Abstract
Restricted bioavailability of copper in certain environments can interfere with cellular respiration because copper is an essential cofactor of most terminal oxidases. The global response of the metabolically versatile bacterium and opportunistic pathogen Pseudomonas aeruginosa to copper limitation was assessed under aerobic conditions. Expression of cioAB (encoding an alternative, copper-independent, cyanide-resistant ubiquinol oxidase) was upregulated, whereas numerous iron uptake functions (including the siderophores pyoverdine and pyochelin) were expressed at reduced levels, presumably reflecting a lower demand for iron by respiratory enzymes. Wild-type P. aeruginosa was able to grow aerobically in a defined glucose medium depleted of copper, whereas a cioAB mutant did not grow. Thus, P. aeruginosa relies on the CioAB enzyme to cope with severe copper deprivation. A quadruple cyo cco1 cco2 cox mutant, which was deleted for all known heme-copper terminal oxidases of P. aeruginosa, grew aerobically, albeit more slowly than did the wild type, indicating that the CioAB enzyme is capable of energy conservation. However, the expression of a cioA′-′lacZ fusion was less dependent on the copper status in the quadruple mutant than in the wild type, suggesting that copper availability might affect cioAB expression indirectly, via the function of the heme-copper oxidases.
Copper is an essential micronutrient for most living organisms, as it participates in electron transport and in many biologically important redox reactions. The ability of copper to cycle between an oxidized Cu(II) state and a less stable reduced Cu(I) state makes it an important catalytic cofactor of cytochrome oxidases, the terminal enzymes in cellular respiration, and of other oxidases utilizing dioxygen (18). However, copper can become highly cytotoxic if allowed to accumulate in excess of cellular needs, as it is involved in the production of reactive oxygen species, including hydroxyl radicals (20). Therefore, both prokaryotes and eukaryotes must tightly regulate copper homeostasis (44).
Bacteria have evolved different strategies to maintain the intracellular copper concentration at a low level and within a narrow range (8). Two major types of mechanisms that prevent a copper overload in gram-negative bacteria have been described. One type involves periplasmic multicopper oxidases and copper-sequestering proteins, which are expressed under the control of two-component systems sensing periplasmic copper ions. For example, the proteins of such systems are encoded by the copABCD and copRS operons in Pseudomonas syringae (3, 36) and the pcoABCD pcoE pcoRS cluster in Escherichia coli (44). The other type of mechanism relies mainly on P-type ATPases that actively pump Cu(I) ions out of the cytoplasm and that are positively regulated by cytoplasmic transcription factors. As examples, we may cite the products of the cueAR operon in Pseudomonas putida and in Pseudomonas fluorescens (1, 19) and the similar copA-cueR system in E. coli (39). In Pseudomonas aeruginosa, mutation in either copR or cueA results in increased sensitivity to toxic copper concentrations (49).
Whereas many studies have focused on how bacterial cells avoid copper toxicity, less is known about how microorganisms react to and cope with copper deficiency. In aqueous solutions under oxic conditions, copper is present in its cupric Cu(II) form. Above pH 7.4, Cu(II) can form poorly soluble carbonates and hydroxides (27). In biological fluids, copper is mostly bound to organic molecules. In human serum, the concentration of free Cu(II) is estimated to be about 10−13 M, mainly due to the complexation of copper with plasma proteins such as albumin, ceruloplasmin, and transcuprein (24, 32). The fact that copper can be poorly bioavailable raises the question of how environmental and pathogenic microorganisms adapt to copper limitation. There are scattered reports in the literature on this issue. For instance, in the marine bacterium Pseudomonas perfectomarina (now called Pseudomonas stutzeri), a lack of copper interferes with the last step of denitrification, i.e., the reduction of nitrous oxide to dinitrogen, which is catalyzed by a copper-containing enzyme (33). In the cyanobacterium Synechocystis sp., copper deprivation causes an arrest of respiratory metabolism because cytochrome c oxidase fails to function, whereas photoautotrophic growth remains possible (17). Some methane-oxidizing bacteria can scavenge copper ions by producing specific chalkophores (copper chelators); chalkophores are akin to siderophores, which are iron chelators and provide iron to iron-starved cells (29). In the yeast Saccharomyces cerevisiae, copper starvation results in the downregulation of respiratory functions and reveals a link between copper and iron metabolism (50).
We have begun to study the adaptation of P. aeruginosa to copper limitation. P. aeruginosa is a widely occurring environmental bacterium and a pathogen in compromised hosts (25, 41); as such, it is likely to encounter situations of limited copper availability. The ability of P. aeruginosa to cause disease is based not only on its capacity to produce a large variety of virulence factors but also on its great metabolic versatility. P. aeruginosa is an aerobic, facultatively anaerobic organism which preferentially obtains its metabolic energy via aerobic respiration and is well adapted to low oxygen concentrations. By controlling the expression of multiple cytochrome oxidases, P. aeruginosa appears to exploit the best-suited electron transport chain in response to the available oxygen supply. The genome sequence reveals gene clusters for three cytochrome c oxidases (ccoNOQP1, ccoNOQP2, and coxBA-coIII) and one quinol oxidase (cyoABCDE), all of which belong to the heme-copper superfamily (9, 10, 11). Heme-copper oxidases can be inhibited by cyanide, resulting in a block of electron transport via these oxidases. Interestingly, P. aeruginosa is one of the few bacteria capable of producing cyanide at concentrations that can inhibit its own heme-copper oxidases (7). To prevent self-intoxication, P. aeruginosa has a cytochrome bd-type cyanide-insensitive oxidase (CIO) (the product of the cioAB cluster), which apparently lacks copper in its active site and allows the bacterium to respire oxygen when the other oxidases are inhibited (14).
Here we investigated the adaptation of P. aeruginosa to copper limitation under aerobic conditions. The organism's global transcriptional response reveals that a range of genes involved in iron metabolism and respiration is affected. In a copper-depleted environment, P. aeruginosa entirely relies on CIO for aerobic respiration, and CIO expression is markedly induced. Genetic analysis suggests that CIO induction is a consequence of reduced aerobic respiration via the four cyanide-sensitive terminal oxidases.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains and plasmids used in this study are listed in Table 1. All media and solutions were prepared with deionized, double-distilled water. Bacteria were routinely grown on nutrient agar and in nutrient yeast broth (NYB) (48) at 37°C. When required, antibiotics were added to these media at the following concentrations: 100 μg/ml for ampicillin, 12.5 μg/ml for tetracycline, 25 μg/ml for kanamycin for E. coli, and 300 μg/ml for carbenicillin and 100 μg/ml for tetracycline for P. aeruginosa. Growth and β-galactosidase experiments were performed in a minimal medium (OS-glucose) containing 0.5% (wt/vol) glucose, 0.1% (wt/vol) ammonium sulfate, 0.01% (wt/vol) Triton X-100, and salt solutions (38), from which CuSO4 was omitted unless stated otherwise. All glassware was rendered copper free by a 24-h treatment with 0.1 M HCl and rinsed once in double-distilled water before sterilization. A freshly prepared ascorbic acid solution (final concentration, 1 mM) and the copper chelator bathocuproine disulphonic acid (BCS; final concentration, 150 μM; Sigma-Aldrich) were added to OS-glucose when appropriate. Control experiments in which ascorbic acid had been omitted showed that under these conditions the complexation of copper by BCS was incomplete. Growth in OS-glucose medium was obtained in 100-ml Erlenmeyer flasks filled with 20 ml of medium, under conditions of good aeration (shaking at 180 rpm) at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169 [φ80dlacZΔΜ15] F− Nalr | 45 |
| HB101 | proA2 hsdS20(rB− m B−) recA13 ara-14 lacY1 galK2 rpsL20 supE44 xyl-5 mtl-1 F− | 45 |
| P. aeruginosa | ||
| PAO1 (ATCC 15692) | Wild type | American Type Culture Collection |
| PAO6437 | PAO1 containing a 2,400-bp deletion in the cioAB locus | This study |
| PAO6573 | PAO1 containing a 1,933-bp deletion in the copRS locus | This study |
| PAO6593 | PAO1 containing a 4,109-bp deletion in the coxBA-PA0107-coIII locus | This study |
| PAO6594 | PAO1 containing a 1,688-bp deletion in the roxSR locus | This study |
| PAO6597 | PAO6593 containing a 4,830-bp deletion in the cyoABCDE operon | This study |
| PAO6650 | PAO6597 containing a 6,445-bp deletion in the two adjacent ccoNOQP1 and ccoNOQP2 operons | This study |
| Plasmids | ||
| pRK2013 | Helper plasmid; Tra+ Kmr | 16 |
| pME3087 | Suicide vector for allelic replacement; Tcr; ColE1 replicon | 54 |
| pME3641 | Plasmid carrying a translational proC′-′lacZ fusion; Cbr | 46 |
| pME6013 | Cloning vector for translational lacZ fusions; Tcr | 47 |
| pME6015 | Cloning vector for translational lacZ fusions; Tcr | 47 |
| pME6016 | Cloning vector for transcriptional lacZ fusions; Tcr | 47 |
| pME6031 | Expression vector carrying ptac lacIQ; Tcr | 23 |
| pME7226 | Plasmid carrying a translational pchR′- ′lacZ fusion; Tcr | 34 |
| pME7541 | Suicide construct used for deletion of the cioAB operon; Tcr | This study |
| pME7554 | Plasmid carrying a translational cioA′- ′lacZ fusion; Tcr | This study |
| pME7576 | Suicide construct used for deletion of the copRS operon; Tcr | This study |
| pME9301 | Plasmid carrying a translational pvdS′- ′lacZ fusion; Tcr | This study |
| pME9302 | Suicide construct used for deletion of the coxB-coIII cluster; Tcr | This study |
| pME9303 | Suicide construct used for deletion of the cyoABCDE operon; Tcr | This study |
| pME9305 | pME6031 derivative carrying the cioAB genes; Tcr | This study |
| pME9306 | Plasmid carrying a transcriptional cioA-lacZ fusion; Tcr | This study |
| pME9307 | Suicide construct used for deletion of the roxSR operon; Tcr | This study |
| pME9308 | Suicide construct used for deletion of the two adjacent ccoNOQP operons; Tcr | This study |
Determination of cellular copper concentrations.
Total copper in whole cells was measured by inductively coupled plasma mass spectrometry (Hewlett-Packard 4500; Agilent Technologies, Palo Alto, CA). P. aeruginosa PAO1 was grown in OS-glucose medium with vigorous shaking at 37°C for 14 h. This culture was diluted 1:200 in 200 ml of the same medium supplemented with ascorbic acid plus BCS, ascorbic acid alone, or 1.5 μM CuSO4. The cultures were harvested in exponential growth phase (optical density at 600 nm [OD600] ≅ 1) by centrifugation at 15,000 × g for 15 min, washed twice with 5 ml 0.9% NaCl, digested with 500 μl of low-metal-content concentrated HNO3 (Baker instra grade) at 95°C for 1 h, and then diluted with 4.5 ml of double-distilled water before the measurements were performed. The instrument was calibrated using a standard CuSO4 solution. In parallel, viable counts (CFU/ml) were measured for the bacterial cultures. This allowed us to estimate the number of copper atoms/viable P. aeruginosa cell.
Construction of plasmids and gene replacement mutants.
DNA cloning and plasmid preparations were performed according to standard methods (45). Large-scale preparations of plasmid DNA were performed using JETstar 2.0 (Genomed). Restriction and DNA-modifying enzymes were used following the instructions of the manufacturers. All oligonucleotide primers used below are listed in Table S1 in the supplemental material. A transcriptional cioA-lacZ fusion, in which the +1 nucleotide of lacZ was fused to the major +1 start site of the cioA promoter (14), was constructed by cloning a 410-bp fragment containing the cioAB promoter region into the EcoRI-BamHI sites of pME6016. This fragment was generated by PCR using the P. aeruginosa PAO1 genome as the template and primers cioA-Pa1 and cioRV2. A translational cioA′-′lacZ fusion was constructed by inserting a 621-bp BglII-EcoRI fragment carrying the proximal part of cioA into the BamHI-EcoRI sites of pME6015. This fragment was generated by PCR amplification of the PAO1 genome by use of primers cioA-Pa1 and cioA-Pa2. A translational pvdS′-′lacZ fusion was constructed similarly by fusing a 0.65-kb EcoRI-BamHI fragment carrying the proximal part of pvdS with its own promoter (amplified from the PAO1 genome with primers PpvdSFW and PpvdSRV) to ′lacZ in pME6013.
For the inactivation of the cioAB operon in the P. aeruginosa PAO1 chromosome, a 624-bp fragment overlapping cioA and a 617-bp fragment overlapping cioB were amplified by PCR using the primer couples cioA-Pa1/cioA-Pa2 and cioB-Pa1/cioB-Pa2, respectively. These products were digested with EcoRI-BglII and BglII-HindIII, respectively, and cloned into the corresponding sites of the suicide vector pME3087, giving plasmid pME7541. Plasmid pME7541 was then introduced into P. aeruginosa PAO1 by triparental mating, using the helper strain E. coli HB101(pRK2013). Merodiploids were resolved as previously described (58). The resulting strain, P. aeruginosa PAO6437, carried an in-frame ΔcioAB mutation. To complement this mutation, a 4-kb fragment containing the cioAB operon with its own promoter region was PCR amplified from the PAO1 genome by use of primers cioA-Pa1 and cioB-Pa2. The product was then digested with EcoRI and HindIII and cloned into the corresponding sites of the shuttle vector pME6031, giving plasmid pME9305.
For the deletion of coxB (PA0105), coxA (PA0106), PA0107, and coIII (PA0108), a 1,316-bp fragment overlapping coxB and a 1,084-bp fragment overlapping coIII were amplified by PCR using primers coxupFW/coxupRV and coxdwFW/coxdwRV, respectively. These products were digested with BamHI-EcoRI and EcoRI-HindIII, respectively, and cloned into pME3087, giving plasmid pME9302. Plasmid pME9302 was then used as described above to produce strain PAO6593.
A double mutant (PAO6597) deleted for the cox and cyoABCDE clusters was obtained as follows. A 1,060-bp fragment overlapping cyoA and a 1,085-bp fragment overlapping cyoE were amplified by PCR using primers cyoupFW/cyoupRV and cyodwFW/cyodwRV, respectively. These products were digested with BamHI-EcoRI and EcoRI-HindIII, respectively, and cloned into pME3087, giving plasmid pME9303, which served to construct strain PAO6597. A quadruple mutant carrying a deletion of all four operons encoding heme-copper terminal oxidases was derived from PAO6597 as follows. A 1,287-bp fragment overlapping ccoN2 (PA1557) and a 1,280-bp fragment overlapping ccoP1 (PA1552) were amplified by PCR using primers ccoupFW/ccoupRV and ccodwFW/ccodwRV, respectively. These products were digested with BamHI-EcoRI and EcoRI-HindIII, respectively, and cloned into pME3087, resulting in plasmid pME9308. Plasmid pME9308 was crossed into PAO6597 as described above, giving strain PAO6650 [Δ(coxBA-PAO107-coIII) ΔcyoABCDE ΔccoNOQP1 ccoNOQP2].
For the deletion of roxS (PA4494) and roxR (PA4493) from the PAO1 chromosome, an 802-bp fragment overlapping roxS and an 896-bp fragment overlapping roxR were amplified by PCR using primers mroxSFW/mroxSRV and mroxRFW/mroxRRV, respectively. These products were digested with BamHI-XbaI and XbaI-HindIII, respectively, and cloned into pME3087, giving plasmid pME9307. Plasmid pME9307 was then introduced into P. aeruginosa PAO1 as described above; after the excision of the integrated plasmid strain, PAO6594 (ΔroxSR) was obtained.
A copRS (PA2809-PA2810) mutant of PAO1 was constructed by amplifying a 993-bp fragment carrying copR′ and a 615-bp fragment carrying ′copS with primers copupFW/copupRV and copdwFW/copdwRV, respectively. These products were cut with EcoRI-BamHI and BamHI-HindIII, respectively, and cloned into pME3087, resulting in plasmid pME7576, which was used to generate strain PAO6573 (ΔcopRS) as described above. For all mutants described here, the deletions were confirmed by PCR and PCR fragments were checked by sequencing.
β-Galactosidase assays and pyoverdine determination.
β-Galactosidase assays (35) were performed with P. aeruginosa cultures grown in triplicate in OS-glucose medium. Data are mean values for three independent samples ± standard deviations. PyoverdinePAO1 was quantified by measuring the absorbance at 405 nm of culture supernatants diluted 9:1 in 100 mM Tris-HCl (pH 8.0) per cell population density (in OD600 units) as previously described (53).
RNA isolation, generation of cDNA probes, and transcriptome analysis.
P. aeruginosa PAO1 was inoculated at an OD600 of 0.01 into 20 ml of OS-glucose medium supplemented with 1 mM of ascorbic acid, with or without 150 μM BCS. The cultures were grown at 37°C with vigorous shaking, until they reached an OD600 of approximately 1, and then cells were harvested and RNAProtect Bacteria (Qiagen) were added. Total RNA was isolated by the hot phenol method as described elsewhere (31), followed by DNase I treatment (Roche). The integrity of total RNA was confirmed by agarose gel electrophoresis and an RNA 6000 Nano LabChip in an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). Next, 10 μg of total RNA was used with random primers and Superscript II reverse transcriptase (Invitrogen Corp., Carlsbad, CA) to perform cDNA synthesis. cDNA fragmentation, labeling, hybridization, staining, and washing steps were performed according to the manufacturer's protocol for the Affymetrix P. aeruginosa GeneChip arrays (Affymetrix, Inc., Santa Clara, CA). Finally, the arrays were scanned with the Affymetrix GeneChip scanner 3000. Processing of the P. aeruginosa GeneChip (Affymetrix) was performed at the University of Lausanne Center for Integrative Genomics. For each condition, cultures were grown in triplicate, and RNAs from these cultures were pooled before proceeding to cDNA synthesis. In addition, biological replicates for each condition were performed on a separate day and run on a different microarray chip. We refer to the “most strongly induced or repressed genes” as those genes meeting the following criteria: (i) the P value obtained for each transcript analyzed is less than 0.05 and (ii) the absolute change in the transcript level is equal to or greater than twofold.
RESULTS
Copper depletion affects the expression of genes involved in iron metabolism of P. aeruginosa.
To assess the effect of a copper-depleted environment on the transcriptional expression of P. aeruginosa genes, we developed a defined medium (OS-glucose) containing BCS, a specific, high-affinity Cu(I) chelator (Kd, 10−20 M [6, 43]). BCS has previously been used to create copper limitation in a Synechocystis sp. and in yeast (17, 50). Unless stated otherwise, OS-glucose medium was not amended with copper salts, contained an excess of iron (10 μM FeSO4), and was supplemented with 1 mM ascorbic acid (freshly prepared) to reduce any trace of Cu(II) present to Cu(I) (18). Various BCS concentrations were tested to determine the highest level of BCS that could be added to liquid OS-glucose without affecting the growth rate of the wild-type P. aeruginosa PAO1. When strain PAO1 was grown in this medium containing 150 μM BCS, its growth rate was indistinguishable from that observed for the unamended medium (data not shown). Ascorbate was omitted in those experiments where CuSO4 was added because ascorbate would have exacerbated copper toxicity. Growth was achieved in shake flasks under conditions of good aeration; therefore, oxygen was not a growth-limiting factor.
Inductively coupled plasma mass spectrometry analysis revealed that wild-type P. aeruginosa PAO1 cells, when grown in the presence of BCS for six generations, displayed a significant decrease in the cellular copper concentration (1,000 ± 400 Cu atoms per viable cell) compared with that found for the unamended culture (38,000 ± 7,000 Cu atoms per viable cell) and with that found in a culture supplemented with 1.5 μM CuSO4 (360,000 ± 70,000 Cu atoms per viable cell). The value measured for cells grown in BCS medium was close to the detection limit and represents a maximal estimate. These results, obtained from three independent experiments with strain PAO1, show that BCS can be used to reduce the cellular copper content to very low levels and also support previous observations that P. aeruginosa has homeostatic regulatory mechanisms preventing an overload of cellular copper (49). In a control experiment, we verified that the complexation of copper by BCS and the addition of excess copper did not cause significant changes in cellular iron, zinc, nickel, manganese, molybdenum, and cobalt (data not shown).
To search for genes differentially regulated during copper starvation versus copper sufficiency, we performed a transcriptomic analysis of P. aeruginosa cells grown to exponential phase (OD600 ≈ 1.0) in the presence or absence of BCS. Among the 5,901 genes represented on the Affymetrix chip, 132 genes exhibited a ≥2-fold decrease in transcript levels, whereas only 10 genes showed a ≥2-fold increase in mRNA levels (Table 2). Copper starvation resulted in a strong decrease of the expression of many genes and of operons that play a role in siderophore-mediated iron acquisition (13, 42, 52), including genes for pyoverdine biosynthesis (pvdS for the master regulator and sigma factor PvdS, pvdA, pvdQ, pvdP, pvdNO, pvdF, pvdE, pvdIJD, pvdH, pvdLG), pyochelin biosynthesis (pchR for the pathway-specific regulator PchR, pchDCBA), the corresponding siderophore receptors (fpvA and fpvB; fptA), and a TonB protein (PA5531) (Table 2). Copper starvation also diminished the expressions of genes involved in heme uptake and metabolism (hasA, hasR, phuT, phuR, hxuC, and hemO) and heterologous siderophore uptake (pfeR, pfeS, and pfeA; pirA; chtA; foxA, foxR, and foxI) and of the manganese-cofactored superoxide dismutase (sodA) and fumarase (fumC1) genes (Table 2). In general, the expression of these genes is known to be repressed by the ferric uptake regulator Fur in the presence of iron (13, 22, 42, 52). Surprisingly, few genes were expressed at elevated levels during copper starvation. Among such genes, we noted bfrB, encoding the iron storage protein bacterioferritin (Table 2). Altogether, there was a strong overlap between P. aeruginosa genes responding to copper starvation (Table 2) and those regulated by iron depletion (12, 37, 40), although copper and iron limitations have opposite effects on the expression of these genes.
TABLE 2.
List of 142 P. aeruginosa genes most strongly induced or repressed in response to copper starvation
| Gene (name) | Fold change | Protein (function) |
|---|---|---|
| Induced genes | ||
| PA0460 | 3.0 | Hypothetical protein |
| PA0918 | 2.8 | Cytochrome b561 |
| PA1562 (acnA) | 2.1 | Aconitate hydratase 1 |
| PA1761 | 2.3 | Hypothetical protein |
| PA2953 | 2.2 | Electron transfer flavoprotein-ubiquinone oxidoreductase |
| PA3235 | 3.8 | Conserved hypothetical protein |
| PA3531 (bfrB) | 2.8 | Bacterioferritin |
| PA3602 | 2.0 | Conserved hypothetical protein |
| PA3923 | 2.3 | Hypothetical protein |
| PA5300 (cycB) | 2.1 | Cytochrome c5 |
| Repressed genes | ||
| PA0149 | −2.1 | Probable sigma-70 factor, ECF subfamilya |
| PA0150 | −2.1 | Probable transmembrane sensor |
| PA0423 | −2.1 | Conserved hypothetical protein |
| PA0471 | −4.1 | Probable transmembrane sensor for ferrioxamine |
| PA0472 | −4.6 | Probable sigma-70 factor, ECF subfamily |
| PA0524 (norB) | −2.1 | Nitric oxide reductase subunit B |
| PA0672 (hemO) | −27.7 | Heme oxygenase |
| PA0802 | −2.7 | Hypothetical protein |
| PA0929 | −2.8 | Two-component response regulator |
| PA0931 (pirA) | −13.4 | Receptor protein for enterobactin |
| PA1134 | −2.1 | Hypothetical protein |
| PA1245 | −2.8 | Hypothetical protein |
| PA1300 | −6.6 | Probable sigma-70 factor, ECF subfamily |
| PA1301 | −5.7 | Probable transmembrane sensor |
| PA1302 | −2.4 | Probable heme utilization protein precursor |
| PA1318 (cyoB) | −2.7 | Cytochrome o ubiquinol oxidase subunit I |
| PA1319 (cyoC) | −2.8 | Cytochrome o ubiquinol oxidase subunit III |
| PA1363 | −3.6 | Probable sigma-70 factor, ECF subfamily |
| PA1364 | −2.3 | Probable transmembrane sensor |
| PA1365 | −3.0 | Probable siderophore receptor |
| PA1701 | −2.3 | Conserved hypothetical protein in type III secretion |
| PA1706 (pcrV) | −2.3 | Type III secretion protein PcrV |
| PA1707 (pcrH) | −2.5 | Regulatory protein PcrH |
| PA1708 (popB) | −2.2 | Translocator protein PopB |
| PA1709 (popD) | −2.1 | Translocator outer membrane protein PopD |
| PA1710 (exsC) | −2.3 | Type III secretion regulator ExsC |
| PA1711 (exsE) | −2.0 | Type III secretion protein ExsE |
| PA1713 (exsA) | −2.2 | Type III secretion transcriptional regulator ExsA |
| PA1714 (exsD) | −2.5 | Type III secretion regulator ExsD |
| PA1715 (pscB) | −2.0 | Type III secretion apparatus protein |
| PA1716 (pscC) | −2.1 | Type III secretion outer membrane protein PscC precursor |
| PA1718 (pscE) | −6.1 | Type III secretion protein PscE |
| PA1719 (pscF) | −3.8 | Type III secretion protein PscF |
| PA1720 (pscG) | −2.4 | Type III secretion protein PscG |
| PA1721 (pscH) | −2.7 | Type III secretion protein PscH |
| PA1722 (pscI) | −2.7 | Type III secretion protein PscI |
| PA1723 (pscJ) | −2.1 | Type III secretion protein PscJ |
| PA1909 | −2.3 | Hypothetical protein |
| PA1910 (ufrA) | −2.3 | Probable TonB-dependent receptor protein |
| PA1911 | −3.6 | Probable transmembrane sensor |
| PA1912 | −3.9 | Probable sigma-70 factor, ECF subfamily |
| PA2033 | −10.6 | Hypothetical protein |
| PA2034 | −5.0 | Hypothetical protein |
| PA2384 | −16.7 | Hypothetical protein |
| PA2385 (pvdQ) | −31.8 | Acylase |
| PA2386 (pvdA) | −111.4 | l-Ornithine N5-oxygenase |
| PA2389 | −6.1 | Conserved hypothetical protein |
| PA2390 | −6.8 | Probable ATP-binding/permease fusion ABC transporter |
| PA2391 (ompQ) | −4.6 | Probable outer membrane protein |
| PA2392 (pvdP) | −7.6 | Periplasmic protein PvdP |
| PA2393 | −57.6 | Probable dipeptidase |
| PA2394 (pvdN) | −69.5 | Periplasmic protein PvdN |
| PA2395 (pvdO) | −25.7 | Membrane protein PvdO |
| PA2396 (pvdF) | −36.5 | Hydroxy-ornithine formylase |
| PA2397 (pvdE) | −42.3 | ABC transporter PvdE |
| PA2398 (fpvA) | −35.9 | Ferripyoverdine receptor |
| PA2399 (pvdD) | −21.6 | Nonribosomal peptide synthetase PvdD |
| PA2400 (pvdJ) | −19.3 | Nonribosomal peptide synthetase PvdJ |
| PA2401 (pvdJ) | −23.2 | |
| PA2402 (pvdI) | −17.5 | Nonribosomal peptide synthetase PvdI |
| PA2403 | −6.7 | Hypothetical protein |
| PA2404 | −6.9 | Hypothetical protein |
| PA2405 | −8.0 | Hypothetical protein |
| PA2406 | −2.7 | Hypothetical protein |
| PA2407 | −5.4 | Probable adhesion protein |
| PA2408 | −6.6 | Probable ATP-binding component of ABC transporter |
| PA2409 | −5.0 | Probable permease of ABC transporter |
| PA2410 | −4.3 | Hypothetical protein |
| PA2411 | −34.3 | Probable thioesterase |
| PA2412 | −81.5 | Conserved hypothetical protein MbtH |
| PA2413 (pvdH) | −40.8 | l-2,4-Diaminobutyrate:2-ketoglutarate 4-aminotransferase |
| PA2424 (pvdL) | −26.6 | Nonribosomal peptide synthetase PvdL |
| PA2425 (pvdG) | −11.6 | Thioesterase PvdG |
| PA2426 (pvdS) | −64.5 | Sigma factor PvdS |
| PA2427 | −7.3 | Hypothetical protein |
| PA2451 | −5.4 | Hypothetical protein |
| PA2452 | −14.5 | Hypothetical protein |
| PA2466 (foxA) | −2.6 | Ferrioxamine receptor FoxA |
| PA2467 (foxR) | −3.2 | Anti-sigma factor FoxR |
| PA2468 (foxI) | −3.3 | ECF sigma factor FoxI |
| PA2664 (fhp) | −2.6 | Flavohemoprotein |
| PA2686 (pfeR) | −3.3 | Two-component response regulator PfeR |
| PA2687 (pfeS) | −3.6 | Two-component sensor PfeS for enterobactin |
| PA2688 (pfeA) | −5.2 | Ferric enterobactin receptor, outer membrane protein PfeA |
| PA3392 (nosZ) | −2.1 | Nitrous oxide reductase |
| PA3407 (hasAp) | −5.4 | Heme acquisition protein HasAp |
| PA3408 (hasR) | −2.5 | Heme uptake outer membrane receptor HasR |
| PA3409 | −2.3 | Probable transmembrane sensor |
| PA3410 | −6.4 | Probable sigma-70 factor, ECF subfamily |
| PA3446 | −2.2 | Conserved hypothetical protein |
| PA3530 | −9.5 | Conserved hypothetical protein |
| PA3601 | −2.0 | Conserved hypothetical protein |
| PA3690 | −3.1 | Probable metal-transporting P-type ATPase |
| PA3841 (exoS) | −2.1 | Exoenzyme S |
| PA3842 | −2.4 | Probable chaperone |
| PA3866 | −3.1 | Pyocin protein |
| PA3899 | −9.2 | Probable sigma-70 factor, ECF subfamily |
| PA3900 | −3.5 | Probable transmembrane sensor for Fe(III) citrate |
| PA3938 | −2.1 | Probable periplasmic taurine-binding protein |
| PA4156 | −2.5 | Probable TonB-dependent receptor |
| PA4168 (fpvB) | −25.6 | Second ferric pyoverdine receptor FpvB |
| PA4218 (fptX) | −4.1 | Fe(III)-pyochelin inner membrane permease |
| PA4220 (fptB) | −5.1 | Hypothetical protein |
| PA4221 (fptA) | −4.5 | Fe(III)-pyochelin outer membrane receptor |
| PA4227 (pchR) | −12.4 | Transcriptional regulator PchR |
| PA4228 (pchD) | −4.0 | Salicylate adenylase |
| PA4229 (pchC) | −4.3 | Thioesterase PchC |
| PA4230 (pchB) | −4.7 | Isochorismate pyruvate-lyase |
| PA4231 (pchA) | −3.0 | Isochorismate synthase |
| PA4359 | −2.3 | Conserved hypothetical protein |
| PA4467 | −11.4 | Hypothetical protein |
| PA4468 (sodA) | −34.9 | Superoxide dismutase |
| PA4469 | −56.1 | Hypothetical protein |
| PA4470 (fumC) | −68.9 | Fumarate hydratase FumC1 |
| PA4471 | −40.6 | Hypothetical protein |
| PA4513 | −3.8 | Probable oxidoreductase |
| PA4514 | −5.8 | Probable outer membrane receptor for iron transport |
| PA4515 | −2.2 | Conserved hypothetical protein |
| PA4570 | −17.4 | Hypothetical protein |
| PA4675 (chtA) | −2.5 | Probable TonB-dependent receptor for aerobactin |
| PA4705 | −3.4 | Hypothetical protein |
| PA4706 | −5.4 | Probable ATP-binding component of ABC transporter |
| PA4707 | −7.6 | Probable permease of ABC transporter |
| PA4708 (phuT) | −13.8 | Heme transport protein PhuT |
| PA4709 | −13.6 | Probable hemin degrading factor |
| PA4710 (phuR) | −113.9 | Heme/hemoglobin uptake outer membrane receptor PhuR |
| PA4711 | −2.0 | Hypothetical protein |
| PA4895 | −3.2 | Probable transmembrane sensor |
| PA4896 | −8.6 | Probable sigma-70 factor, ECF subfamily |
| PA5304 (dadA) | −2.0 | d-Amino acid dehydrogenase, small subunit |
| PA5530 | −2.1 | Probable dicarboxylate transporter |
| PA5531 (tonB) | −2.8 | TonB protein |
ECF, extracytoplasmic function.
Copper starvation downregulates siderophore expression.
To validate the transcriptional profiling data, we investigated the effect of copper starvation on the expression of two major siderophore regulators, PvdS and PchR, in P. aeruginosa PAO1. To this end, we measured the expression of translational pvdS′-′lacZ and pchR′-′lacZ fusions (carried by plasmids pME9301 and pME7226, respectively). As for the microarray analysis, cells were grown in OS-glucose medium with or without the copper chelator BCS. Under the usual iron-replete conditions and in the presence of BCS, both fusions were repressed in wild-type PAO1 (Table 3), confirming the transcriptome data. Due to the high iron concentration used in this experiment, pyoverdine could be not be detected in the culture supernatant.
TABLE 3.
Pyoverdine and pyochelin gene expression and pyoverdine production in P. aeruginosa PAO1
| Strain | Fusion | Fe statusa | Cu statusb | OD600 | β-Galactosidase activity (avg Miller units ± SD) | Pyoverdine concn (avg A405/OD600 ± SD) |
|---|---|---|---|---|---|---|
| PAO1(pME9301) | pvdS′-′lacZ | + | + | 1.15 ± 0.19 | 126.6 ± 15.2 | NDc |
| + | − | 1.11 ± 0.08 | 33.7 ± 5.4 | ND | ||
| − | + | 0.10 ± 0.03 | 440.0 ± 148.9 | 1.27 ± 0.40 | ||
| − | − | 0.11 ± 0.01 | 224.9 ± 9.6 | 0.58 ± 0.04 | ||
| PAO1(pME7226) | pchR′-′lacZ | + | + | 1.22 ± 0.10 | 40.5 ± 5.3 | ND |
| + | − | 0.92 ± 0.07 | 3.3 ± 0.7 | ND | ||
| − | + | 0.13 ± 0.02 | 331.0 ± 29.1 | 1.07 ± 0.09 | ||
| − | − | 0.14 ± 0.01 | 240.7 ± 27.4 | 0.59 ± 0.09 | ||
| PAO6650(pME9301) | pvdS′-′lacZ | + | + | 1.56 ± 0.09 | 51.7 ± 1.4 | ND |
| + | − | 1.38 ± 0.06 | 10.3 ± 0.5 | ND | ||
| PAO6650(pME7226) | pchR′-′lacZ | + | + | 1.56 ± 0.01 | 94.0 ± 28.8 | ND |
| + | − | 1.38 ± 0.04 | 6.1 ± 0.5 | ND |
OS-glucose medium with (+) or without (−) 10 μM FeSO4; 1 mM ascorbate was always present.
OS-glucose medium supplemented (−) or not (+) with 150 μM BCS; 1 mM ascorbate was always present.
ND, not determined.
We next measured pyoverdine and the activities of the pvdS′-′lacZ and pchR′-′lacZ fusions in cells grown in OS-glucose medium without added FeSO4. In this medium, cells had difficulty growing, and therefore the assays were performed at low cell population densities (Table 3). Nevertheless, both pyoverdine production and pvdS′-′lacZ expression were downregulated by the addition of BCS under these conditions (Table 3). BCS addition also caused a significant reduction in the expression of the pchR′-′lacZ fusion (Table 3). Thus, copper limitation also had a negative impact on siderophore expression in P. aeruginosa under conditions of poor iron availability. Previous work indicates that both pyoverdine and pyochelin of P. aeruginosa are able to chelate not only Fe(III) but also Cu(II) (53, 57), although it is questionable whether these siderophores can actually promote copper uptake. Our data argue against a scenario in which copper-depleted cells would induce siderophore production to enhance copper uptake.
Copper availability influences the expression of aerobic respiratory pathways in P. aeruginosa.
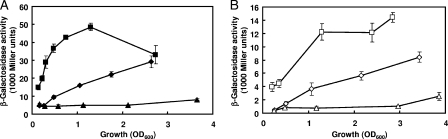
The transcriptome data (Table 2) reveal that two genes of the quinol oxidase operon cyoABCDE, cyoB (PA1318) and cyoC (PA1319), were downregulated, whereas two genes encoding uncharacterized cytochromes (PA0918 and PA5300) were upregulated during conditions of copper starvation. The cyoABCDE cluster encodes a putative bo3-type quinol oxidase (55), which appears to function as a low-affinity terminal oxidase under high-oxygen conditions (2). Furthermore, for copper-starved cells we observed a two- to threefold downregulation in the expression of 16 genes involved in the type III secretion apparatus (pcrV, pcrH, popB, popD, exsC, exsE, exsA, exsD, and pscBCEFGHIJ) (Table 2), which can probably be attributed to the reduced expression of the PvdS sigma factor and, consequently, of the major ExsA regulator (12). While we did not follow up the effect on type III secretion genes, we decided to investigate the impact of copper starvation on respiratory functions in more detail. Assuming that copper is an essential cofactor of the cyo, cox, cco1, and cco2 terminal oxidases, we reasoned that during copper starvation these oxidases would not function properly and that P. aeruginosa would mainly rely on the CIO—which does not contain Cu—to respire oxygen. Given that the transcriptional expression of cyo oxidase was downregulated (Table 2), we expected to see a compensating upregulation of the expression of the alternative quinol oxidase CIO. Although such an effect was not evident from the microarray data (taken at an early growth phase), we found that a transcriptional cioA-lacZ fusion carried by pME9306 was upregulated in strain PAO1 grown in OS-glucose medium during conditions of copper limitation, especially during early growth phases (Fig. 1A). As the β-galactosidase expression of this fusion was extremely high, this experiment may not reflect the full extent of upregulation during later growth phases. A translational cioA′-′lacZ fusion (on pME7556), which specified lower β-galactosidase activities, showed upregulated expression throughout growth under copper limitation (Fig. 1B). In unsupplemented OS-glucose medium, the expression of both fusion constructs increased in a growth-phase-dependent manner, as previously observed by Cooper et al. (11). An excess (1.5 μM) of CuSO4 strongly repressed the expression of both fusions (Fig. 1A and B). To show that this regulation was specific, we measured the expression of a constitutive housekeeping gene, proC (46), under the same conditions. We found that the expression level of a translational proC′-′lacZ fusion (on pME3641) in strain PAO1 remained constant at 800 ± 150 Miller units throughout growth, with or without the addition of BCS.
FIG. 1.
Activities of β-galactosidase reporter plasmids containing either a cioA-lacZ transcriptional fusion (A) or a cioA′-′lacZ translational fusion (B) in wild-type strain PAO1. Cultures were grown aerobically in OS-glucose medium containing 1 mM ascorbate (diamonds), the ascorbate medium amended with 150 μM BCS (squares), or medium with 1.5 μM CuSO4 but without ascorbate (triangles). Each value is the average of three different cultures ± standard deviation.
CIO is crucial for aerobic growth of P. aeruginosa during copper limitation.
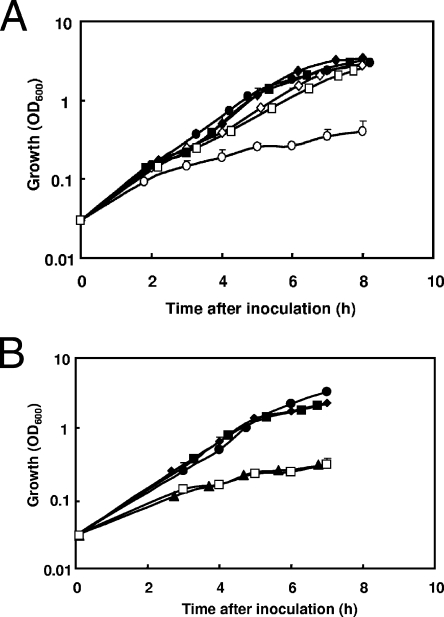
To demonstrate the pivotal function of CIO in aerobic respiration during copper limitation, we constructed a cioAB deletion mutant of P. aeruginosa PAO1 (PAO6437) and tested its ability to grow in OS-glucose medium with or without the addition of BCS. Without BCS, the mutant PAO6437 and the wild-type PAO1 showed similar growth rates (Fig. 2A). The addition of BCS strongly inhibited the growth of strain PAO6437 but had little effect on wild-type PAO1 and on the complemented mutant PAO6437/pME9305 (Fig. 2A). In the complementing plasmid, pME9305, the cioAB operon is under the control of its own promoter. The addition of 2 μM CuSO4 to the medium containing BCS fully restored the growth of the cioAB mutant PAO6437. By contrast, the addition of 10 μM ZnSO4 did not restore growth in the presence of BCS and had no effect in the absence of BCS (Fig. 2B). Taken together, these results show that CIO is essential for aerobic growth of P. aeruginosa during copper limitation.
FIG. 2.
(A) Effect of copper starvation on the growth of the wild-type PAO1, the cioAB mutant PAO6437, and the complemented mutant PAO6437/pME9305. (A) Growth of the wild-type PAO1 (diamonds), PAO6437 (circles), and PAO6437/pME9305 (squares) in OS-glucose containing 1 mM ascorbate was measured by turbidimetry. Cultures were untreated (filled symbols) or contained 150 μM BCS (open symbols). Each value is the average of three different cultures ± standard deviation. (B) Relief of BCS-induced copper starvation by copper but not by zinc. Strain PAO6347 (cioAB) was grown in untreated OS-glucose containing 1 mM ascorbate (filled diamonds), supplemented with 150 μM BCS (filled triangles), 10 μM ZnSO4 (filled squares), 150 μM BCS plus 10 μM ZnSO4 (open squares), or 150 μM BCS plus 2 μM CuSO4 (filled circles). Each value is the average of three different cultures ± standard deviation.
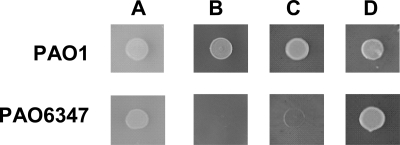
We confirmed the copper requirement of the cioAB mutant on solid OS-glucose medium by using another specific Cu(I) chelator, tetrathiomolybdate (TTM) (5, 30). In the presence of 1 mM TTM, the wild-type PAO1 grew, whereas the cioAB mutant was completely inhibited. The addition of CuSO4 overcame this inhibition partially at 30 μM and entirely at 300 μM (Fig. 3).
FIG. 3.
Specific growth inhibition of a P. aeruginosa cioAB mutant by the copper chelator TTM. The wild-type PAO1 and the cioAB mutant PAO6437 were grown in OS-glucose to an OD600 of approximately 2.5 and then 10 μl of each culture was spotted onto OS-glucose plates (A) supplemented with 1 mM TTM (B), with 1 mM TTM and 30 μM CuSO4 (C), or with 1 mM TTM and 300 μM CuSO4 (D). Incubation was at 37°C for 18 h.
Copper-mediated regulation of CIO does not rely on the two-component systems RoxSR and CopRS.
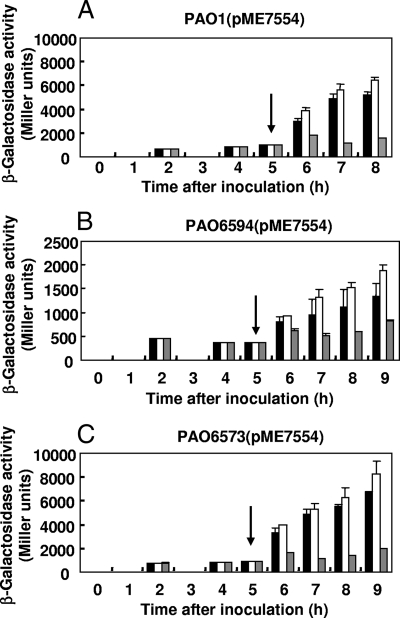
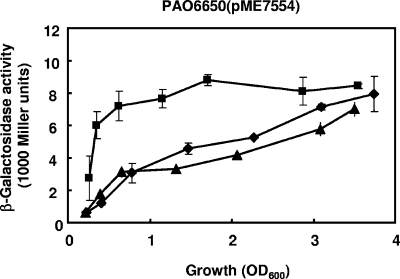
Previous studies revealed that CIO expression is positively controlled by the two-component system RoxSR (9). Moreover, another two-component system, CopRS, is known to be involved in resistance to copper stress in P. aeruginosa (21, 49). To test a potential role of these two-component systems in the regulation of CIO by copper, we constructed roxSR and copRS deletion mutants of P. aeruginosa termed PAO6594 and PAO6573, respectively. The roxSR mutant, similar to the cioAB mutant, showed reduced growth in OS-glucose in the presence of BCS (data not shown). Therefore, to study CIO expression in the roxSR mutant during copper limitation, we performed a shift experiment. The wild-type PAO1, the roxSR mutant PAO6594, and the copRS mutant PAO6573 were transformed with pME7554 (carrying the translational cioA′-′lacZ fusion) and grown in OS-glucose to an OD600 of approximately 0.8. Then, cultures were split and challenged with BCS or with 1.5 μM CuSO4 (Fig. 4) or left untreated. For the roxSR mutant, we observed reduced CIO expression, confirming results previously reported by Comolli and Donohue (9). In the copRS strain, CIO expression was comparable to that observed for the wild-type strain PAO1. In all three strains, the addition of copper caused a 2- to 3-fold decrease in CIO expression compared to what was seen for the control, whereas the addition of BCS caused a minor (1.5-fold) upregulation of CIO expression. These data indicate that the mechanism controlling cioAB expression in relation to copper availability is independent of the two-component systems RoxSR and CopRS. The fact that the BCS effect was less pronounced in the shift experiment (Fig. 4) than in the batch experiment (Fig. 2B) suggests that BCS depleted the cellular copper reserves more slowly in the dense populations prior to the shift compared to what was seen for the initial very low population densities used in the batch experiment.
FIG. 4.
Activity from a β-galactosidase reporter plasmid containing a cioA′-′lacZ translational fusion (pME7554) in P. aeruginosa PAO1 (A), the ΔroxSR mutant PAO6594 (B), or the ΔcopRS mutant PAO6573 (C). Cells were grown in OS-glucose without ascorbate to an OD600 of approximately 0.8, and then each culture was split and challenged with 150 μM BCS plus 1 mM ascorbate (white bars), with 1.5 μM CuSO4 (gray bars), or with 1 mM ascorbate (black bars). Arrows indicate the time points when BCS, ascorbate, or copper were added to the cultures. Each value is the average of three different cultures ± standard deviation.
Copper-mediated regulation of CIO appears to be indirect.
It has been proposed that the electron flow toward the four terminal oxidases belonging to the heme-copper family indirectly regulates the expression of CIO (9). One signal involved in this regulation may be hydrogen cyanide, which acts as an inhibitor of the heme-copper oxidases (11). We reasoned that a lack of copper might act like cyanide. In the absence of copper, the heme-copper oxidases would be unable to transfer electrons to oxygen and the resulting reduced electron flow might activate CIO expression. To test this hypothesis, we constructed a quadruple mutant, PAO6650 (ccoNOQP1 ccoNOQP2 coxBA-coIII cyoABCDE), lacking all four heme-copper oxidases. The growth of PAO6650 (doubling time, ∼80 min) was slower than that of the wild-type PAO1 (doubling time, ∼30 min) in NYB, and after 9 h of incubation, the growth yield of PAO1 was 1.5-fold higher than that of PAO6650. Nevertheless, it is remarkable that the CIO enzyme alone can support aerobic growth of P. aeruginosa.
In the quadruple mutant, cioA′-′lacZ expression increased in a growth-phase-dependent manner (Fig. 5), much like what was seen for the parental strain PAO1 (Fig. 2B). Interestingly and unlike the wild type, the quadruple mutant did not show any negative effect on CIO expression upon the addition of copper (Fig. 5). The addition of BCS caused an initial and transient increase of CIO expression relative to what was seen for the untreated culture (Fig. 5). However, this unexplained effect disappeared at later growth phases. At high population densities (OD600 ≥ 3), the availability of copper had no significant effect on cioA′-′lacZ expression in the quadruple mutant (Fig. 5), whereas in the wild type there was a 10-fold expression difference between copper-replete and copper-limited conditions (Fig. 2B). These data show that copper-mediated regulation of CIO depends to a large extent on the function of the heme-copper oxidases.
FIG. 5.
Cell population density-dependent β-galactosidase expression of a cioA′-′lacZ translational fusion (pME7554) in strain PAO6650 (cco1 cco2 cox cyo). Cultures were grown in OS-glucose containing 1 mM ascorbate (diamonds), in the ascorbate medium supplemented with 150 μM BCS (squares), or in medium with 1.5 μM CuSO4 but without ascorbate (triangles). Each value is the average of three different cultures ± standard deviation.
We wondered whether siderophore regulation by copper availability might be altered in the quadruple oxidase-negative mutant PAO6650. However, the expression of the pvdS′-′lacZ and pchR′-′lacZ fusions was still downregulated by copper limitation, as in the wild-type PAO1 (Table 3). We conclude that respiratory functions are not involved in the link between iron metabolism and copper availability.
DISCUSSION
We have shown here that the P. aeruginosa wild-type PAO1 can perform aerobic respiration when very little, if any, copper is bioavailable. Only when the cioAB genes were inactivated by mutation did the organism require copper for aerobic growth, and this requirement could not be satisfied by iron or zinc. These results are consistent with genomic data which predict that aerobic respiration of P. aeruginosa depends on four heme-copper terminal oxidases (encoded by the ccoNOQP1, ccoNOQP2, coxBA-coIII, and cyoABCDE clusters) and one copper-free, cyanide-resistant oxidase (encoded by the cioAB genes). Thus, when copper is limiting, P. aeruginosa essentially relies on CIO for aerobic growth. Interestingly, as shown by the quadruple cco1 cco2 cox cyo mutant, the CIO pathway alone seems to allow fairly good growth of P. aeruginosa, implying that protons are translocated and that ATP is generated effectively in this pathway. Alternative cyanide-resistant ubiquinol oxidases also exist in plants and some fungi. Whereas in plants these enzymes do not conserve energy, those in fungi appear to be able to do so (4, 26).
BCS probably does not penetrate cells and therefore depletes them of copper progressively during growth. Therefore, we conducted a serial transfer experiment (not shown) in which we grew P. aeruginosa PAO1 in OS-glucose medium with BCS. As growth continued normally for at least 20 generations, we believe that copper is not essential for aerobic growth. We confirmed this finding by using the potent permeable copper chelator TTM. On defined medium containing 1 mM TTM, the wild-type PAO1 was able to grow, whereas the cioAB mutant was not (Fig. 3). We did not assess the effect of copper limitation during anaerobic respiration with nitrate or nitrite, which P. aeruginosa can use as alternative electron acceptors (60). In P. aeruginosa, as in P. stutzeri, the last enzyme of denitrification, N2O reductase, is a copper protein (33, 60), and it is conceivable that a truncated form of denitrification ending with N2O might operate under conditions of copper limitation. In the absence of respiration, P. aeruginosa is also capable of marginal anaerobic growth on arginine by fermenting arginine via the arginine deiminase pathway (51). Further experiments will be needed to see how copper availability affects these processes.
In P. aeruginosa, stress imposed by high copper concentrations induces the expression of copper resistance genes regulated by the two-component system CopSR and several efflux genes as well as the pyoverdine biosynthetic genes (49). Our transcriptomic data (Table 2) show that, conversely, copper deficiency results in the downregulation of the pyoverdine biosynthetic genes and pyoverdine production. It is possible that pyoverdine, by chelating Cu(II) in culture media, might alleviate copper toxicity to some extent, although there is no experimental evidence for this. We also found that pyochelin biosynthetic genes were downregulated during copper deprivation. These data are difficult to reconcile with a previous study (49) showing that downregulation of the same genes occurs during copper stress. It is striking that many P. aeruginosa genes whose expression is low under copper limitation (Table 2) are involved in iron metabolism, suggesting mechanistic links between iron and copper metabolism. Such links have been noted before for E. coli (28), yeast (50), and mammals (56). In P. aeruginosa, we verified that the genes for two key regulators of iron uptake, the sigma factor PvdS and the pyochelin regulator PchR, were very poorly expressed during copper limitation, both in high- and low-iron media (Table 3). We do not know at this stage what causes this effect. Although the E. coli Fur protein binds Cu2+ ions and thereby is converted to a repressor in vitro (15), it is unlikely that such a mechanism operates in P. aeruginosa in vivo. If it did, we would expect to find derepression of the Fur-repressible pvdS and pchR genes under copper-limiting conditions. However, the opposite effect was observed.
We found amazingly few genes that were upregulated by copper deprivation in P. aeruginosa (Table 2). These results argue against the existence in P. aeruginosa of an inducible, chalkophore-dependent copper uptake system of the kind that delivers copper to copper-starved cells of some methylotrophs (29). As long as P. aeruginosa can rely on CIO function for respiration, this bacterium may not need an expensive copper-scavenging system. When respiration is curtailed because of a lack of copper, the cell has a reduced requirement for iron. This is reflected, on the one hand, by the enhanced expression of the bacterioferritin gene bfrB and, on the other hand, by the downregulation of multiple iron uptake systems. A potential link between copper and iron regulation might be provided by the PA2384 gene, whose expression was decreased 17-fold by copper limitation (Table 2). This gene is presumed to code for a DNA-binding protein which positively regulates PvdS and PchR expression (59).
Copper availability affected the expression of several respiratory enzymes. In particular, we found that both transcriptional and translational cioA-lacZ fusions were markedly upregulated during copper deprivation and downregulated by excess copper (Fig. 1). It is not clear why this effect was not revealed by our transcriptomic data. A possible explanation could be that in the control culture grown without BCS the cioAB transcript levels were already very high (reflected by the high β-galactosidase activities of the transcriptional lacZ fusion), such that a further enhancement of these mRNA levels upon BCS addition might be difficult to pick up by hybridization. In fact, the plasmid-borne transcriptional lacZ fusion also seemed to arrive at a ceiling during exponential growth with BCS and did not increase further at later growth phases, whereas the less strongly expressed translational lacZ fusion did. Neither the CopSR nor the RoxSR two-component system appeared to be important for copper-dependent regulation of the cioAB cluster. By contrast, in the quadruple cco1 cco2 cox cyo mutant, the extent of this regulation was strongly diminished, especially at high cell population densities, suggesting that some function of the heme-copper oxidases accounts for the copper-dependent regulation of the cioAB genes, at least in part. Whatever signal might be emitted by the heme-copper oxidases, this signal does not appear to be sensed by the CopSR and RoxSR two-component systems. From a physiological perspective, it makes sense that the wild type should respond to copper deprivation by cioAB overexpression, as this optimizes the potential for aerobic respiration.
Supplementary Material
Acknowledgments
We thank Geneviève Goy and Karine Lapouge for invaluable help with preliminary experiments and for supplying pME7554; Otto Hagenbüchle, Alexandra Paillusson, and Sylvain Pradervand for performing microarray analysis; and Cristina Lamelas and Isabelle Worms for expert help with Cu content measurements.
This study was supported in part by a genomics project of the University of Lausanne.
Footnotes
Published ahead of print on 15 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adaikkalam, V., and S. Swarup. 2002. Molecular characterization of an operon, cueAR, encoding a putative P1-type ATPase and a MerR-type regulatory protein involved in copper homeostasis in Pseudomonas putida. Microbiology 1482857-2867. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Ortega, C., and C. Harwood. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65153-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnesano, F., L. Banci, I. Bertini, S. Mangani, and A. R. Thompsett. 2003. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc. Natl. Acad. Sci. USA 1003814-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthold, D. A., M. E. Andersson, and P. Nordlund. 2000. New insight into the structure and function of the alternative oxidase. Biochim. Biophys. Acta 1460241-254. [DOI] [PubMed] [Google Scholar]
- 5.Bissig, K. D., T. C. Voegelin, and M. Solioz. 2001. Tetrathiomolybdate inhibition of the Enterococcus hirae CopB copper ATPase. FEBS Lett. 507367-370. [DOI] [PubMed] [Google Scholar]
- 6.Blair, D., and H. Diehl. 1961. Bathophenanthrolinedisulphonic acid and bathocuproinedisulphonic acid, water soluble reagents for iron and copper. Talanta 7163-174. [Google Scholar]
- 7.Blumer, C., and D. Haas. 2000. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173170-177. [DOI] [PubMed] [Google Scholar]
- 8.Changela, A., K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran, and A. Mondragón. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 3011383-1387. [DOI] [PubMed] [Google Scholar]
- 9.Comolli, J. C., and T. J. Donohue. 2002. Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol. Microbiol. 45755-768. [DOI] [PubMed] [Google Scholar]
- 10.Comolli, J. C., and T. J. Donohue. 2004. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol. Microbiol. 511193-1203. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, M., G. R. Tavankar, and H. D. Williams. 2003. Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology 1491275-1284. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis, P. 2008. The ‘core’ and ‘accessory’ regulons of Pseudomonas-specific extracytoplasmic sigma factors. Mol. Microbiol. 68810-812. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis, P., C. Baysse, and S. Matthijs. 2008. Iron uptake in Pseudomonas, p. 213-235. In P. Cornelis (ed.), Pseudomonas genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 14.Cunningham, L., M. Pitt, and H. D. Williams. 1997. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24579-591. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 1692624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditta, G., T. Schmidhauser, E. Yacobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helsinki. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13149-153. [DOI] [PubMed] [Google Scholar]
- 17.Durán, R. V., M. Hervás, M. A. De La Rosa, and J. A. Navarro. 2004. The efficient functioning of photosynthesis and respiration in Synechocystis sp. PCC 6803 strictly requires the presence of either cytochrome c6 or plastocyanin. J. Biol. Chem. 2797229-7233. [DOI] [PubMed] [Google Scholar]
- 18.Frieden, E., S. Osaki, and H. Kobayashi. 1965. Copper proteins and oxygen. Correlations between structure and function of the copper oxidases. J. Gen. Physiol. 49213-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giddens, S. R., R. W. Jackson, C. D. Moon, M. A. Jacobs, X. X. Zhang, S. M. Gehrig, and P. B. Rainey. 2007. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc. Natl. Acad. Sci. USA 10418247-18252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutteridge, J. M., and B. Halliwell. 2000. Free radicals and antioxidants in the year 2000. A historical look to the future. Ann. N. Y. Acad. Sci. 899136-147. [DOI] [PubMed] [Google Scholar]
- 21.Ha, U., J. Kim, H. Badrane, J. Jia, H. V. Baker, D. Wu, and S. Jin. 2004. An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system. Mol. Microbiol. 54307-320. [DOI] [PubMed] [Google Scholar]
- 22.Hassett, D. J., M. L. Howell, U. A. Ochsner, M. L. Vasil, Z. Johnson, and G. E. Dean. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 1791452-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13232-237. [DOI] [PubMed] [Google Scholar]
- 24.Hellman, N. E., and J. D. Gitlin. 2002. Ceruloplasmin metabolism and function. Annu. Rev. Nutr. 22439-458. [DOI] [PubMed] [Google Scholar]
- 25.Høiby, N. 2006. P. aeruginosa in cystic fibrosis patients resists host defenses, antibiotics. Microbe 1571-577. [Google Scholar]
- 26.Joseph-Horne, T., D. W. Hollomon, and P. M. Wood. 2001. Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta 1504179-195. [DOI] [PubMed] [Google Scholar]
- 27.Karthikeyan, K. G., H. A. Elliott, and F. S. Cannon. 1997. Adsorption and coprecipitation of copper with hydrous oxides of iron and aluminum. Environ. Sci. Technol. 312721-2725. [Google Scholar]
- 28.Kershaw, C. J., N. L. Brown, C. Constantinidou, M. D. Patel, and J. L. Hobman. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 1511187-1198. [DOI] [PubMed] [Google Scholar]
- 29.Kim, H. J., D. W. Graham, A. A. DiSpirito, M. A. Alterman, N. Galeva, C. K. Larive, D. Asunskis, and M. A. Sherwood. 2004. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 3051612-1615. [DOI] [PubMed] [Google Scholar]
- 30.Laurie, S. H. 2000. Thiomolybdates—simple but very versatile reagents. Eur. J. Inorg. Chem. 112443-2450. [Google Scholar]
- 31.Leoni, L., A. Ciervo, N. Orsi, and P. Visca. 1996. Iron regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of Fur and PvdS on promoter activity. J. Bacteriol. 1782299-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linder, M. C., and M. Hazegh-Azam. 1996. Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 63797S-811S. [DOI] [PubMed] [Google Scholar]
- 33.Matsubara, T., K. Frunzke, and W. G. Zumft. 1982. Modulation by copper of the products of nitrite respiration in Pseudomonas perfectomarinus. J. Bacteriol. 149816-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michel, L., N. González, S. Jagdeep, T. Nguyen-Ngoc, and C. Reimmann. 2005. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol. Microbiol. 58495-509. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Mills, S. D., C. A. Jasalavich, and D. A. Cooksey. 1993. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J. Bacteriol. 1751656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 451277-1287. [DOI] [PubMed] [Google Scholar]
- 38.Ornston, L. N., and R. Y. Stanier. 1966. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. J. Biol. Chem. 2413776-3786. [PubMed] [Google Scholar]
- 39.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 27630670-30677. [DOI] [PubMed] [Google Scholar]
- 40.Palma, M., S. Worgall, and L. E. Quadri. 2003. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 180374-379. [DOI] [PubMed] [Google Scholar]
- 41.Pier, G. B., and R. Ramphal. 2005. Pseudomonas aeruginosa, p. 2587-2615. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas and Bennett's principles and practice of infectious diseases, 6th ed., vol. 2. Elsevier, Philadelphia, PA. [Google Scholar]
- 42.Poole, K., and G. A. McKay. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8d661-d686. [DOI] [PubMed] [Google Scholar]
- 43.Rae, T. D., P. J. Schmidt, R. A. Pufahl, V. C. Culotta, and T. V. O'Halloran. 1999. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284805-808. [DOI] [PubMed] [Google Scholar]
- 44.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27197-213. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Savioz, A., A. Zimmermann, and D. Haas. 1993. Pseudomonas aeruginosa promoters which contain a conserved GG-N10-GC motif but appear to be RpoN-indipendent. Mol. Gen. Genet. 23874-80. [DOI] [PubMed] [Google Scholar]
- 47.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 1821215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanisich, V. A., and B. W. Holloway. 1972. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. 1991-108. [DOI] [PubMed] [Google Scholar]
- 49.Teitzel, G. M., A. Geddie, S. K. De Long, M. J. Kirisits, M. Whiteley, and M. R. Parsek. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 1887242-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Bakel, H., E. Strengman, C. Wijmenga, and F. C. Holstege. 2005. Gene expression profiling and phenotype analyses of S. cerevisiae in response to changing copper reveals six genes with new roles in copper and iron metabolism. Physiol. Genomics 22356-367. [DOI] [PubMed] [Google Scholar]
- 51.Vander Wauven, C., A. Piérard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visca, P. 2004. Iron regulation and siderophore signalling in virulence by Pseudomonas aeruginosa, p. 69-123. In J. L. Ramos (ed.), Pseudomonas, vol. 2. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 53.Visca, P., L. Serino, and N. Orsi. 1992. Isolation and characterization of Pseudomonas aeruginosa mutants blocked in the synthesis of pyoverdin. J. Bacteriol. 1745727-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, U. Schnider, G. Défago, and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Publishers, Weinheim, Germany.
- 55.Williams, H. D., J. E. Zlosnik, and B. Ryall. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 521-71. [DOI] [PubMed] [Google Scholar]
- 56.Winzerling, J. J., and J. H. Law. 1997. Comparative nutrition of iron and copper. Annu. Rev. Nutr. 17501-526. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, R., and W. S. Kisaalita. 1995. Purification of pyoverdines of Pseudomonas fluorescens 2-79 by copper-chelate chromatography. Appl. Environ. Microbiol. 613769-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye, R. W., D. Haas, J. O. Ka, V. Krishnapillai, A. Zimmermann, C. Baird, and J. M. Tiedje. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 1773606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng, P., J. Sun, R. Geffers, and A. P. Zeng. 2007. Functional characterization of the gene PA2384 in large-scale gene regulation in response to iron starvation in Pseudomonas aeruginosa. J. Biotechnol. 132342-352. [DOI] [PubMed] [Google Scholar]
- 60.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.