Abstract
Clostridium difficile infection is increasing in both frequency and severity, with the emergence of new highly virulent strains highlighting the need for more rapid and effective methods of control. Here, we show that bacteriophage endolysin can be used to inhibit and kill C. difficile. The genome sequence of a novel bacteriophage that is active against C. difficile was determined, and the bacteriophage endolysin gene was subcloned and expressed in Escherichia coli. The partially purified endolysin was active against 30 diverse strains of C. difficile, and importantly, this group included strains of the major epidemic ribotype 027 (B1/NAP1). In contrast, a range of commensal species that inhabit the gastrointestinal tract, including several representatives of the clostridium-like Firmicutes, were insensitive to the endolysin. This endolysin provides a platform for the generation of both therapeutic and detection systems to combat the C. difficile problem. To investigate a method for the protected delivery and production of the lysin in the gastrointestinal tract, we demonstrated the expression of active CD27L endolysin in the lactic acid bacterium Lactococcus lactis MG1363.
Clostridium difficile is rapidly gaining notoriety as a cause of nosocomial diarrhea and colitis, with both the incidence and severity of infections increasing in the last few years (4, 10, 28). The pathogen is an obligately anaerobic gram-positive bacterium that has the capacity to form spores that resist heating, drying, and exposure to disinfectants, thus contributing to the persistence of the pathogen in the hospital environment (28). The pathogenic potential of virulent C. difficile strains is realized when the gastrointestinal (GI) tract microbiota becomes impaired or unbalanced, a common consequence of antibiotic therapy, leading to C. difficile-associated disease (CDAD) (4). Pathogenesis occurs as a result of the production of toxins, including the two major exotoxins A and B, with severe infections leading to pseudomembranous colitis (10). CDAD is currently treated with antibiotics, but posttreatment relapse is frequent (28). The increased incidence of CDAD and the emergence of more-virulent strains highlight the need for new approaches to the control of C. difficile. The association of CDAD with the disruption of the normal microbiota emphasizes the desirability of treatments which better target the pathogen without collateral damage to the protective commensal species. In this regard, bacteriophage therapy (21, 33, 34) has potential, and this approach has been investigated previously in the hamster disease model (35). Several temperate bacteriophages that are active against C. difficile have been identified and described in detail (12, 17-19, 27, 29, 42, 43). Also, a recent study (17) confirmed the ability of the prophages in the sequenced genome of the C. difficile strain 630 (42) to release active bacteriophage particles.
While bacteriophage therapy has potential, there are some limitations. All of the studied C. difficile bacteriophages appear to have narrow host ranges (12, 27, 29), which may be problematic for application in CDAD therapy. The selection of resistant mutants is likely to occur, and in the case of temperate bacteriophages, there is the potential for lysogenization to generate an immune subpopulation. We and others have previously investigated the potentials of bacteriophage endolysins as antimicrobial agents (11, 14-16, 26). This phenomenon of “lysis from without,” utilizing bacteriophage endolysins to attack pathogens, is well-established. Several authors have demonstrated impressive therapeutic results using bacteriophage endolysins both to treat infection and to prevent colonization with, for example, Streptococcus pneumoniae (23, 25); group A, C, and E streptococci (30); Staphylococcus aureus (36); and Bacillus anthracis (40), and endolysins have also been shown previously to act on biofilms (39). Endolysins specifically bind to and hydrolyze bacterial cell walls, and in the case of bacteriophage release, access is facilitated by the disruption of the cell membrane by an associated holin (11, 26). Endolysins consist of two domains—a peptidoglycan hydrolase activity is commonly located at the N terminus and can consist of one or more of a variety of glycosidases, amidases, or peptidases, while the endolysin's specificity is achieved by a cell wall binding domain that recognizes cell surface features that are specific to the bacteria that it targets (26). This targeting ability of the C-terminal region can provide an ideal combination of broad-spectrum activity against a range of strains from a given species and limited activity against organisms outside of that taxonomic group. Thus, an endolysin that is active against C. difficile but inactive against the commensal microbiota would provide a potential tool for therapy and a framework for genetic engineering to improve and/or extend activity (8), as well as for the basis of a method for C. difficile detection (24).
In this study, we discovered and sequenced the genome of a novel temperate bacteriophage from C. difficile and identified the endolysin gene, which we designated cd27l, by homology searches. We describe the expression of the endolysin as a histidine-tagged protein and show that this enzyme has activity against diverse C. difficile strains in vitro but no activity against clostridium-like members of the GI tract microbiota.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. difficile strains were obtained from the National Collection of Type Cultures, Central Public Health Laboratory, London, United Kingdom (NCTC 11204, NCTC 11205, NCTC 11206, NCTC 11207, NCTC 11208, NCTC 11209, NCTC 11223, NCTC 12726, NCTC 12727, NCTC 12728, NCTC 12731, NCTC 11382, NCTC 13287, NCTC 13307, and NCTC 13366), and Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany (DSMZ 12056 and DSMZ 12057), or were kindly donated by Jonathan Brazier, Anaerobe Reference Unit, Department of Medical Microbiology and Public Health Laboratory Service, University Hospital of Wales, Cardiff (R23 521, R23 524, R23 613, R23 614, R23 621, R23 635, R23 639, R23 642, R23 720, R23 727, R23 732, R23 737, and G83/03). Strains were grown anaerobically at 37°C in BHI+C (brain heart infusion [BHI] supplemented with vitamin K [10 μl; 50 mg/liter], hemin [5 mg/liter], resazurin [1 mg/liter], and l-cysteine [0.5 g/liter]) and were maintained in Robertson's cooked meat medium (SGL Ltd.) at room temperature. Escherichia coli strains were grown in Luria broth with shaking (200 rpm) at 37°C. Lactococcus lactis UKLc10 (46) was grown at 30°C in M17 (Difco) supplemented with 0.5% (wt/vol) glucose. Commensal, environmental, and clostridial strains were obtained from the culture collection of the Institute of Food Research (IFR), Norwich, United Kingdom, or DSMZ and were grown as recommended by DSMZ or in BHI+C. The following strains were used: Anaerococcus hydrogenalis (cluster XIII) DSMZ 7454, Bifidobacterium adolescentis DSMZ 20083, Bifidobacterium angulatum DSMZ 20098, Bifidobacterium bifidum DSMZ 20082, Bifidobacterium longum DSMZ 20219, Bifidobacterium pseudocatenulatum DSMZ 20438, Clostridium cellobioparum (cluster III) DSMZ 1351, Clostridium coccoides (cluster XIVa) NCTC 11035, Clostridium colinum (cluster XIVb) DSMZ 6011, Clostridium innocuum (cluster XIVb) DSMZ 1286, Clostridium leptum (cluster IV) DSMZ 753, Clostridium nexile (cluster XIVa) DSMZ 1787, Clostridium perfringens (cluster I) NCTC 3110, Clostridium ramosum (cluster XVIII) DSMZ 1402, Enterococcus faecalis FI10734, Enterococcus faecium FI10735, Enterococcus hirae FI10477, E. coli K-12, Eubacterium barkeri (cluster XV) DSMZ 1223, Lactobacillus bulgaricus FI10643, Lactobacillus casei FI10736, Lactobacillus gasseri NCIMB1171, Lactobacillus johnsonii FI9785, Lactobacillus plantarum FI108595, Lactobacillus rhamnosus FI10737, Lactobacillus sakei FI10645, L. lactis MG1363, Lactococcus garvieae FI08174, Listeria innocua NCTC 11288, Listeria monocytogenes NCTC 5412, Micrococcus luteus FI10640, Pediococcus pentosaceus FI10642, Pediococcus acidilactici FI10738, Salmonella enterica serovar Typhimurium FI10739, Salmonella enterica serovar Enteritidis FI10113, Staphylococcus aureus FI10139, Streptococcus anginosus FI10740, and Veillonella atypica FI10741.
Bacteriophage propagation and sequencing.
Bacteriophage particles were induced with mitomycin C (Sigma), harvested, and assayed as described by Sell et al. (43). Plaques were picked with a sterile Pasteur pipette into 250 μl of BHI+C and stored at 4°C. Supernatants were concentrated by polyethylene glycol precipitation (38), negatively stained in saturated uranyl acetate, and examined by electron microscopy (19). The novel bacteriophage referred to hereinafter as ΦCD27 was propagated by the infection of C. difficile type strain NCTC 11204 to increase the titer, and bacteriophage genomic DNA was extracted from cleared filtered lysate by using a λ midikit (Qiagen), giving a yield of ca. 160 μg. The presence of cohesive ends was tested by the heating of restricted DNA as described by Govind et al. (19).
The sequencing of the bacteriophage ΦCD27 genome and the assembly of the genome sequence were performed at the DNA sequencing facility of the Department of Biochemistry, University of Cambridge. The DNA sequences were assembled with the Phred-Phrap program. Open reading frames (ORFs) were identified by Artemis (37) with BlastP searches (1) using the UniProtKB/TrEMBL database (http://www.ebi.ac.uk/trembl/). Start sites were selected on the basis of the best match to the consensus ribosome binding site sequence AGGAGG or by comparison to homologous sequences. Genome comparisons were performed with ACT (6), and domain and family assignment searches were performed using InterProScan (49). Amino acid and nucleotide alignments were performed using the Clustal W algorithm in Vector NTI (Invitrogen).
Genomic Southern blot analysis.
A partial holin gene sequence was amplified by PCR from ΦCD27 genomic DNA by using Phusion DNA polymerase (Finnzymes) and primers CD27-HOL1 (5′-ATG GAT AAT TTA ATA AG) and CD27-HOL2 (5′-TCC TTC AAY TGT TTG TAA G), giving a product of 245 bp. C. difficile genomic DNA was extracted as described previously (41) and was digested with either HindIII or DraI, electrophoresed, and transferred onto a Hybond N+ membrane (Amersham) by conventional techniques (38). The holin probe was labeled, hybridized, and detected using the ECL kit (Amersham) with high-stringency washes.
Subcloning of bacteriophage endolysin genes into E. coli and L. lactis.
The ΦCD27 endolysin gene cd27l was amplified from genomic DNA by using primers designed to create an NdeI site at the 5′ end (5′-TTA CAT ATG AAA ATA TGT ATA ACA GTA GG, where underlining indicates the bases corresponding to the restriction endonuclease site) and a XhoI site downstream of the coding sequence (5′-CAA CCA CCT CGA GTT GAT AAC). NdeI- and XhoI-restricted PCR products were ligated into the expression vector pET15b (Novagen) by using Fast-Link DNA ligase (Epicentre), and the resulting construct, pET15b-cd27l, was introduced into chemically competent E. coli TOP10 (Invitrogen). Positive transformants were selected with ampicillin (100 μg/ml), gene identity was confirmed by sequencing, and chemically competent E. coli BL21(DE3) (Invitrogen) was transformed with the plasmid constructs or the original vector pET15b for protein expression.
For expression in L. lactis, the cd27l coding sequence was subcloned into the vector pUK200 (46), which had been modified to include a sequence encoding a six-histidine tag, yielding pUK200His (N. Horn, unpublished data). Electrocompetent E. coli MC1022 was transformed with the construct pUK200His-cd27l, and transformants were selected on chloramphenicol (15 μg/ml). After the confirmation of the plasmid sequence, electrocompetent L. lactis UKLc10 was transformed with pUK200His-cd27l or the empty vector, and transformants were selected with 5 μg of chloramphenicol/ml (46). Electroporation was performed as described previously for Lactobacillus johnsonii (22) by using a pulse of 2.5 kV, 200 Ω, and 25 μF.
Protein expression, analysis, and partial purification.
Endolysin expression in E. coli BL21(DE3) was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 to 4 h. Crude protein lysates from induced and uninduced cultures containing pET15b-cd27l or pET15b were produced by cell disruption with 0.1-mm acid-washed glass beads (Sigma) in TN buffer (20 mM Tris-HCl, pH 8, 50 mM NaCl) by using a FastPrep FP120 cell disrupter (Savant) with four 30-s bursts on speed setting 10, with incubation on ice for 5 min between bursts. Cell debris was pelleted by centrifugation at 13,000 × g for 20 min at 4°C, and the supernatants were stored at −20°C. Protein expression of the recombinant L. lactis strains containing pUK200His-cd27l or pUK200His was induced for 5 h with 1 ng of nisin/ml (46), and crude proteins were extracted as described above for E. coli. His-tagged endolysins from E. coli were partially purified under native conditions by using the nickel-nitrilotriacetic acid (Ni-NTA) Fast Start kit (Qiagen), which eluted the protein in a final buffer of 50 mM Na phosphate, 300 mM NaCl, and 250 mM imidazole, pH 8.0 (referred to hereinafter as EB).
Proteins were quantified using Bradford reagent (Bio-Rad) and visualized on 10% NuPage Novex bis-Tris gels in MOPS (morpholinepropanesulfonic acid) buffer stained with Simply Blue Safestain (Invitrogen). For Western blot analysis, the electrophoresed proteins were transferred onto a polyvinylidene difluoride membrane by using NuPage buffer (Invitrogen). His-tagged endolysins were detected as described in the Ni-NTA Fast Start kit handbook (Qiagen) by using an anti-His tag monoclonal antibody (Novagen) with alkaline phosphatase-linked anti-mouse immunoglobulin G (Sigma) as the secondary antibody and colorimetric detection with Sigma Fast BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium as the substrate.
Lysis assays.
The lysis of C. difficile cells was assessed based on the method described by Yoong et al. (47). Cells of C. difficile strain NCTC 11204 were grown to mid-log phase, 1.8-ml aliquots were harvested by centrifugation (13,000 × g for 2 min) into screw-cap tubes, and pellets were either used immediately or flash-frozen in liquid nitrogen and stored at −20°C. Pellets were resuspended in 1.3 ml of phosphate-buffered saline (PBS), pH 7.3, on ice, and 270-μl aliquots were added to 30 μl of either endolysin or a buffer. For crude protein lysate, the buffer control was TN, and for Ni-NTA-purified protein (protein E1), the buffer control was EB. The drop in the optical density at 600 nm (OD600) at 37°C was measured in duplicate in multiwell plates by using the Bioscreen C system (Labsystems), with shaking and measurement every 2 min. The activity profiles under different pH conditions were examined by adjusting the PBS to pHs between 4.5 and 8.3 (47). To assess activities against different bacterial species, 7 μg of partially purified protein (E1) was used with an incubation time of 1 h. For viable-cell counts, replicate assays under anaerobic conditions were set up by using 100 μg of partially purified endolysin with prereduced PBS and continuous gentle shaking. Assays were carried out in duplicate, with cell counts determined at time zero to allow the estimation of the number of cells in each assay mixture. Cells were assayed in 10-fold dilutions from ca. 108 cells to ca. 103 cells. At 2 h, 30-μl samples were taken for 10-fold serial dilutions in PBS; 10-μl aliquots of these dilutions were spotted onto BHI agar, and the remaining 270-μl assay mixture from one of each duplicate pair was plated to allow cell enumeration.
RESULTS
ΦCD27 genome analysis.
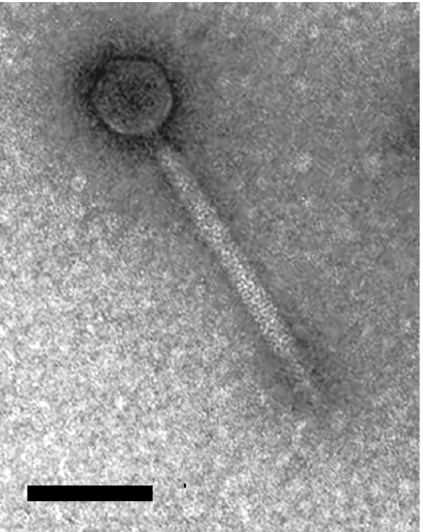
Mitomycin C induction of C. difficile strain NCTC 12727 yielded a bacteriophage, designated ΦCD27, which produced plaques on 4 of the 27 strains tested, including the type strain NCTC 11204 and strains NCTC 11205, NCTC 11207, and NCTC 11209. Electron microscopy analysis of ΦCD27 (Fig. 1) revealed a bacteriophage with an icosahedral head of ca. 60 nm and a tail with a width of ca. 17 nm and a length of between 158 and 210 nm. This bacteriophage shows a clear resemblance to other C. difficile bacteriophages ΦCD119 (19) and ΦC2 (17); this similarity and the presence of a tail sheath suggest that it belongs to the family Myoviridae of the order Caudovirales. The induction and cross testing of an additional 26 C. difficile strains failed to identify any new bacteriophage particles.
FIG. 1.
Electron micrograph of bacteriophage ΦCD27. The bar represents 100 nm.
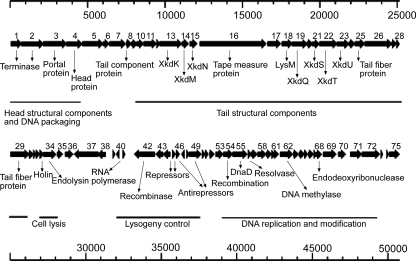
The ΦCD27 genome comprises double-stranded DNA with an average GC content of 29.4%. DNA sequencing of the 50,930-bp genome revealed no distinct termini, and the restriction and electrophoresis of heated and unheated samples of genomic DNA failed to identify potential cohesive ends. Artemis analysis of the genomic sequence identified 75 proposed ORFs (Fig. 2; also see the supplemental material). Many of these ORFs showed significant homology to identified bacteriophage ORFs, including those from C. difficile bacteriophages ΦCD119 (orf1 to orf79) (19) and ΦC2 (p1 to p84) (17) and the two prophages in the sequenced C. difficile strain 630 genome (prophage 1 [CD0904 to CD0979] and prophage 2 [CD2889 to CD2952]) (42). Matches were also found in sequences from bacterial genomes of members of the Clostridiaceae, which may be further prophage sequences, and in the sequences of various ORFs in the C. difficile strain 630 chromosome outside of the determined prophage areas; these sequences may represent evolutionary leftovers or bacterial genes with similar functions. Putative functions were assigned on the basis of BLAST results where present and/or the results of domain searches (see the supplemental material).
FIG. 2.
ΦCD27 genome map showing predicted ORFs. Arrows indicate the directions of transcription. Proposed functional modules are marked based on BLAST results and similarity to published sequences of ΦCD119, ΦC2, and C. difficile strain 630 prophages (17, 19, 42) (see the supplemental material).
ACT analysis of both nucleotide and translated sequences indicated large areas of homology to the published bacteriophage sequences (17, 19, 42). The sequences of ΦC2 (17) showed the highest degrees of similarity, particularly in the area of the DNA replication and modification module and the region encompassing orf27 to orf42, which includes the cell lysis module. However, unlike those in ΦC2, the genes for holin and endolysin in ΦCD27 are immediately adjacent to each other. The proposed area of DNA replication and modification retains roughly the same order of genes as that in ΦC2 from p62 (orf51) to p80 (orf70), with the apparent loss of both p74 and p75 and the addition of three short putative ORFs (orf65, orf66, and orf67) in ΦCD27; this module and the proposed lysogeny control module showed the highest levels of homology to sequences from ΦC2 or C. difficile strain 630 prophage 2 (42). In contrast, in the region encoding tail structural components, the highest degrees of similarity were those to sequences, presumably of prophage regions, from genomes of other members of the Clostridiales, particularly Clostridium beijerinckii, Clostridium botulinum, and “Alkaliphilus metalliredigens.”
The sequence of the proposed ΦCD27 endolysin gene (cd27l, or orf34) is 813 bp, coding for a predicted protein of 270 amino acid residues. The nucleotide and amino acid sequences aligned with N-acetylmuramoyl-l-alanine amidase gene and protein sequences from C. difficile phages, with the greatest levels of homology being those to the endolysin protein sequence from ΦC2 (95.9% amino acid identity) and to the sequence of a hypothetical protein (ZP_01803398) corresponding to the chromosome of C. difficile QCD-32g58 (ca. 94.4% amino acid identity). Goh et al. (17) also found some bacteriophage-related sequences in this strain, but a complete prophage genome was not identified. A holin probe covering the first 245 bp of orf33 showed 86 to 89% nucleotide homology to holin genes from published C. difficile phage genome sequences. This probe hybridized to sequences from 16 other strains of C. difficile, supporting previous observations that the carriage of prophage sequences in C. difficile is widespread (17, 29) (data not shown).
Cloning and expression of a ΦCD27 gene for biologically active endolysin in E. coli.
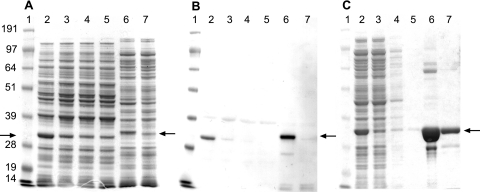
The ΦCD27 endolysin gene cd27l was expressed with a sequence encoding a six-histidine tag in E. coli, and the tagged protein was visible in electrophoresed crude extracts (Fig. 3A), with a size of ca. 32 kDa. The identity of the tagged protein was confirmed by Western analysis using a His tag antibody, and the results showed that the endolysin was present in both induced and uninduced samples (Fig. 3B). Nickel-NTA columns were used to partially purify the His-tagged endolysin, designated CD27L, from E. coli extracts (Fig. 3C), and this protein was subsequently used in lysis assays with the sensitive C. difficile strain 11204.
FIG. 3.
(A) Gel analysis of crude protein lysates from E. coli and L. lactis expressing ΦCD27 endolysin and empty vector controls. Lanes: 1, SeeBlue marker (Invitrogen); 2 to 5, lysates from L. lactis containing pUK200His-cd27l (2 and 3) or pUK200His (4 and 5) with (2 and 4) or without (3 and 5) nisin induction; and 6 and 7, lysates from E. coli containing pET15b-cd27l (6) or pET15b (7), both with IPTG induction. Arrows indicate the His-tagged endolysin—when it is expressed from pET15b-cd27l with the His tag MGSSHHHHHHSSGLVPRGSH, the size is ca. 32 kDa, and when it is expressed from pUK200His-cd27l with the His tag MSHHHHHHA, the size is ca. 31 kDa. (B) Western analysis of the gel in panel A with a six-His tag antibody. (C) Gel analysis of Ni-NTA column-purified His-tagged endolysin from E. coli expressing pET15b-cd27l. Lanes: 1, SeeBlue marker; 2 to 5, E. coli(pET15b-cd27l) total protein extracts after 5 h of induction with IPTG; 2, crude lysate; 3, column flowthrough; 4, primary wash effluent; 5, secondary wash effluent; 6, primary eluate (E1); and 7, secondary eluate (E2). Crude protein lysates were loaded at 10 μg of total protein per lane, while lanes loaded with samples from Ni-NTA column fractionation contained 6.5 μl per lane.
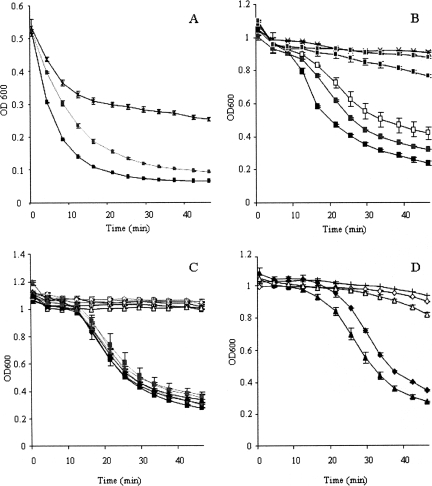
Partially purified CD27L rapidly lysed C. difficile cells that had been flash-frozen in liquid nitrogen. In these assays, 7 μg of protein reduced the OD600 by half within 6 min. However, the frozen cells also showed gradual lysis in buffer-only controls (Fig. 4A). The use of unfrozen cells gave more stable controls, although the endolysin-induced lysis was slower, with a lag of 5 to 15 min compared to frozen cells before the lysis was observed. As little as 350 ng of protein produced a drop in the OD compared to that of controls, and progressively more efficient lysis was achieved with increasing endolysin concentrations (Fig. 4B). CD27L was shown to maintain similar levels of activity over a fairly wide range of pHs, between 4.5 and 8.3 (Fig. 4C). Crude protein extracted in TN buffer also produced an effective drop in cell density (Fig. 4D). The extraction and assay of crude protein in alternative buffers (20 mM sodium phosphate, pH 8, or 50 mM Tris, pH 7.5) gave no significant effect on the enzyme activity, nor did increasing the NaCl concentration to 100 mM affect the activity (data not shown).
FIG. 4.
Lysis assays with cells of C. difficile strain 11204. Cells were grown to mid-exponential phase, harvested by centrifugation, and then either flash-frozen in liquid nitrogen and stored at −20°C before being assayed (A) or used immediately (B to D). Cells were incubated with Ni-NTA column-purified endolysin (E1) from E. coli expressing CD27L from pET15b-cd27l (A to C) or with crude protein extracts (D). (A) Effect of CD27L (7 μg, black squares, or 0.7 μg, gray squares) on frozen cells compared to that of the buffer control (EB; x's). (B) Effects of different amounts of CD27L (10.5 μg, black rectangles; 3.5 μg, gray rectangles; 0.7 μg, white rectangles; 0.35 μg, black squares; and 0.07 μg, gray squares) on fresh cells compared to that of the buffer control (EB; x's). (C) Activity profile of CD27L (black symbols) compared to that of the buffer control (EB; white symbols) tested at pH 4.5 (black squares), 5.8 (gray squares), 6.5 (black diamonds), 7.0 (gray diamonds), 7.6 (black triangles), and 8.3 (gray triangles). (D) Lysis of cells incubated with 50 μg of crude protein extracts from E. coli expressing pET15b-cd27l (black triangles) or pET15b (white triangles) or from L. lactis expressing pUK200His-cd27l (black diamonds) or pUK200His (white diamonds) or with the TN buffer control (plus signs). Values are the means of results from duplicate assays ± standard deviations.
Replicate assays under anaerobic conditions were performed to examine the effect on cell viability. With the addition of 100 μg of partially purified CD27L, assay mixtures containing ca. 108 cells at time zero showed a drop of 1 log after 2 h of incubation, while assay mixtures to which 107 or 106 cells had been added showed a drop of 2 log compared to buffer controls. In assays with lower initial cell numbers, the lysin was more effective, with only four viable colonies being recovered from an assay mixture inoculated with 105 cells and no live cells remaining in assay mixtures inoculated with 104 cells or fewer.
Specificity of the ΦCD27 endolysin.
Of the 30 different C. difficile strains tested, all were sensitive to the partially purified CD27L endolysin, and importantly, this group included two strains of the hypervirulent ribotype 027. CD27L failed to decrease the ODs of cultures of a range of other Clostridium spp. or examples of the major clostridial clusters commonly represented in the GI tract (clusters III, IV, XIVa, XIVb, XV, and XVI [9]), as well as a selection of other gut commensals and bacteria from different environments (the strains tested are listed in Materials and Methods).
Expression of endolysin CD27L in L. lactis.
The delivery of bacteriophage endolysin to the GI tract represents a distinct challenge, and one approach is to use a genetically modified lactic acid bacterium. The potential of L. lactis MG1363 for GI tract delivery of vaccines (20, 31) and immunomodulatory cytokines (5, 44, 48) has already been established. Thus, we cloned the gene encoding histidine-tagged bacteriophage endolysin CD27L into L. lactis under the control of the well-established nisin-inducible promoter PnisA. The production of a protein of the appropriate size was demonstrated with crude extracts (Fig. 3A). Western analysis with a His tag antibody confirmed the presence of the histidine-tagged protein, and elevated production following induction with nisin was demonstrated (Fig. 3B). Crude protein extracts from L. lactis expressing CD27L lysed fresh cells (Fig. 4C) to a degree similar to that observed with E. coli as the host for gene expression.
DISCUSSION
The cross screening of mitomycin C-induced supernatants from 27 strains of C. difficile identified only one bacteriophage. The rarity of the detection of bacteriophages in C. difficile following mitomycin C induction has been noted in previous publications (12, 18, 27), and it has been suggested that this rarity is due to the limited host range of temperate phages, possibly due to high-frequency carriage of prophages (17, 29). Our observation that more than half of the strains examined contained sequences which hybridized to the ΦCD27 holin sequence supports this premise. The ΦCD27 genome sequence shows large areas of homology to published C. difficile bacteriophage sequences, notably those of ΦC2 and prophage 2 of C. difficile strain 630. The similarity among prophages has been cited previously as an evolutionary benefit, allowing the exchange of modules (7). The analysis of the ΦCD27 genome lends weight to this idea, with certain modules showing closer homology to sequences (presumably prophage sequences) from environmental Clostridiales than to those from the C. difficile bacteriophages.
In this work, we identified the bacteriophage ΦCD27 gene that encodes an endolysin, expressed the endolysin gene in both E. coli and L. lactis, and demonstrated that the endolysin enzyme was biologically active, lysing cells of C. difficile. The ΦCD27 bacteriophage had a narrow host range, infecting only four of the tested strains. The host range of the endolysin was significantly broader, as the enzyme affected all 30 strains tested—this phenomenon has been noted before (14-16) and emphasizes the improved therapeutic potential of endolysins compared to those of their parent bacteriophages. From the viewpoint of potential exploitation as a novel antimicrobial agent, the significant observation is that an endolysin derived from a C. difficile bacteriophage can have a broad spectrum of lytic activity against strains of C. difficile but little activity against a range of clostridium-like commensal species that are a significant part of the GI tract microbiota. These properties offer the potential to develop a targeted antimicrobial that destroys C. difficile without causing collateral damage to the protective GI tract microbiota. Although therapeutic results for endolysin activity in vivo have been impressive (23, 30, 36, 40), the use of an endolysin in the chemically complex and microbially diverse GI tract is demanding. Delivery to the lower GI tract represents a particular challenge, and we have demonstrated that biologically active C. difficile endolysin can be expressed in an L. lactis strain that has an established capacity for the delivery of therapeutically effective vaccines (20, 31) and cytokines (5, 44, 48) to the GI tract. It is encouraging that the C. difficile ΦCD27 endolysin was biologically active over a relatively wide pH range, suggesting that it would remain functional in the GI tract environment.
We have demonstrated that CD27L is effective against C. difficile both in a cell lysis assay and in an analysis of cell viability, where complete killing of 104 cells was achieved. However, this activity is not as rapid or devastating as those of some other bacteriophage endolysins that are active against other host bacterial species (25, 40). This result may reflect the properties of this endolysin, but it is also possible that the heterologous expression of a correctly folded protein is suboptimal. Other workers have noted problems with heterologous expression and the formation of inclusion bodies or a requirement for rare codons (32, 50). Our characterization of CD27L provides a start point from which to investigate the improvement of activity by genetic engineering or by the coexpression of other enzyme activities from the same C-terminal binding domain. Gaeng et al. (13) found an unexpected increase in lytic activity from the truncation of a Listeria monocytogenes endolysin, while Donovan et al. (8) produced an effective antimicrobial by fusing endolysin sequences to lysostaphin. We plan to investigate these approaches, including the definition and separate characterization of a cell wall binding domain and the exploitation of hybrid endolysins.
One potential problem with the use of an endolysin to destroy cells of C. difficile is that bacterial lysis in the gut may cause a short-term increase in the symptoms of CDAD due to the release of internalized toxins (45). This issue could be addressed by combining the endolysin treatment with developing therapies that aim to neutralize the toxins, including the use of antibodies (2) and toxin-absorbing agents such as tolevamer (3, 28). These approaches in isolation have the potential for symptom relief by neutralizing the toxin, but they do not eliminate the pathogen. In contrast to conventional antibiotic treatments, treatment with the CD27L C. difficile endolysin, with its exquisite specificity, offers a novel approach to the destruction of the pathogen with the maintenance of an effective GI tract microbiota.
Supplementary Material
Acknowledgments
We are grateful to Jon Brazier (Public Health Laboratory, Cardiff, United Kingdom) for the generous donation of C. difficile strains, Nikki Horn (IFR) for providing the plasmid pUK200His and for valuable expert advice, Mary Parker (IFR) for electron microscopy, Carmen Nueno-Palop and Carl Harrington (IFR) for providing commensal strains, John Lester and Shilo Dickens (University of Cambridge) for bacteriophage sequencing, Udo Wegmann (IFR) for plasmid sequencing and advice, Andrew Warry (BBSRC Bioscience IT Services, Harpenden, United Kingdom) for expert computational biology support, and Claire Shearman (IFR) for helpful discussions.
This work was supported by funding from the Biotechnology and Biological Science Research Council.
Footnotes
Published ahead of print on 15 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, Z. Zhang, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babcock, G. J., T. J. Broering, H. J. Hernandez, R. B. Mandell, K. Donahue, N. Boatright, A. M. Stack, I. Lowy, R. Graziano, D. Molrine, D. M. Ambrosino, and W. D. Thomas. 2006. Human monoclonal antibodies directed against toxins A and B prevent Clostridium difficile-induced mortality in hamsters. Infect. Immun. 746339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, R. H., R. Dagher, D. M. Davidson, and J. K. Marquis. 2006. Tolevamer, a novel toxin-binding polymer: overview of preclinical pharmacology and physicochemical properties. Aliment. Pharmacol. Ther. 241525-1534. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, J. G. 2006. The new epidemic of Clostridium difficile-associated enteric disease. Ann. Intern. Med. 145758-764. [DOI] [PubMed] [Google Scholar]
- 5.Braat, H., P. Rottiers, D. W. Hommes, N. Huyghebaert, E. Remaut, J. P. Remon, S. J. H. Van Deventer, S. Neirynck, M. P. Peppelenbosch, and L. Steidler. 2006. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn's disease. Clin. Gastroenterol. Hepatol. 4754-759. [DOI] [PubMed] [Google Scholar]
- 6.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 213422-3423. [DOI] [PubMed] [Google Scholar]
- 7.Casjens, S. R. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8451-458. [DOI] [PubMed] [Google Scholar]
- 8.Donovan, D. M., S. Dong, W. Garrett, G. M. Rousseau, S. Moineau, and D. G. Pritchard. 2006. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl. Environ. Microbiol. 722988-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 3081635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, B., B. J. Chang, C. L. Golledge, and T. V. Riley. 2007. Clostridium difficile-associated diarrhoea. Intern. Med. J. 37561-568. [DOI] [PubMed] [Google Scholar]
- 11.Fischetti, V. A. 2005. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13491-496. [DOI] [PubMed] [Google Scholar]
- 12.Fortier, L. C., and S. Moineau. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl. Environ. Microbiol. 737358-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaeng, S., S. Scherer, H. Neve, and M. J. Loessner. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 662951-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasson, M. J. October 1992. Bacteriophage lysins and their applications in destroying and testing for bacteria. European patent EP0510907.
- 15.Gasson, M. J. April 1991. Lysins from bacteriophages. United Kingdom patent GB2255561B.
- 16.Gasson, M. J. June 1998. Viral products. U.S. patent 5,763,251.
- 17.Goh, S., P. F. Ong, K. P. Song, T. V. Riley, and B. J. Chang. 2007. The complete genome sequence of Clostridium difficile phage ΦC2 and comparisons to ΦCD119 and inducible prophages of CD630. Microbiology 153676-685. [DOI] [PubMed] [Google Scholar]
- 18.Goh, S., T. V. Riley, and B. J. Chang. 2005. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl. Environ. Microbiol. 711079-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govind, R., J. A. Fralick, and R. D. Rolfe. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage ΦCD119. J. Bacteriol. 1882568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanniffy, S. B., A. T. Carter, E. Hitchin, and J. M. Wells. 2007. Mucosal delivery of a pneumococcal vaccine using Lactococcus lactis affords protection against respiratory infection. J. Infect. Dis. 195185-193. [DOI] [PubMed] [Google Scholar]
- 21.Hermoso, J. A., J. L. Garcia, and P. Garcia. 2007. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 10461-472. [DOI] [PubMed] [Google Scholar]
- 22.Horn, N., U. Wegmann, A. Narbad, and M. J. Gasson. 2005. Characterisation of a novel plasmid p9785S from Lactobacillus johnsonii FI9785. Plasmid 54176-183. [DOI] [PubMed] [Google Scholar]
- 23.Jado, I., R. Lopez, E. Garcia, A. Fenoll, J. Casal, and P. Garcia. 2003. Phage lytic enzymes as therapy for antibiotic-resistant Streptococcus pneumoniae infection in a murine sepsis model. J. Antimicrob. Chemother. 52967-973. [DOI] [PubMed] [Google Scholar]
- 24.Kretzer, J. W., R. Lehmann, M. Schmelcher, M. Banz, K. P. Kim, C. Korn, and M. J. Loessner. 2007. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl. Environ. Microbiol. 731992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 2942170-2172. [DOI] [PubMed] [Google Scholar]
- 26.Loessner, M. J. 2005. Bacteriophage endolysins—current state of research and applications. Curr. Opin. Microbiol. 8480-487. [DOI] [PubMed] [Google Scholar]
- 27.Mahony, D. E., P. D. Bell, and K. B. Easterbrook. 1985. Two bacteriophages of Clostridium difficile. J. Clin. Microbiol. 21251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monaghan, T., T. Boswell, and Y. R. Mahida. 2008. Recent advances in Clostridium difficile-associated disease. Gut 57850-860. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, E., and J. Foldes. 1991. Electron microscopic investigation of lysogeny of Clostridium difficile strains isolated from antibiotic-associated diarrhea cases and from healthy carriers. APMIS 99321-326. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, D., L. Loomis, and V. A. Fischetti. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 984107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nouaille, S., L. A. Ribeiro, A. Miyoshi, D. Pontes, Y. Le Loir, S. C. Oliveira, P. Langella, and V. Azevedo. 2003. Heterologous protein production and delivery systems for Lactococcus lactis. Genet. Mol. Res. 2102-111. [PubMed] [Google Scholar]
- 32.O'Flaherty, S., A. Coffey, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2005. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1877161-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parisien, A., B. Allain, J. Zhang, R. Mandeville, and C. Q. Lan. 2008. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. Microbiol. 1041-13. [DOI] [PubMed] [Google Scholar]
- 34.Petty, N. K., T. J. Evans, P. C. Fineran, and G. P. Salmond. 2007. Biotechnological exploitation of bacteriophage research. Trends Biotechnol. 257-15. [DOI] [PubMed] [Google Scholar]
- 35.Ramesh, V., J. A. Fralick, and R. D. Rolfe. 1999. Prevention of Clostridium difficile-induced ileocecitis with bacteriophage. Anaerobe 569-78. [Google Scholar]
- 36.Rashel, M., J. Uchiyama, T. Ujihara, Y. Uehara, S. Kuramoto, S. Sugihara, K. Yagyu, A. Muraoka, M. Sugai, K. Hiramatsu, K. Honke, and S. Matsuzaki. 2007. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage ΦMR11. J. Infect. Dis. 1961237-1247. [DOI] [PubMed] [Google Scholar]
- 37.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16944-945. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sass, P., and G. Bierbaum. 2007. Lytic activity of recombinant bacteriophage Φ11 and Φ12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl. Environ. Microbiol. 73347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418884-889. [DOI] [PubMed] [Google Scholar]
- 41.Sebaihia, M., M. W. Peck, N. P. Minton, N. R. Thomson, M. T. Holden, W. J. Mitchell, A. T. Carter, S. D. Bentley, D. R. Mason, L. Crossman, C. J. Paul, A. Ivens, M. H. Wells-Bennik, I. J. Davis, A. M. Cerdeno-Tarraga, C. Churcher, M. A. Quail, T. Chillingworth, T. Feltwell, A. Fraser, I. Goodhead, Z. Hance, K. Jagels, N. Larke, M. Maddison, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, B. White, S. Whithead, and J. Parkhill. 2007. Genome sequence of a proteolytic (group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 171082-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38779-786. [DOI] [PubMed] [Google Scholar]
- 43.Sell, T. L., D. R. Schaberg, and F. R. Fekety. 1983. Bacteriophage and bacteriocin typing scheme for Clostridium difficile. J. Clin. Microbiol. 171148-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2891352-1355. [DOI] [PubMed] [Google Scholar]
- 45.Voth, D. E., and J. D. Ballard. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18247-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wegmann, U., J. R. Klein, I. Drumm, O. P. Kuipers, and B. Henrich. 1999. Introduction of peptidase genes from Lactobacillus delbrueckii subsp. lactis into Lactococcus lactis and controlled expression. Appl. Environ. Microbiol. 654729-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoong, P., R. Schuch, D. Nelson, and V. A. Fischetti. 2004. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 1864808-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuvaraj, S., M. P. Peppelenbosch, and N. A. Bos. 2007. Transgenic probiotica as drug delivery systems: the golden bullet? Expert Opin. Drug Deliv. 41-3. [DOI] [PubMed] [Google Scholar]
- 49.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17847-848. [DOI] [PubMed] [Google Scholar]
- 50.Zimmer, M., N. Vukov, S. Scherer, and M. J. Loessner. 2002. The murein hydrolase of the bacteriophage Φ3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl. Environ. Microbiol. 685311-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.