Abstract
Soluble lytic transglycosylase B1 from Pseudomonas aeruginosa was coupled to Sepharose and used to immobilize interaction partners from membrane protein extracts. Penicillin-binding protein 2 (PBP2) was identified as a binding partner, suggesting that the two proteins function together in the biosynthesis of peptidoglycan. By use of an engineered truncated derivative, the N-terminal module of PBP2 was found to confer the binding properties.
Enlargement and growth of the peptidoglycan (PG) sacculus is effected by the coordination of both synthetic and lytic enzymes. The penicillin-binding proteins (PBPs) are biosynthetic enzymes that catalyze transglycosylation and/or transpeptidation reactions for the incorporation of new material into the PG layer (11). The lytic transglycosylases (LTs), on the other hand, cleave the β-1,4-glycosidic bond between MurNAc and GlcNAc and are understood to function as space-makers for the incorporation of new material (reviewed in reference 29). These conflicting enzymatic activities are thought to be controlled by the association of the respective enzymes in multienzyme complexes (14, 29). Support for this hypothesis is confined to only a few studies involving a limited number of bacterial species. With Escherichia coli, a protein-protein interaction network between high-molecular-weight (HMW) PBPs, HMW LTs, and low-molecular-weight PBPs (specifically, d,d-endopeptidases) has been demonstrated by affinity chromatography experiments (3, 26, 34, 35). Affinity chromatography was also used to indicate that membrane-bound LT A (MltA) interacts with PBP2 in Neisseria meningitidis serogroup B (15). Both affinity chromatography and immunoprecipitation approaches demonstrated the existence of multienzyme complexes in Caulobacter crescentus membranes involving PBP1a, -2, and -3a and the morphogenic protein MreC (7, 9). That multienzyme complexes exist in vivo has been demonstrated with both E. coli and Haemophilus influenzae using cross-linking reagents (1, 4, 14, 27, 28).
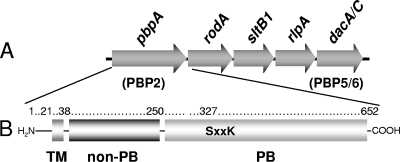
Despite the importance of Pseudomonas aeruginosa as an opportunistic pathogen in humans, very little is known about the biosynthesis of its PG, the target of a number and variety of antibiotics. We have initiated studies of both the LTs and PBPs of this bacterium. Of the nine LTs that appear to be encoded on the P. aeruginosa genome (6), we have been characterizing the four family 3 enzymes. One, MltB, is a lipoprotein, whereas three other homologues are produced in soluble form; these include soluble LT B1 (SltB1), SltB2, and SltB3 (5). Of these, SltB1 is encoded by a gene that is clustered at a locus with others involved in PG synthesis, including PBP2 (Fig. 1) (18, 19), suggesting that they may represent binding partners. To test this possibility, recombinant SltB1 was used as the ligand in affinity chromatography of P. aeruginosa preparations.
FIG. 1.
Organization of the gene cluster harboring pbpA and modular structure of PBP2 from P. aeruginosa. (A) The cluster of genes at the pbpA (PBP2) locus on the P. aeruginosa chromosome includes dacA-dacC (dacA/C) and sltB1, encoding PBP5-PBP6 and SltB1, respectively. (B) The structure of PBP2 is comprised of an N-terminal transmembrane anchor (TM; residues 1 to 38), a non-PB module (residues 39 to 250), and a C-terminal PB module (residues 256 to 652) connected by a short linker sequence. Depicted within the PB module is the Ser-X-X-Lys consensus sequence involving Ser327 as the catalytic residue and site of penicillin binding.
Preparation of proteins and affinity chromatography media.
Cells of E. coli BL21 harboring pNBAC258-2 or pACKD16 were induced to overproduce P. aeruginosa SltB1-His6 or sPBP2-His6, respectively, for purification as described previously (5, 18). Membrane protein extraction from a 10-liter culture of P. aeruginosa PAO1 grown to mid-exponential phase (optical density at 600 nm, ∼ 0.6) was performed using a procedure adapted from Vollmer et al. (34). Harvested cells were resuspended in 10 mM Tris-HCl and 10 mM MgCl2 (pH 6.8) with Complete Mini EDTA-free protease inhibitor cocktail tablets (Roche), 5 μg/ml DNase, and 10 μg/ml RNase and broken by passage through a French press. Unlysed cells were removed by low-speed centrifugation (8,000 × g, 10 min, 4°C), and membranes were then collected by ultracentrifugation (100,000 × g, 1 h, 4°C). Membrane proteins were extracted by stirring overnight at 4°C in 40 ml of extraction buffer (10 mM Tris-HCl, 10 mM MgCl2, 150 mM NaCl, 0.02% NaN3 [pH 6.8], 2% [wt/vol] Triton X-100). Insoluble material was removed by subsequent ultracentrifugation (100,000 × g, 1 h, 4°C), and the preparation was dialyzed against equilibration buffer (10 mM Tris-HCl, 10 mM MgCl2, 50 mM NaCl, 0.02% NaN3 [pH 6.8], 1% [wt/vol] Triton X-100). This gave 5.75 mg/ml, for a total yield of 421 mg of membrane protein, which was stored at −20°C until needed. The non-penicillin-binding (non-PB) module of PBP2 was prepared from E. coli BL21 harboring pACBL17 (18) in a similar manner except it was found that the use of 2% (wt/vol) N-laurylsarcosine for extraction and dialysis instead of 1% Triton X-100 provided maximum yields.
Purified SltB1 (∼2.5 mg) was covalently linked to CNBr-activated Sepharose 4B (GE Healthcare, Piscataway, NJ) in coupling buffer (0.1 M NaHCO3 and 0.5 M NaCl [pH 8.3]) at 4°C overnight. A second control column was also prepared, but it was lacking SltB1. Following a wash with 20 ml coupling buffer, uncoupled sites were blocked with 0.1 M Tris-HCl (pH 8.0) for 2 h at 4°C. The gel suspensions were then washed successively with 0.1 M Tris-HCl and 0.5 M NaCl (pH 8.0); 0.1 M sodium acetate and 0.5 M NaCl (pH 4.8); and 100 ml of 1% Triton X-100 (wt/vol) in equilibration buffer (10 mM Tris-HCl [pH 6.8] containing 10 mM MgCl2, 50 mM NaCl, and 0.02% NaN3).
Affinity chromatography of membrane proteins on SltB1-Sepharose 4B.
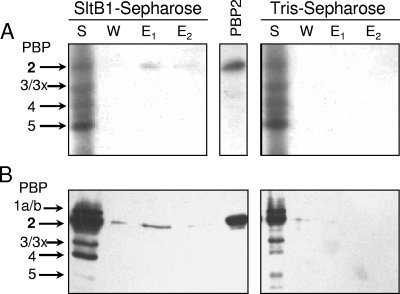
Approximately 100 mg of P. aeruginosa PAO1 membrane proteins were applied to the test and control columns and mixed at 4°C overnight. Unbound proteins were removed with 500 ml equilibration buffer followed by 500 ml of column buffer (equilibration buffer with 0.05% [wt/vol] Triton X-100). Proteins retained on the columns were recovered in three consecutive 1.5-ml elutions which were separated by washes with 50 ml of the respective elution buffers. These elution buffers were the column buffer containing 150 mM NaCl, 1 M NaCl, and 0.5 M NaCl with 5 mM EGTA, respectively. Each of the three fractions was stored at −20°C prior to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12.5% acrylamide (17) at 20 mA. Despite the relatively high protein load used in these affinity chromatographies, very little protein was detected in the lanes loaded with elution fractions following staining with either Coomassie brilliant blue or silver (data not shown). In the latter case, one unidentified protein with an apparent molecular mass of ∼70 kDa was detected in elutions from the test column but not from the control column loaded with an identical protein sample.
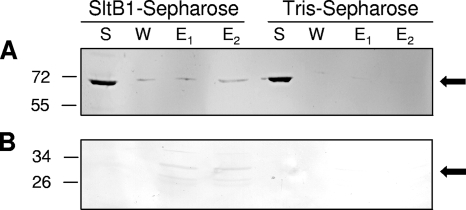
The low level of protein detection was expected given the relatively low concentration of PG-metabolizing enzymes, including the PBPs, within bacterial cells (8). Consequently, each of the collected fractions were assayed specifically from the presence of PBPs by using both biotinylated ampicillin as described previously (18, 19) and [phenyl-4(n)-3H]benzylpenicillin (25 Ci/mmol and 1 mCi/ml; TRK 779; GE Healthcare) according to the method described by Spratt (30, 31) as modified by Preston et al. (25). Prior to autoradiography of the latter, the gels were soaked in Amplify Scintillant (GE Healthcare) for 15 to 30 min and dried in vacuo on Whatman filter paper. The gels were exposed to Bioflex MSI film (Clonex, Markham, Ontario, Canada) with two Hyperscreen intensifying screens (Amersham) at −70°C for 2 to 6 weeks and then developed. Both of these PBP assays demonstrated that P. aeruginosa PBP2 was retained by the SltB1 column but not by the control column lacking bound SltB1 (Fig. 2). Some PBP2 was detected in the 150 mM NaCl fraction from the SltB1 column by using the biotinylated ampicillin assay, but both assays showed that the majority of the PBP2 was eluted from the affinity matrix in 1 M NaCl. The [3H]penicillin assay also showed some PBP2 eluting with 5 mM EGTA. This interaction was more obvious when membrane preparations from E. coli harboring pACKD16 (19) overproducing a soluble derivative of P. aeruginosa PBP2 (sPBP2) were loaded onto the columns (data not shown). With the recombinant sPBP2, it was also possible to assay the SDS-PAGE gels by Western immunoblot analysis, taking advantage of the presence of a C-terminal His6 tag. The Western immunoblot analysis used a commercially available mouse anti-His antibody (His Probe H-3; Santa Cruz Biotechnology, Santa Cruz, CA) as previously described (5) and confirmed the identity of the detected protein as P. aeruginosa PBP2. Unequivocal proof that PBP2 interacts with SltB1 was obtained in an experiment using a purified preparation of P. aeruginosa sPBP2. Again, 1 M NaCl was required to recover the majority of the applied sPBP2 from the affinity matrix (Fig. 3). The requirement for the relatively high salt concentration to elute either PBP2 or sPBP2 from the SltB1 matrix suggests that the nature of the interaction is likely ionic, involving one or more salt bridges.
FIG. 2.
PBP assays of fractions from affinity chromatography of membrane proteins on SltB-Sepharose. A P. aeruginosa PA01 membrane-protein extract (S) was applied to either SltB1-Sepharose or a control column of Tris-Sepharose in 10 mM Tris-HCl, 10 mM MgCl2, 50 mM NaCl, and 0.05% Triton X-100. After the flowthrough fraction was collected, the resins were washed with the same buffer containing 150 mM NaCl (W) and then eluted with buffer containing 1 M NaCl (E1) and 5 mM EGTA (E2). Samples (30 μl) of the fractions were incubated with [3H]penicillin G (A) or biotinylated ampicillin (B) and analyzed by SDS-PAGE as described in Materials and Methods. The PBPs were visualized by autoradiography (A) or chemiluminescence (B). A sample of purified P. aeruginosa PBP2 (PBP2) was applied as a positive control, and the assignment of the other PBPs was based on apparent molecular masses.
FIG. 3.
Immunoblots of fractions from affinity chromatography of PBP2 applied to SltB1-Sepharose and Tris-Sepharose. Purified preparations of recombinant PBP2 (A) and the isolated non-PB module (B) were applied to either SltB1-Sepharose or a control Tris-Sepharose column. Conditions of the affinity chromatography and SDS-PAGE were as described in the legend to Fig. 2. Following electrophoresis, the recombinant proteins were detected by Western immunoblot analysis using an anti-six-His antibody as described in Materials and Methods. The positions of molecular mass markers (in kilodaltons) are indicated on the left, and the arrows denote the positions of PBP2 (top) and the non-PB module (bottom).
SPR analysis of the SltB1-PBP2 interaction.
To obtain a quantitative measurement of the SltB1-PBP2 interaction, approximately 130 resonance units of purified SltB1 was coupled to a Biacore CM5 sensor chip (Biacore, Inc., Piscataway, NJ) through its free amine groups by following the procedure recommended by the manufacturer. Unreacted active groups were blocked by an injection of 70 μl of 1 M ethanolamine, pH 8.5. Surface plasmon resonance (SPR) studies were performed with a Biacore 2000 instrument. Purified sPBP2 in 10 mM HEPES, 150 mM NaCl, and 0.005% Tween 20 (pH 7.4) (HPS-P buffer) was used as the analyte at concentrations of 3,690, 1,840, 921, 460, and 230 nM. Data were recorded during injection of 200 μl analyte at a flow rate of 100 μl/min, followed by 150 s of dissociation. Regeneration was achieved by injecting 100 μl of HPS-P buffer containing 1 M NaCl. The evaluation of the sensorgrams was performed with BIAevaluation software 3.1. The baselines of the sensorgrams were normalized to zero at the time of the start of the injection, and the sensorgram value for a control lane (with only ethanolamine immobilized) was subtracted from that for the flow cells containing immobilized protein to remove any effects of nonspecific binding to the matrix. This was followed by subtracting a blank buffer injection from the sensorgrams to compensate for any buffer effects. BIAevaluation software was then used to fit the kinetics of the curves to estimate binding affinity constants.
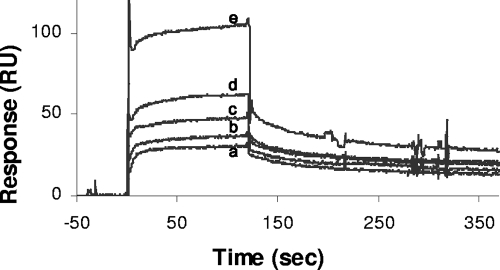
A dose-dependent interaction between injected sPBP2 and immobilized SltB1 was observed (Fig. 4). An analysis of the sensorgrams provided a Kd (dissociation constant) of 133 ± 26 nM for this interaction, which is consistent with those determined for other interactions involving PBPs. For example, Vollmer et al. (34) calculated a Kd by SPR for the E. coli PBP1b-MipA interaction to be in the range of 200 to 2000 nM, depending on the concentration of analyte, while Bertsche et al. (3) determined a Kd of approximately 100 nM for the E. coli PBP1b-PBP3 interaction for lower concentrations of PBP1b and 400 nM for higher concentrations.
FIG. 4.
SPR analysis of interaction between P. aeruginosa SltB1 and PBP2. The soluble derivative of PBP2, sPBP2, in 10 mM HEPES, 150 mM NaCl, and 0.005% Tween 20 (pH 7.4) was applied to SltB1 immobilized on a CM5 sensor chip at a flow rate of 100 μl/min. The representative sensorgrams show the responses to injections of sPBP2 at concentrations of 230 nM (a), 460 nM (b), 920 nM (c), 1,840 nM (d), and 3,690 nM (e) using a Biacore 2000 spectrometer.
Role of N-terminal module of PBP2 in protein-protein interactions.
That immobilized SltB1 could retain sPBP2, a variant of PBP2 lacking its transmembrane helix, demonstrated that the site of interaction is clearly neither within the transmembrane region nor dependent upon its presence. As with other class B HMW PBPs, P. aeruginosa PBP2 is bimodular, with its C-terminal module functioning as the transpeptidase and possessing PB capacity (11). The purpose and function of the N-terminal module for any class B HMW PBPs remain unknown, but recently we have postulated that this module may contribute to protein-protein interactions (18). The same morphological change from rods to large spheres was observed when either the complete recombinant PBP2s from E. coli or P. aeruginosa or constructs encoding only their respective N-terminal, non-PB modules were overproduced in E. coli. Furthermore, these morphological changes were not observed when each of the proteins were overproduced in E. coli MHD79 (18), a mutant strain lacking six LTs (MltA, MltB, MltC, MltD, Slt70, and EmtA) (12). These observations thus suggested a link between the LTs and the non-PB module of PBP2.
To test this hypothesis, membrane preparations were obtained from E. coli BL21 harboring pACBL17 after induction with 1 mM isopropyl-β-d-thiogalactopyranoside for the overproduction of the N-terminal, non-PB module of P. aeruginosa PBP2. This module includes the N-terminal 250 amino acids of P. aeruginosa PBP2, from Met1 to Ser250, and it has a molecular mass of 30.3 kDa (Fig. 1). Its coding sequence was cloned into pET30a(+), thereby endowing the overproduced protein with a C-terminal six-His tag (19). The presence of the His tag is especially important for detection of the protein by Western immunoblot analysis using the anti-six-His tag antibody, because if the C-terminal PB module were lacking, PBP assays could not be used. The membrane fraction containing the non-PB module was subjected to chromatography on SltB1-Sepharose as described above. Like the full-length protein, it was retained by the affinity matrix, and 1 M NaCl was required to completely remove the module from the column. Hence, it appears that the N-terminal, non-PB module does confer binding properties for complexation of PBP2 with SltB1. This thus represents the first report demonstrating a specific function for the N-terminal module present in the class B HMW PBPs, beyond simply conferring proper folding and/or stability of the C-terminal PB module and serving as a membrane anchor (10).
Unfortunately, quantitative data for this interaction could not be obtained by SPR due to a lack of purified protein. Numerous attempts to obtain either the complete N-terminal module or a truncated derivative lacking the 38-residue signal sequence in soluble and homogeneous form failed; the majority of the isolated protein forms insoluble aggregates. Whereas the addition of chaotropic reagents, such as 8 M urea, do permit purification of the module (unpublished results), the lack of an appropriate assay precludes the ability to monitor and assess refolding. Indeed, an analysis of the SPR sensorgrams obtained using the N-terminal module purified in this manner (data not shown) indicated an instability of the protein, thus making the sensorgrams uninterpretable.
The three-dimensional structures of the PBP2s from E. coli, P. aeruginosa, and closely related homologs are not known, thus preventing a rational analysis of the specific regions or amino acid residues potentially involved in binding interactions. However, the structure of E. coli Slt35, a close homolog of P. aeruginosa SltB1, has been solved (33), and it has been shown to possess a single helix-loop-helix Ca2+-binding motif analogous to the EF-hand motif of Ca2+-binding proteins such as troponin C (13). This EF-hand motif is conserved among the family 3 LTs, suggesting its functional importance (6). The motif has been shown to be essential for thermal stability in Slt35 and it may be necessary for proper folding of the protein in the periplasm. The exposed portion of the EF hand has also been proposed to participate in interactions with other proteins (33). The importance of EF hands in protein-protein interactions has been well documented for several eukaryotic systems (for examples, see references 20 and 24) and also for at least one prokaryotic system, the cellulosome (2, 21). Hence, we have initiated studies to probe the importance of the EF hand in SltB1 for its interaction with PBP2.
Concluding remarks.
Investigations of PG metabolism are complicated by the facts that the product is totally insoluble, that the majority of the enzymes involved are membrane associated, and that there is an apparent redundancy in their production, at least for the PBPs and LTs. Hence, despite its historic and continuing importance as a major target for antibacterials, details of its biosynthesis and maturation are only now beginning to emerge. From the limited studies to date, it is becoming apparent that differences in the organization and association of the enzymatic machinery do exist. For example, differences exist between the complement of both the PBPs (11, 19) and the LTs (6) present in E. coli and P. aeruginosa. Another significant difference between these two bacteria pertains to the scaffolding protein MipA, which has been shown to mediate the interaction between LTs and PBP1b in E. coli (34). A BLAST search using E. coli MipA as the probe revealed no homologs in the P. aeruginosa genome (http://www.pseudomonas.com). In the apparent absence of a scaffolding-type protein, it would appear that P. aeruginosa might be more dependent upon the specific interactions localized to individual proteins for their assembly into biosynthetic complexes. Alternatively, there may exist paralogs of MipA which have yet to be discovered.
The finding that P. aeruginosa SltB1 and PBP2 form binding partners was not entirely surprising, as their encoding genes are clustered on the bacterium's chromosome along with others associated with PG synthesis (Fig. 1). Thus, their immediate genetic proximity provides support for the idea that these two enzymes may cooperate in vivo. Furthermore, while experimental evidence is currently lacking, it is conceivable that the proteins encoded by the other two genes of this cluster, RodA and PBP5, may associate with PBP2 and SltB1 in a larger multienzyme complex. Indeed, both RodA and PBP2 are known to be essential for the rod shape of both P. aeruginosa and E. coli (18 and references therein), while PBP5 has been shown to direct cell morphology, at least in E. coli (23, 32). A question that arises, however, is why PBP5 was not detected in the PBP assays of the eluants from the affinity chromatography if all the proteins in this gene cluster form a complex. PBP5 is the most abundant PBP in E. coli, representing between 32 and 64% of the PB activity (8, 30). Assuming this PBP exists in P. aeruginosa at similar concentrations, it should have been readily detected in the PBP assays if present. It is possible that the affinity, if any, of PBP5 for the complex could be very weak and the enzyme was lost in the washing steps of the chromatography. Alternatively, and perhaps more likely, the orientation of the bound SltB1 and PBP2 on the Sepharose matrix may sterically preclude the association of PBP5 and other potential interaction partners, such as RodA, MurG, and MreBCD (16, 22).
Only one isoform of the family 3 LTs is produced in E. coli (viz., MltB) compared to the four found in P. aeruginosa (MltB, SltB1, SltB2, SltB3) (6). It is not yet known why the P. aeruginosa genome encodes the four homologs, but this might reflect its metabolic diversity, which is greater than that of E. coli, given the role that the LTs play in the insertion of secretion systems within the PG sacculus (29). It is also unclear at this time why PBP2 in E. coli does not appear to complex with MltB (35), while a close association of the functional equivalents in P. aeruginosa exists at both the protein and genetic levels. This divergence underscores the differences in the metabolism of PG across the bacteria and illustrates the need to exercise caution when attempting to generalize the observations and interpretations made with a specific bacterium. This is especially important given that the majority of the investigations to date on multienzyme complexes involved in PG metabolism have been focused on a limited number of bacterial species. Nonetheless, it is expected that the more general properties of homologous enzymes, such as the role of the N-terminal, non-PB module in the class 2 PBP2s in providing the site(s) for binding interaction partners as determined here for P. aeruginosa PBP2, would be shared.
Acknowledgments
We thank A. Marini for expert technical assistance.
This study was supported by an operating grant (MOP49623) to A.J.C. from the Canadian Institutes of Health Research and a postgraduate scholarship (PGS B) to B.A.L. from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Alaedini, A., and R. A. Day. 1999. Identification of two penicillin-binding multienzyme complexes in Haemophilus influenzae. Biochem. Biophys. Res. Commun. 264191-195. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., J. P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58521-554. [DOI] [PubMed] [Google Scholar]
- 3.Bertsche, U., T. Kast, B. Wolf, C. Fraipont, M. E. G. Aarsman, K. Kannenberg, M. von Reschenberg, M. Nguyen-Distèche, T. den Blaauwen, J.-V. Höltje, and W. Vollmer. 2006. Interaction between two murein (peptidoglycan) synthases, PBP 3 and PBP 1B, in Escherichia coli. Mol. Microbiol. 61675-690. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj, S., and R. A. Day. 1997. Detection of intra-cellular protein-protein interactions: penicillin interactive proteins and morphogene proteins, p. 469-480. In D. R. Marshak (ed.), Techniques in protein chemistry, vol. 8. Academic Press, San Diego, CA. [Google Scholar]
- 5.Blackburn, N. T., and A. J. Clarke. 2002. Characterization of soluble and membrane-bound family 3 lytic transglycosylases from Pseudomonas aeruginosa. Biochemistry 411001-1013. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, N. T., and A. J. Clarke. 2001. Identification of four families of peptidoglycan lytic transglycosylases. J. Mol. Evol. 5278-84. [DOI] [PubMed] [Google Scholar]
- 7.Divakaruni, A. V., R. R. Loo, Y. Xie, J. A. Loo, and J. W. Gober. 2005. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 10218602-18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty, T. J., K. Kennedy, R. E. Kessler, and M. J. Pucci. 1996. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J. Bacteriol. 1786110-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 511321-1332. [DOI] [PubMed] [Google Scholar]
- 10.Goffin, C., C. Fraipont, J. Ayala, M. Terrak, M. Nguyen-Distèche, and J.-M. Ghuysen. 1996. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J. Bacteriol. 1785402-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goffin, C., and J.-M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 621079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J.-V. Höltje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 1846093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herzberg, O., and M. N. James. 1988. Refined crystal structure of troponin C from turkey skeletal muscle at 2.0 Å resolution. J. Mol. Biol. 203761-779. [DOI] [PubMed] [Google Scholar]
- 14.Höltje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings, G. T., S. Savino, E. Marchetti, B. Arico, T. Kast, L. Baldi, A. Ursinus, J.-V. Höltje, R. A. Nicholas, R. Rappuoli, and G. Grandi. 2002. GNA33 from Neisseria meningitidis serogroup B encodes a membrane-bound lytic transglycosylase (MltA). Eur. J. Biochem. 2693722-3731. [DOI] [PubMed] [Google Scholar]
- 16.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 5578-89. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 18.Legaree, B. A., C. B. Adams, and A. J. Clarke. 2007. Overproduction of penicillin-binding protein 2 and its inactive variants causes morphological changes and lysis in Escherichia coli. J. Bacteriol. 1894975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legaree, B. A., K. E. Daniels, J. T. Weadge, D. Cockburn, and A. J. Clarke. 2007. Function of penicillin-binding protein 2 in viability and morphology of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 59411-424. [DOI] [PubMed] [Google Scholar]
- 20.Lin, P., T. Fischer, T. Weiss, and M. G. Farquhar. 2000. Calnuc, an EF-hand Ca2+ binding protein, specifically interacts with the C-terminal α5-helix of Gαi3. Proc. Natl. Acad. Sci. USA 97674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lytle, B. L., B. F. Volkman, W. M. Westler, and J. H. D.Wu. 2000. Secondary structure and calcium-induced folding of the Clostridium thermocellum dockerin domain determined by NMR spectroscopy. Arch. Biochem. Biophys. 379237-244. [DOI] [PubMed] [Google Scholar]
- 22.Mohammadi, T., A. Karczmarek, M. Crouvoisier, A. Bouhss, D. Mengin-Lecreulx, and T. den Blaauwen. 2007. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 651106-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson, D. E., and K. D. Young. 2001. Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 1833055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson, M. R., and W. J. Chazin. 1998. Structures of EF-hand Ca2+-binding proteins: diversity in the organization, packing and response to Ca2+ binding. Biometals 11297-318. [DOI] [PubMed] [Google Scholar]
- 25.Preston, D. A., C. Y. Wu, L. C. Blaszczak, D. E. Seitz, and N. G. Halligan. 1990. Biological characterization of a new radioactive labeling reagent for bacterial penicillin-binding proteins. Antimicrob. Agents Chemother. 34718-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romeis, T., and J.-V. Höltje. 1994. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J. Biol. Chem. 26921603-21607. [PubMed] [Google Scholar]
- 27.Satta, G., R. Fontana, and P. Canepari. 1994. The two-competing site (TCS) model for cell shape regulation in bacteria: the envelope as an integration point for the regulatory circuits of essential physiological events. Adv. Microb. Physiol. 36181-245. [DOI] [PubMed] [Google Scholar]
- 28.Scheffers, D.-J., and M. G. Pinho. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheurwater, E., C. W. Reid, and A. J. Clarke. 2008. Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40586-591. [DOI] [PubMed] [Google Scholar]
- 30.Spratt, B. G. 1977. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur. J. Biochem. 72341-352. [DOI] [PubMed] [Google Scholar]
- 31.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 722999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spratt, B. G., A. Boyd, and N. Stoker. 1980. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J. Bacteriol. 143569-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Asselt, E. J., A. J. Dijkstra, K. H. Kalk, B. Takacs, W. Keck, and B. W. Dijkstra. 1999. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure 71167-1180. [DOI] [PubMed] [Google Scholar]
- 34.Vollmer, W., M. von Rechenberg, and J.-V. Höltje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 2746726-6734. [DOI] [PubMed] [Google Scholar]
- 35.von Rechenberg, M., A. Ursinus, and J.-V. Höltje. 1996. Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb. Drug Resist. 2155-157. [DOI] [PubMed] [Google Scholar]