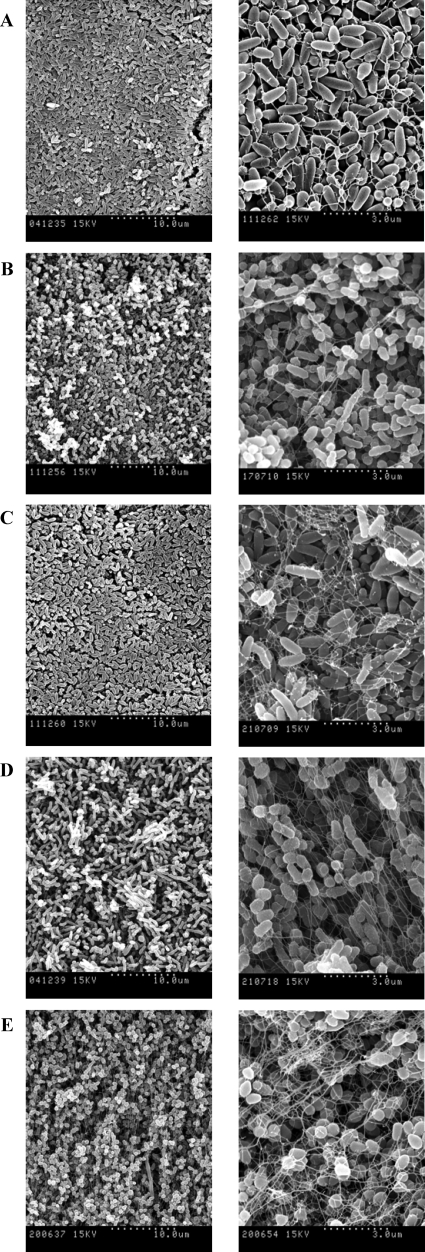
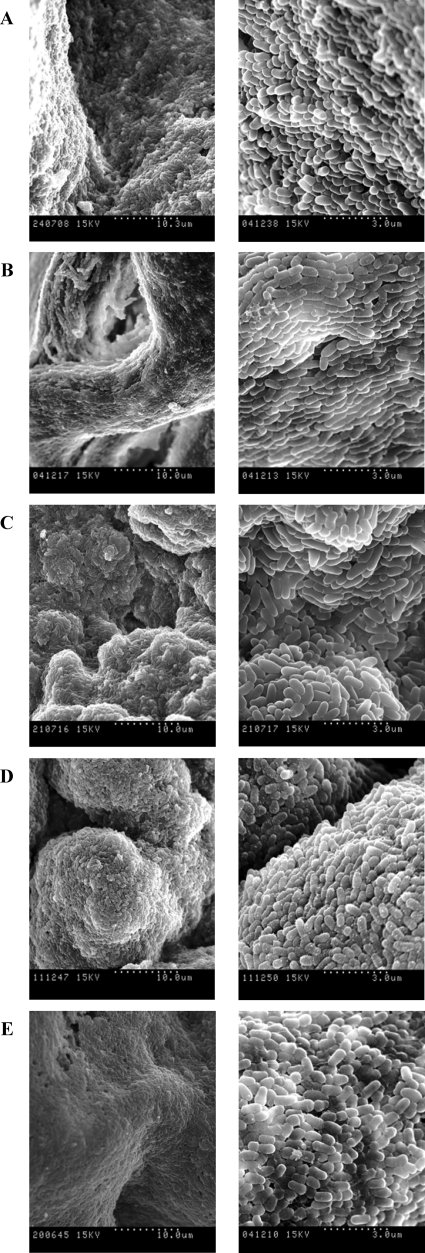
SEM showed that bacterial cells in smooth colonies appeared to be without orientation, either being isolated or forming little groups (Fig. 2). In contrast, cells in the rough colonies were positioned very closely to each other to form curved structures arranged in a definite order (Fig. 3). Unlike with the rough colonies, empty spaces were clearly visible among single cells of the smooth colonies and a network of long fibers was present (Fig. 2). Short fibers that seem to connect one cell to another were also visible in rough M. vaccae colonies (Fig. 3E, right).
FIG. 2.
SEM images of smooth colonies of M. chubuense (A), M. gilvum (B), M. obuense (C), M. parafortuitum (D), and M. vaccae (E). The right column shows the same sample as the left column but at greater magnification. These images are representative of the studies performed with six colonies of each species.
FIG. 3.
SEM images of rough colonies of M. chubuense (A), M. gilvum (B), M. obuense (C), M. parafortuitum (D), and M. vaccae (E). The right column shows the same sample as the left column but at greater magnification. These images are representative of the studies performed with six colonies of each species.
Only smooth colonies were able to widely spread on the surface of motility medium.
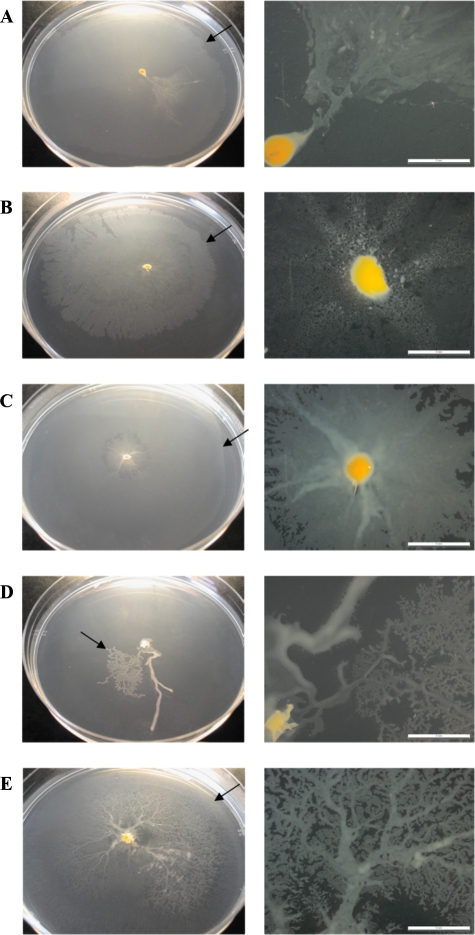
As is clearly shown in Fig. 4 and 5, the smooth colonies spread more efficiently in the motility medium than the rough variants. The images correspond to 6-day-old cultures and are representative of the growth patterns observed during the 3 years that the study lasted. The observation of colonies with binocular stereomicroscopy showed that those with smooth morphotypes form different kinds of extensions from the initial inoculation point to the periphery of the colony (Fig. 4, right panels). Ziehl-Neelsen staining confirmed the presence of mycobacterial cells in these prolongations and in the periphery of the smooth colonies (data not shown). In contrast, the rough variants did not form any kind of prolongations and extended just a few millimeters from the inoculation point (Fig. 5).
FIG. 4.
Spreading of smooth colonies on motility medium after 6 days of growth. (A) M. chubuense; (B) M. gilvum; (C) M. obuense; (D) M. parafortuitum; (E) M. vaccae. Arrows indicate the external margins of the colonies. The right column shows pictures obtained with a binocular stereomicroscopy. These images are representative of the studies performed with 10 colonies of each species.
FIG. 5.
Spreading of rough colonies on motility medium after 6 days of growth. (A) M. chubuense; (B) M. gilvum; (C) M. obuense; (D) M. parafortuitum; (E) M. vaccae. Arrows indicate the external margins of the colonies. In the right column are pictures obtained with a binocular stereomicroscopy. These images are representative of the studies performed with 10 colonies of each species.
The strains studied did not contain GPLs or other species-specific cell wall surface lipids.
TLC revealed the absence of GPLs in all the strains studied (see Fig. S1 in the supplemental material). All strains contained ubiquitous dimycoloyl trehaloses (DMTs) and phosphatidylinositol mannosides (PIMs) as major cell wall glycolipids. Minor spots with a chromatographic behavior similar to that of monomycoloyl trehaloses (MMTs) were observed in all the strains. No differences between smooth and rough variants in the content of cell wall glycolipids, nor in the content of phospholipids or mycolic acids, were found by TLC (see Fig. S1 to S6 in the supplemental material). The specific apolar lipids phthiocerol dimycocerosates were not detected, and only trace amounts of compounds that migrate, such as menaquinones or triacylglycerols, were detected in all the strains (data not shown).
All the smooth colonies contained a compound that migrates on TLC as the SP previously characterized in M. vaccae.
Chloroform-methanol extracts of all smooth colonies contained a red compound that migrates on TLC as the SP previously characterized in M. vaccae (26). It can be seen in Fig. 6 that SP migrates on TLC with the front of the solvent, showing a characteristic red color and adopting a flamelike form. By TLC, all smooth colonies presented a spot that migrates like SP (SP-L), and all rough variants were devoid of it. SP and SP-L were also easily extractable from bacterial cells with butanol or with a water solution containing Triton X-114 at room temperature with continuous stirring (data not shown).
FIG. 6.
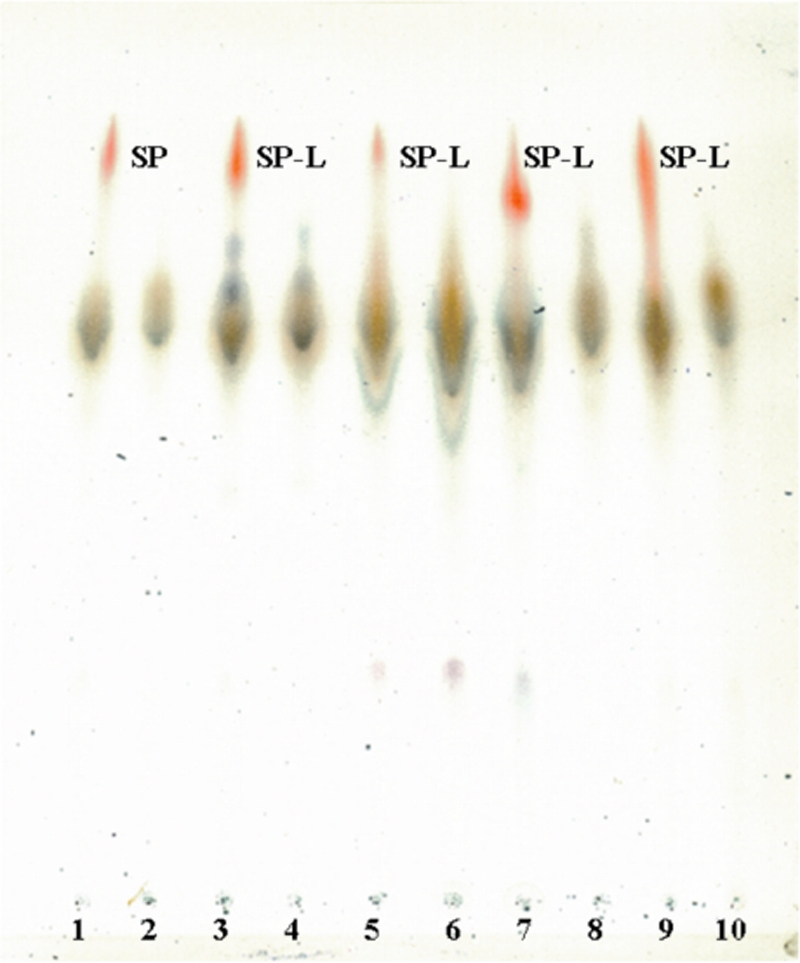
TLC images of chloroform-methanol extracts from M. vaccae (lanes 1 and 2), M. gilvum (lanes 3 and 4), M. obuense (lanes 5 and 6), M. chubuense (lanes 7 and 8), and M. parafortuitum (lanes 9 and 10). In the odd-numbered lanes, extracts are from smooth colonies, and in the even-numbered lanes, extracts are from rough colonies. TLC was developed with methanol-chloroform (90:10, vol/vol) and revealed with anthrone. Red spots indicate SP and SP-L.
1-Tetradecanol was detected only in the chloroform-methanol extracts of the smooth colonies.
The SP previously characterized in M. vaccae is a complex polyester formed by C16 to C19 fatty acids esterified by C14 to C18 alcohols (26). Unlike the other alcohols that also form part of mycolic acids, 1-tetradecanol (C14 alcohol) seems to be present only in SP (26, 29). So, the detection of 1-tetradecanol can be considered a marker of the presence of SP and SP-L compounds. The saponification breaks the ester linkage, releasing in the medium free alcohols that can be recovered using organic solvents. This fraction is named the neutral compound fraction to distinguish it from free acids that remain in the methanolic solution. In the GC-MS analysis of neutral compounds, 1-tetradecanol was detected in extracts of smooth colonies but not in those of their rough variants. These results confirm TLC analyses in which SP and SP-L compounds were observed only in smooth variants. In Fig. S7 in the supplemental material, we show the GC-MS chromatogram of the neutral compounds of smooth and rough colonies of M. chubuense as representative of the rest of the strains. The identification of 1-tetradecanol was made by comparing the retention time and mass spectra with those of authentic 1-tetradecanol (Merck). Complete chromatograms of neutral and acidic compounds from purified SP can be found in the report of Rodríguez-Güell et al. (26).
Both smooth motile and rough nonmotile variants formed biofilms on glass and polystyrene surfaces.
We evaluated comparatively the abilities of smooth and rough variants of M. chubuense, M. gilvum, M. obuense, M. parafortuitum, and M. vaccae strains to attach to glass and polystyrene tubes at 1, 2, 3, 4, and 7 days of incubation.
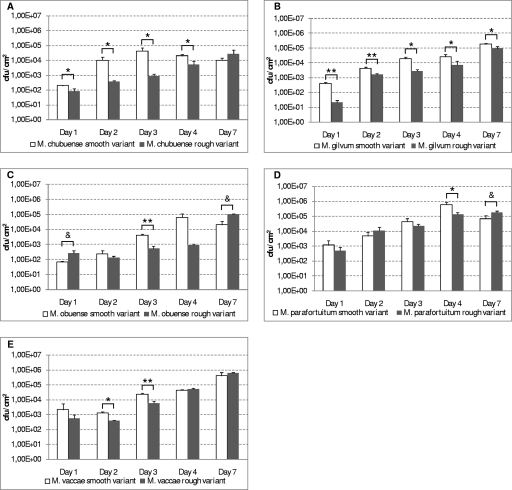
The assay of cellular adhesion to glass showed that the adhesion levels of smooth variants were higher than those of rough variants (Fig. 7). This difference was statistically significant in M. chubuense (days 1 to 4) and M. gilvum (all days) and in M. obuense and M. vaccae only on day 3 and days 2 and 3, respectively. However, we observed that the rough variant of M. obuense presented an increased capacity to adhere to the glass compared to the smooth variant on days 1 and 7. In M. parafortuitum, smooth and rough variants did not differ quantitatively in their levels of adherence to glass during the different times analyzed, with the exception of days 4 and 7. At those times, we observed significant differences.
FIG. 7.
Cellular adhesion of smooth and rough variants of M. chubuense (A), M. gilvum (B), M. obuense (C), M. parafortuitum (D), and M. vaccae (E) strains to glass tubes at 1, 2, 3, 4, and 7 days of incubation. The cellular adhesion level is expressed as the number of CFU/cm2. The results are expressed as means ± standard deviations obtained from triplicates. The results are representative of one out of three independent experiments. Cellular adhesion levels of smooth variants were significantly higher than those of rough variants (*, P < 0.05; **, P < 0.01). Cellular adhesion levels of rough variants were significantly higher than those of smooth variants (&, P < 0.05).
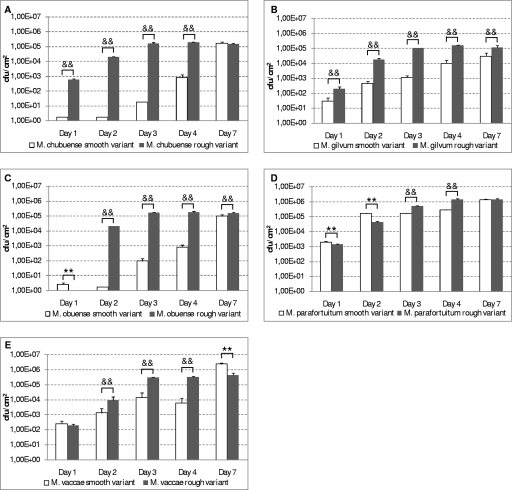
In contrast, as Fig. 8 shows, rough variants presented a great capacity of adhesion to a polystyrene surface compared with that of smooth variants. This difference was even higher in the cases of M. chubuense, M. gilvum, and M. obuense compared to M. vaccae. On the contrary, in the case of M. parafortuitum, the cellular adhesion level of the smooth variant was significantly higher than that of the rough variant on days 1 and 2.
FIG. 8.
Cellular adhesion of smooth and rough variants of M. chubuense (A), M. gilvum (B), M. obuense (C), M. parafortuitum (D), and M. vaccae (E) strains to polystyrene tubes at 1, 2, 3, 4, and 7 days of incubation. The cellular adhesion level is expressed as the number of CFU/cm2. The results are expressed as means ± standard deviations obtained from triplicates. The results are representative of one out of three independent experiments. Cellular adhesion levels of smooth variants were significantly higher than those of rough variants (**, P < 0.01). Cellular adhesion levels of rough variants were significantly higher than those of smooth variants (&&, P < 0.05).
DISCUSSION
NTM have the capacity of displaying different colonial morphologies on the surface of solid medium. That is, after various passages of one strain on the surface of solid medium, it is not unusual to obtain natural mutants that show a colonial morphology that is clearly different from that shown by the original strain. As recent examples, spontaneous morphological mutants of M. smegmatis, M. avium, Mycobacterium fortuitum, M. vaccae, and Mycobacterium abscessus (15-17, 26) have been described. It is not always possible to isolate morphological mutants. After working with smooth and rough strains of Mycobacterium kansasii, Belisle and Brennan could not isolate smooth colony variants from rough strains, and rough colony variants could not be isolated from smooth ones (1). The authors of that study concluded that colony morphology was a more stable character in M. kansasii than in M. avium. Taking into account that differences exist among Mycobacterium species in their capacities to give spontaneous morphological colony mutations, one must also consider another important aspect, the solid media used to test this capacity of change. Older studies, performed mainly on substrains of Mycobacterium bovis BCG, showed that a clear observation of the differences in colonial morphologies among some substrains was possible only in specific solid media (22, 23). We have obtained a good differentiation of the morphotypes by using TSA for growth. This is not a conventional medium for mycobacterial culture. Classical and more widely used media are Löwenstein-Jensen, Middlebrook 7H10, 7H11 agar, and Sauton's. In a previous work, we compared Middlebrook 7H10, Sauton's, and TSA in their abilities to show differences in the colonial morphology of M. vaccae, and we found that, at least for the strain studied, TSA was the most discriminative medium (26). The present work confirms that TSA is a good medium to use to obtain rough colony variants and to discriminate between smooth and rough colony morphologies in rapidly growing pigmented mycobacteria (Fig. 1).
SEM images of smooth colonies show that bacilli are connected by a matrix of an unknown fibrous material; in addition, many empty spaces are observed among the bacillary cells (Fig. 2). In contrast, SEM images of rough variants showed very closely packed cells with a specific orientation (Fig. 3). SEM studies of the arrangement of cells in mycobacterial colonies grown on the surface of solid medium were performed mainly in the 1970s and 1980s. In 1972, Drucker (11) reported the presence of extracellular material in colonies of Mycobacterium phlei, although no image was supplied in the article. Two years later, very nice SEM images of entire colonies of M. phlei showed that the colonies were covered with ridges made of clustered cells (14). SEM studies of smooth and rough colonies of Mycobacterium lepraemurium indicated the presence of filamentous strands in the rough type, and only short bridges were found between the bacilli of smooth colonies (21). We have also observed short fibers in rough colonies of M. vaccae but not in the rough colonies of the other species studied, so this is a characteristic that can vary among species. Furthermore, it is necessary to take into account that the conditions of the culture can modify the external surface of Mycobacterium cells. For example, it has been reported that the appearance of cell surface blebs and fibrils in aged Mycobacterium tuberculosis cells grown in liquid medium (8) and the development of a more irregular surface in Mycobacterium paratuberculosis cells grown in liquid medium not supplemented with Tween 80 compare with those of cells grown in liquid medium supplemented with 1% of Tween 80 (30). Thus, comparative studies among SEM structures should be done between strains grown under the same conditions as we have done in this work.
The relationship between the morphology and the microstructure of the colonies was also studied in substrains of BCG. Interestingly, rough colonies were seen as a group of bacilli arranged end to end and in parallel rows, whereas in smooth colonies the bacilli showed a lack of orientation (13). Other authors reported that BCG colonies of some substrains were completely covered in an amorphous material (10). To date, the nature of the fibers or the other extracellular materials seen by SEM in mycobacteria is still unknown.
Previous motility studies performed on M. smegmatis, M. avium, and M. vaccae showed that motility was limited to smooth colonies (17, 26). Accordingly, we have found that motility was present only in the smooth colonies of M. chubuense, M. gilvum, M. obuense, and M. parafortuitum. They were able to spread widely on the surface of the motility medium (Fig. 4). This motility medium described by Martínez et al. (17) allows a clear visualization of the capacity of mycobacteria to spread on the surface of solid medium.
In M. smegmatis and M. avium, motility was related to the presence of GPLs (17, 25). The mycobacterial cell envelope is composed of three layers: a plasma membrane; a cell wall skeleton consisting of two covalently attached macromolecules, peptidoglycan, and mycoloyl arabinogalactan; and an outer layer made up of polysaccharides and proteins (3, 6, 18). Characteristic lipids and glycolipids have been located in this outer layer. Some of these glycolipids are species specific; these are phenolic glycolipids, lipooligosaccharides, GPLs, and diacyl, triacyl, and polyacyl trehaloses. Others, such as PIMs, MMTs, and DMTs, are ubiquitous glycolipids, found in most species of the Mycobacterium genus (3, 6, 18). In addition to those of M. smegmatis and M. avium, GPLs are also found in Mycobacterium intracellulare, Mycobacterium scrofulaceum, Mycobacterium simiae, “Mycobacterium habana,” M. paratuberculosis, Mycobacterium xenopi, “Mycobacterium butyricum,” Mycobacterium peregrinum, Mycobacterium chelonae, M. abscessus, Mycobacterium senegalense, and Mycobacterium porcinum (5). Hence, GPLs have not been described for M. vaccae and the other pigmented species studied in this work. Furthermore, it has previously been reported that M. vaccae ATCC 15483T contained only the ubiquitous glycolipids PIMs, MMTs, and DMTs (2, 26). We found this same pattern of glycolipids in M. chubuense, M.gilvum, M. obuense, and M. parafortuitum. DMTs and PIMs were the main glycolipids, and only small amounts of MMTs were detected (see Fig. S1 to S4 in the supplemental material).
Valero-Guillén and Martín-Luengo (29) reported the presence of 1-tetradecanol in M. chubuense, M. gilvum, M. obuense, M. parafortuitum, and M. vaccae. These mycobacteria contain the wax ester mycolate, a lipid that, after saponification, releases secondary alcohols, such as 2-octadecanol, 2-eicosanol, and 2-docosanol, but not primary alcohols. Thus, they concluded that 1-tetradecanol was a constituent of the cell envelopes of these mycobacteria but not of the wax ester mycolate (29). We have described that the saponification of purified SP isolated from M. vaccae released 1-tetradecanol (26). In the present study, we also found a positive correlation between the presence of SP-L and the presence of 1-tetradecanol. In the saponified chloroform-methanol extracts of all the smooth colonies, 1-tetradecanol was detected by GC-MS (see Fig. S7 in the supplemental material). In the same extracts, before saponification, SP-L was detected by TLC (Fig. 6). These data, together with the TLC behavior of SP-L, suggest that M. chubuense, M. gilvum, M. obuense, and M. parafortuitum produce a polyester similar to the SP previously described in M. vaccae.
Natural rough variants presented the same pattern of glycolipids, phospholipids, mycolic acids, and other apolar lipids as that of the original smooth colonies (see Fig. S1 to S6 in the supplemental material); however, they were devoid of SP-L (Fig. 6). We reported that M. vaccae ATCC 15483T produced a long-chain SP with an estolidelike structural skeleton and a molecular weight of about 5,000 to 7,000 (26). Nuclear magnetic resonance spectra confirmed the absence of carbohydrates and amino acids in this complex structure. This compound (called SP in the present work) has not been described for other mycobacteria before. Although we have not isolated and structurally analyzed the SP-L produced by M. chubuense, M. gilvum, M. obuense, and M. parafortuitum, we provide evidence of their similar natures. These have the same mobility in TLC, the same form of flame, and the same red color in response to antrone. Both SP and SP-L are easily extractable with the chloroform-methanol mixtures, butanol, or Triton X-114-water solution. They are present only in smooth colonies, and only the chloroform-methanol extracts of these colonies release 1-tetradecanol by saponification. We are unaware of the biological properties and the possible roles that SP and SP-L may play in the biology of mycobacteria. From the data obtained in this study, we can only hypothesize that SP and SP-L may be related to colonial morphology and motility; however, a direct link cannot be established between motility and the presence of the SP until genetic analysis of constructed mutants has been performed.
M. smegmatis mc2155 is a smooth sliding strain that contains GPLs in its cell wall surface and is able to form biofilms on polyvinyl chloride (PVC) surfaces. Rough mutants constructed by transposon insertions in the mps gene, which is involved in GPL synthesis, were unable to slide and form biofilms on PVC (25). More recently, one study performed on one clinical isolate of M. abscessus, another species that contains GPLs, showed that rough spontaneous colonies were devoid of GPLs, did not slide, and were unable to form biofilms on the surface of polystyrene pegs in the Calgary biofilm device (15). In M. avium strains, biofilm formation and motility have been analyzed separately (that is, both characteristics have not been studied in the same strain). GPLs have been related to motility in M. avium and also to adherence to PVC. However, these glycolipids were not necessary to form biofilms on other plastic surfaces such as Permanox or glass (9, 12).
In our study, both smooth motile and rough nonmotile variants formed biofilms on polystyrene and glass surfaces, although some differences were observed (Fig. 7 and 8). Perhaps the most significant was that, with the exception of M. parafortuitum, rough variants adhered to polystyrene more efficiently than smooth ones in the first days of incubation.
We can conclude that (i) M. chubuense, M. gilvum, M. obuense, M. parafortuitum, and M. vaccae can spread on the surface of the motility medium; (ii) this capacity is not related to the presence of GPLs; (iii) rough spontaneous colony variants lose motility; (iv) both smooth and rough variants form biofilms on polystyrene and glass surfaces; and (v) by TLC, only the smooth colony type of all the strains shows a compound with a chromatographic behavior similar to that of a previously described M. vaccae SP.
Motility plays an important role in colonization of the environment by bacteria, and we have provided evidence that this property is present in a group of rapidly growing pigmented mycobacteria.
Supplementary Material
Acknowledgments
This work was supported by grants SAF2002-00514 and 2005SGR-00956 from the Spanish Ministerio de Educación y Ciencia and the Generalitat de Catalunya, respectively.
We thank Alejandro Sánchez of the Servei de Microscopia from the Universitat Autònoma de Barcelona for his help in the microscopic analysis.
Footnotes
Published ahead of print on 8 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Belisle, J. T., and P. J. Brennan. 1989. Chemical basis of rough and smooth variation in mycobacteria. J. Bacteriol. 1713465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan, P. J. 1988. Mycobacterium and other actinomycetes, p. 203-298. In C. Ratledge and S. G. Wilkinson (ed.), Mycobacterial lipids. Academic Press, London, United Kingdom.
- 3.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 4.Cangelosi, G. A., C. O. Palermo, and L. E. Bermudez. 2001. Phenotypic consequences of red-white colony type variation in Mycobacterium avium. Microbiology 147527-533. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, D., and K. H. Khoo. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell. Mol. Life Sci. 582018-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daffé, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39131-203. [DOI] [PubMed] [Google Scholar]
- 7.Daffé, M., M. McNeil, and P. J. Brennan. 1991. Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30378-388. [DOI] [PubMed] [Google Scholar]
- 8.Dahl, J. L. 2005. Scanning electron microscopy analysis of aged Mycobacterium tuberculosis cells. Can. J. Microbiol. 51277-281. [DOI] [PubMed] [Google Scholar]
- 9.Deshayes, C., D. Koncíncová, G. Etienne, and J.-M. Reyrat. 2008. Glycopeptidolipids: a complex pathway for small pleiotropic molecules, p. 345-366. In M. Daffé and J.-M. Reyrat (ed.), The mycobacterial cell envelope. ASM Press, Washington, DC.
- 10.Devadoos, P., M. E. Klegerman, and M. J. Groves. 1991. Surface morphology of Mycobacterium bovis BCG: relation to mechanisms of cellular aggregation. Microbios 65111-125. [PubMed] [Google Scholar]
- 11.Drucker, D. B. 1972. Bacterial colonial microstructure. Microbios 629-33. [PubMed] [Google Scholar]
- 12.Freeman, R., H. Geier, K. M. Weigel, J. Do, T. E. Ford, and G. A. Cangelosi. 2006. Roles for cell wall glycopeptidolipid in surface adherence and planktonic dispersal of Mycobacterium avium. Appl. Environ. Microbiol. 727554-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta, K. C. 1981. Scanning electron microscopic studies of spreading and non-spreading type colonies of Mycobacterium bovis (BCG). Ann. Microbiol. (Paris) 132429-440. [PubMed] [Google Scholar]
- 14.Holmqvist, O., and A. Kolman. 1978. Mycobacterium phlei PN-bb colonies: a morphological characterization and transmission electron microscopy. Ann. Microbiol. (Paris) 129341-349. [PubMed] [Google Scholar]
- 15.Howard, S. T., E. Rhoades, J. Recht, X. Pang, A. Alsup, R. Kolter, C. Rick Lyons, and T. F. Byrd. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 1521581-1590. [DOI] [PubMed] [Google Scholar]
- 16.Marques da Silva, T. R., J. Ribeiro de Freitas, Q. Chagas Silva, C. Pereira Figueira, E. Roxo, S. Cardoso Leão, L. A. Rodrigues de Freitas, and P. Sampaio Tavares Veras. 2002. Virulent Mycobacterium fortuitum restricts NO production by a gamma interferon-activated J774 cell line and phagosome-lysosome fusion. Infect. Immun. 705628-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez, A., S. Torello, and R. Kolter. 1999. Sliding motility in mycobacteria. J. Bacteriol. 1817331-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. Ratledge and J. Stanford (ed.), The biology of mycobacteria, vol. 2. Academic Press, London, England. [Google Scholar]
- 19.Minnikin, D. E., G. I. Hutchinson, A. B. Caldicott, and M. Goodfellow. 1980. Thin-layer chromatography of methanolysates of mycolic acid-containing bacteria. J. Chromatogr. 188221-233. [Google Scholar]
- 20.Minnikin, D. E., J. H. Parlett, G. Dobson, M. Goodfellow, M. Magnusson, and M. Ridell. 1985. Lipid profiles of members of the Mycobacterium tuberculosis complex, p. 75-78. In M. Casal (ed.), Mycobacteria of clinical interest. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 21.Okada, S., M. Nishiura, O. Mori, and T. Mori. 1978. Electron microscopic study of colonies of Mycobacterium lepraemurium. Int. J. Lepr. 46364-371. [PubMed] [Google Scholar]
- 22.Osborn, T. W. 1976. A study of some effects of subculture on two BCG strains. Tubercle 57181-195. [DOI] [PubMed] [Google Scholar]
- 23.Osborn, T. W. 1979. Serial subculture of BCG on solid and liquid media. Tubercle 6083-90. [DOI] [PubMed] [Google Scholar]
- 24.Primm, T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 1798-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recht, J., A. Martínez, S. Torello, and R. Kolter. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 1824348-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Güell, E., G. Agustí, M. Corominas, P. J. Cardona, I. Casals, T. Parella, M. A. Sempere, M. Luquin, and E. Julián. 2006. The production of a new extracellular putative long-chain saturated polyester by smooth variants of Mycobacterium vaccae interferes with Th1-cytokine production. Antonie van Leeuwenhoek 9093-108. [DOI] [PubMed] [Google Scholar]
- 27.Thomas, V., and G. McDonell. 2007. Relationship between mycobacteria and amoebae: ecological and epidemiological concerns. Lett. Appl. Microbiol. 45349-357. [DOI] [PubMed] [Google Scholar]
- 28.Tortoli, E. 2006. The new mycobacteria: an update. FEMS Immunol. Med. Microbiol. 48159-178. [DOI] [PubMed] [Google Scholar]
- 29.Valero-Guillén, P. L., and F. Martín-Luengo. 1986. 1-Tetradecanol, a new alcohol found in the cell wall of some rapidly growing chromogenic mycobacteria. FEMS Microbiol. Lett. 3559-63. [Google Scholar]
- 30.van Boxtel, R. M., R. S. Lambrecht, and M. T. Collins. 1990. Effect of polyoxyethylene sorbate compounds (Tweens) on colonial morphology, growth, and ultrastructure of Mycobacterium paratuberculosis. APMIS 98901-908. [DOI] [PubMed] [Google Scholar]
- 31.Wayne, L. G., and G. P. Kubica. 1986. The mycobacteria. p. 1435-1457. In J. G. Holt, P. H. Sneath, N. S. Mair, and M. E. Sharpe (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 32.Zamora, N., J. Esteban, T. J. Kinnari, A. Celdrán, J. J. Granizo, and C. Zafra. 2007. In-vitro evaluation of the adhesion to polypropylene sutures of non-pigmented, rapidly growing mycobacteria. Clin. Microbiol. Infect. 13902-907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.