Abstract
The zinc metalloprotease EmpA is a virulence factor for the fish pathogen Vibrio anguillarum. Previous studies demonstrated that EmpA is secreted as a 46-kDa proenzyme that is activated extracellularly by the removal of an ∼10-kDa propeptide. We hypothesized that a specific protease is responsible for processing secreted pro-EmpA into mature EmpA. To identify the protease responsible for processing pro-EmpA, a minitransposon mutagenesis (using mini-Tn10Km) clone bank of V. anguillarum was screened for reduced protease activity due to insertions in undescribed genes. One mutant with reduced protease activity was identified. The region containing the mini-Tn10Km was cloned, sequenced, and found to contain epp, an open reading frame encoding a putative protease. Further characterization of epp was done using strain M101, created by single-crossover insertional mutagenesis. Protease activity was absent in M101 cultures even when empA protease activity was induced by salmon gastrointestinal mucus. When the epp mutation was complemented with a wild-type copy of epp (M102), protease activity was restored. Western blot analysis of sterile filtered culture supernatants from wild-type (M93Sm) cells, M101 cells, and M102 cells revealed that only pro-EmpA was present in M101supernatants; both pro-EmpA and mature EmpA were detected in M93Sm and M102 supernatants. When sterile filtered culture supernatants from the empA mutant strain (M99) and M101 were mixed, protease activity was restored. Western blot analysis revealed that pro-EmpA in M101 culture supernatant was processed to mature EmpA only after mixing with M99 culture supernatant. These data show that Epp is the EmpA-processing protease.
Vibrio anguillarum, a marine bacterium, is the causative agent of vibriosis, a systemic disease of both wild and cultured marine fish characterized by hemorrhagic septicemia (2). Outbreaks of vibriosis result in high mortalities among infected fish, and this disease continues to be a major obstacle for the aquaculture industry (2). V. anguillarum enters its fish host through the gastrointestinal (GI) tract and quickly colonizes this nutrient-rich environment (21, 22). Garcia et al. (9) have shown that V. anguillarum grows extremely well in salmon intestinal mucus and that mucus-grown cells specifically express a number of different proteins, including several outer membrane proteins (9) and the extracellular metalloprotease EmpA (7).
The zinc metalloprotease EmpA has been identified as a virulence factor for V. anguillarum and is important for virulence during infection of the GI tract of Atlantic salmon (Salmo salar) (7, 16, 19). In V. anguillarum wild-type strain M93Sm, EmpA is expressed during stationary phase when cells are incubated in Atlantic salmon GI mucus (6, 7). The creation of an empA null mutant (M99) by insertional mutagenesis showed that EmpA is responsible for the protease activity observed for wild-type strain M93Sm (7). However, EmpA activity is dependent on successful secretion and processing of the nascent protein to an active mature protease (16, 25).
According to Milton et al. (16), EmpA is synthesized as a 66.7-kDa preproenzyme. Sequence analysis predicts that the removal of both pre- and propeptides during secretion would result in a mature protein with a molecular mass of 44.6 kDa. However, EmpA protease activity was repeatedly associated with a 36-kDa protein (16). This suggests that the 44.6-kDa protein undergoes further processing to a 36-kDa active form. Recently, Staroscik et al. (25) used Western blot analysis to study EmpA secretion in V. anguillarum culture supernatants. Using anti-LasB antibodies for detection, Western blot analysis revealed an ∼46-kDa band in all culture supernatants as well as an ∼36-kDa band that could be detected only in culture supernatants possessing protease activity (25). The ∼46- and ∼36-kDa bands correspond to the sizes of the predicted secreted proenzyme and the mature protein, respectively (16). Staroscik et al. (25) also confirmed the presence of the cytoplasmic preproenzyme. It has been suggested that the activation of the proenzyme occurs after secretion by the removal of an ∼10-kDa peptide. The protease responsible for the posttranslational modification of pro-EmpA to the mature enzyme has yet to be identified.
Multiple processing steps have been proposed for other metalloproteases, such as the hemagglutinin/protease (HA/protease) of V. cholerae (12), the Vvp protease of V. vulnificus (17), and the LasB elastase of Pseudomonas aeruginosa (16). In this study, we sought to further examine the processing of pro-EmpA to mature EmpA metalloprotease. Minitransposon mutagenesis (mini-Tn10Km) was used to create and screen for protease mutants. One mutant that exhibited a reduction in protease activity compared to wild-type protease activity was detected. The region surrounding this mutation in V. anguillarum was cloned and sequenced. One open reading frame was identified and mutated by insertional mutagenesis. The resulting strain was designated M101. Protease assays and Western blots were performed to determine changes in protease activity and processing of EmpA from what was seen for wild-type strain M93Sm. These experiments led to the identification of a putative EmpA-processing protease gene (epp). Our results strongly suggest that epp is the protease responsible for the activation of pro-EmpA to the mature EmpA protease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. V. anguillarum cultures were routinely grown in Luria-Bertani broth plus 2% NaCl (LB20) (9, 27), supplemented with the appropriate antibiotics, on a rotary shaker at 27°C. Experimental media included LB20, nine-salt solution (NSS) (a carbon-, nitrogen-, and phosphorus-free salt solution; formula listed below [15]), and NSS plus 200 μg salmon GI mucus protein/ml (NSSM) (9). For experiments, 16- to 18-h cultures of V. anguillarum (grown at 27°C) were centrifuged (9,000 × g, 10 min, 4°C), washed twice with NSS (9), and resuspended at the appropriate cell densities in either LB20 or NSSM. Cell densities were determined by serial dilution and plating on LB20 agar plates. For V. anguillarum cultures, antibiotics were used at the following concentrations: 5 μg/ml chloramphenicol (Cm5), 200 μg/ml streptomycin (Sm200), 50 μg/ml kanamycin (Km50), 200 μg/ml ampicillin (Ap200), and 1 μg/ml tetracycline (Tc1). All Escherichia coli strains were routinely grown in Luria-Bertani broth plus 1% NaCl (LB10), supplemented with the appropriate antibiotics, on a rotary shaker at 37°C. For E. coli cultures, antibiotics were used at the following concentrations: 20 μg/ml Cm (Cm20) and 20 μg/ml Tc (Tc20).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Vibrio anguillarum | ||
| M93Sm | Spontaneous Smr mutant of M93 (serotype J-O-1) | 6 |
| M99 | Smr CmrempA mutant; pNQEmpA insertion into empA | 7 |
| MSD1 | Smr Kmrepp mutant; mini-Tn10Km insertion into epp | This study |
| M101 | Smr Cmrepp mutant; pMV07 insertion into epp | This study |
| M102 | Smr Cmr Tcr; M101 complemented with pSUP202-epp | This study |
| Escherichia coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| XL1MRF′ | recA1 endA1 gryA96 thi-1 hsdR17 supE44 relA1 (lac-pro) [F′ proABlacI lacZΔM15 Tn10 (Tcr)] | Stratagene |
| SM10 | thi thr leu tonA lacY supE recA RP4-2-Tc::Mu::Km (λpir) | 16 |
| CC118 | λpir pLOFKm | 13 |
| Plasmids | ||
| pBluescript SKII+ | AprlacZ; pUC ORI | Stratagene |
| pNQ705-1 | Suicide vector, requires pir; Cmr | 16 |
| pSUP202 | Shuttle vector; Tcr Apr Cmr | GenBank accession no. AY428809; 7 |
| pMV01 | pBSK+ + MSD1 mini-Tn10Km fragment; Apr Kmr | This study |
| pMV07 | pNQ705 + epp fragment in SacI/XbaI site; Cmr | This study |
| pSUP202-epp | epp inserted into Apr cassette of pSUP202; Cmr Tcr | This study |
Preparation of NSS.
For 1 liter of medium, the following are included: 17.6 g sodium chloride (NaCl), 1.47 g sodium sulfate anhydrous (Na2SO4), 0.08 g sodium bicarbonate (NaHCO3), 0.25 g potassium chloride (KCl), 0.04 g potassium bromide (KBr), 1.87 g magnesium chloride (MgCl2·6H2O), 0.41 g calcium chloride (CaCl2·2H2O), 0.008 g strontium chloride (SrCl2·6H2O), and 0.008 g boric acid (H3BO3). Instructions are to heat while stirring to dissolve salts and to adjust the pH to 6.5 before autoclaving. The final pH is ∼7.5.
Mini-Tn10Km mutagenesis.
Mini-Tn10Km mutagenesis was carried out using the method developed by Herrero et al. (13), as modified by Rock and Nelson (23). Briefly, V. anguillarum M93Sm cells were mated with E. coli CC118 (λpir) (pLOFKm) containing the mini-Tn10Km. Aliquots (100 μl) of the cell suspension were spread plated onto LB20 Sm200 Km50 plates to select for V. anguillarum mutants containing a mini-Tn10Km insertion (6, 13). V. anguillarum colonies able to grow on LB20 Sm200 Km50 were transferred onto replicate LB20 Sm200 Km50 agar plates and NSSM-plus-1% skim milk agar plates, and protease activity was determined by measuring zones of proteolysis after 24 h at 27°C. Colonies that exhibited reduced protease activity on the NSSM-plus-1% skim milk agar plates were picked from LB20 Sm200 Km50 replicate plates and retested for proteolytic activity on a second NSSM-plus-1% skim milk agar plate. Colonies that still showed reduced proteolytic activity were saved for further study.
Preparation of mucus.
GI mucus was harvested from Atlantic salmon as previously described by Garcia et al. (9). Mucus was heat inactivated (100°C, 10 min) to destroy any inherent protease activity. The protein concentration in harvested mucus was determined using the Bradford assay (Bio-Rad Laboratories, Richmond, CA).
DNA isolation.
Genomic DNA was isolated from V. anguillarum strains by use of the Qiagen DNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The purified genomic DNA was quantified spectrophotometrically by measuring absorption at 260 nm and 280 nm using an Ultrospec 4000 spectrophotometer (Pharmacia Biotech, Piscataway, NJ).
Plasmid DNA isolation.
Plasmid DNA was isolated from bacterial strains by use of the Qiagen Qiaprep spin miniprep kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The purified plasmid DNA was quantified spectrophotometrically by measuring absorption at 260 nm and 280 nm using an Ultrospec 4000 spectrophotometer.
Detection and quantification of protease activity.
The protease activity of culture supernatants was quantified using the azocasein method of Windle and Kelleher (28), as modified by Denkin and Nelson (6). Briefly, culture supernatant was incubated with azocasein (6 mg/ml) dissolved in Tris-HCl (50 mM [pH 8.0]) containing 0.04% NaN3. Culture supernatant was prepared by centrifuging 1 ml of cells (12,000 × g, 10 min). Supernatant was removed and filtered through a 0.22-μm-pore-size cellulose-acetate filter. Filtered supernatant (100 μl) was incubated for 30 min at 27°C with 100 μl of azocasein solution. Reactions were terminated by the addition of trichloroacetic acid (TCA) (10% [wt/vol]) to a final concentration of 6.7% (wt/vol). The mixture was allowed to stand for 2 min and centrifuged (12,000 × g, 8 min) to remove unreacted azocasein, and supernatant containing azopeptides was suspended in 700 μl of 525 mM NaOH. The absorbance of the azopeptide supernatant was measured at 442 nm. Protease activity units (U) were calculated with the following equation: U = [1,000 (OD442)/CFU] × 109, where OD442 is the optical density at 442 nm.
Bacterial matings.
Bacterial matings were carried out using the procedure described by Milton et al. (16). Briefly, plasmids were introduced into V. anguillarum M93Sm from E. coli SM10 by conjugation using overnight cultures of V. anguillarum M93Sm and E. coli SM10 prepared and mixed at a ratio of 1:1 (recipient to donor) in NSS plus 10 mM MgSO4. The cell suspension was vacuum filtered onto a 0.22-μm-pore-diameter nylon membrane, placed on an LB15 agar plate (LB-plus-1.5% NaCl), and allowed to incubate overnight at 27°C. Following incubation, the cells were removed from the filter by vigorous vortexing in NSS plus 10 mM MgSO4. Cell suspensions (100 μl) were plated on LB20 Sm200 Cm5 plus any additional antibiotics and incubated at 27°C until V. anguillarum colonies were observed (usually 24 to 48 h).
Cloning methods.
The region containing the gene interrupted by mini-Tn10Km mutagenesis was cloned into pBluescript SKII+. Briefly, genomic DNA from V. anguillarum mini-Tn10Km mutants was digested with SacI restriction endonuclease (Promega, Madison, WI) and then ligated into the SacI restriction site of pBluescript SKII+. The resulting ligated DNA was used to transform E. coli XL1MRF′. Transformants were selected on LB10 agar plates supplemented with Km50 and Ap200. Plasmid DNA was extracted from any resulting clones and checked for the presence of the mini-Tn10Km insertion by PCR, restriction digestion, and gel electrophoresis. Plasmid DNA from appropriate clones was sequenced by the Rhode Island Genomics and Sequencing Center (http://www.uri.edu/research/gsc/).
PCR amplification.
All PCRs were done using Taq DNA polymerase (Qiagen) under the following conditions: 94°C for 5 min, 94°C for 30 s, 55 to 60°C for 1 min, and 70°C for 30 s. These steps were repeated for 30 cycles, with a final extension at 70°C for 10 min. All PCRs were carried out in a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). Modifications to the PCR cycle were dependent on the melting temperatures of the primers used and the lengths of the desired amplicons.
Construction of pMV07.
The construction of pMV07 was carried out using the method described by Milton et al. (16), as modified by Rock and Nelson (23). Briefly, restriction sites (SacI and XbaI) were engineered into the PCR primers SDeppF and SDeppR, respectively (Table 2) and were used to amplify a 430-bp fragment of epp from V. anguillarum M93Sm genomic DNA starting at 479 bp from the 5′ terminus of the epp gene. The amplified PCR product was digested with the restriction enzymes SacI and XbaI (Promega) and ligated using T4 DNA ligase (Promega) into the mobilizable suicide vector pNQ705, previously digested with SacI and XbaI, to yield pMV07. The resulting plasmid was then introduced into E. coli SM10 by electroporation transformation using a gene pulser (Bio-Rad, Richmond, CA). Transformants were incubated for 1 h at 37°C in a shaking water bath and plated onto LB agar plates containing Cm20. To confirm that the insert was successfully ligated into pNQ705, plasmid DNA was harvested from overnight E. coli cultures and then digested with SacI and XbaI, and the resulting DNA fragments were separated by electrophoresis through a 1.0% agarose gel in Tris-acetate EDTA (TAE) (3) buffer containing 0.2-μg/ml ethidium bromide run at 80 V for 1 h. Inserts were further verified by DNA sequencing (Rhode Island Genomics and Sequencing Center).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Target |
|---|---|---|
| KanD-S1 | GGTTTCATTTGATGCTCGATGAG | Kmr gene |
| KanD-S2 | GATGTTGGACGAGTCGGAATCG | Kmr gene |
| KanD-S3 | CGCTACCTTTGCCATGTTTCAG | Kmr gene |
| KanD-S4 | CGAGCAAGACGTTTCCCGTTG | Kmr gene |
| MT3 | GCGCAATTAACCCTCACTAAAGGG | pBluescript |
| MT7 | GCGTAATACGACTCACTATAGGGC | pBluescript |
| EmpA-F1 | TACAAATTAATTCTCATC | empA |
| EmpA-R1 | TGGCAAGAAAAGACTAG | empA |
| EppF3 | CTGCAGTGATTTAGGGGTAGTCAATGAAGATAA | epp |
| EppR3 | CTGCAGGGTTAAGTGATCAATCGCTTTAATTC | epp |
| SDeppF | GCTAGGAGCTCGCTATACAGGCCCTAATGGAGA | epp |
| SDeppR | GCTAGTCTAGAAGCGTACTTTTTGTGCCTTGAA | epp |
| pNQ706-R | GCGTAACGGCAAAAGCACCGCCGGACATCA | pNQ705-1 |
| Epp(complement)F | GCTAGCTGCAGCATTAATCGAACGAGGTGTGA | epp |
| Epp(complement)R | GCTAGCTGCAGTTAGCGCAGCAGCCAGAAT | epp |
Restriction sites within primers are in boldface.
Insertional mutagenesis of epp.
Site-specific insertional mutagenesis was used to create a gene interruption within the structural gene of epp by use of the procedure described by Milton et al. (16). Briefly, the pMV07 mobilizable suicide vector was transferred into V. anguillarum by conjugation with E. coli SM10. Cm-resistant colonies were selected and screened for insertion within epp. PCR was used to confirm the incorporation of pMV07. For PCR analysis, a primer described previously by Milton et al. (16) which was complementary to the pNQ705 vector was utilized (Table 2). The forward primer, SDepp (F) (Table 2), is complementary to a region upstream of the insertion. PCR products were analyzed by gel electrophoresis. The interruption within epp rendered mutants resistant to Cm at 5 μg/ml. The resulting V. anguillarum epp mutant was designated M101 (Table 1).
Complementation of the epp mutation.
V. anguillarum strain M101 was complemented by cloning the wild-type epp gene into the shuttle vector pSUP202 (23, 24) (accession no. AY428809). Restriction sites (PstI) were engineered into the PCR primer set that amplifies epp and its promoter region (Table 2). Amplified PCR products were digested with PstI (Promega) and the fragment was ligated using T4 DNA ligase (Promega) into the pSUP202 vector. Insertion was confirmed using PCR, digestion, and gel electrophoresis. The resulting pSUP202-epp construct was introduced into E. coli SM10 by electrotransformation and transferred to V. anguillarum M101 by conjugation to complement the epp mutation. The resulting complemented strain was designated M102 (Table 1).
Preparation of protein extracts.
Supernatant was collected from V. anguillarum cultures grown for 16 to 18 h in either LB20 or NSSM to be used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis in a modification of the procedure described by Staroscik et al. (25). Briefly, 9-ml cultures were centrifuged (9,000 × g, 10 min, 4°C) to pellet cells, and supernatants were sterile filtered through 0.22-μm-pore-size MillexGP cartridge filters (Millipore, Billerica, MA). Protein was precipitated with 15% TCA (0°C for 1 h), pelleted by centrifugation (12,000 × g, 15 min at 4°C), rinsed twice with acetone, and resuspended in 50 μl of 2× Laemmli sample buffer (Sigma-Aldrich, St. Louis, MO). Any contaminants that could interfere with SDS-PAGE were removed with the SDS-PAGE sample prep kit (Pierce Biotechnology, Inc., Rockford, IL) according to the manufacturer's instructions. Supernatant proteins were stored at −70°C until use.
SDS-PAGE and Western blot analysis.
Supernatant protein samples were separated using a 10% Tris-HCl gel. Prestained Precision Plus protein standards (Bio-Rad) and 2× Laemmli sample buffer (Sigma) were used for all gels. Protein loaded in each lane represents protein precipitated from 1.5 ml of the original culture supernatant (∼80 μg). Protein concentration was determined using the Bradford assay (Bio-Rad). Gels were transferred to nitrocellulose membranes for immunoblotting using the mini-Protean II system (Bio-Rad). Transfers were performed as described by Towbin et al. (26) at 100 V for 1.5 h. Nitrocellulose membranes were blocked, as described by Girouard et al. (11), with the addition of 5% skim milk. EmpA bands were detected with rabbit anti-P. aeruginosa LasB elastase antibodies at a dilution of 1:5,000, as described by Staroscik et al. (25). Antibody was detected with horseradish peroxidase-conjugated goat anti-rabbit antibody (Sigma) at a dilution of 1:2,500 and was visualized with 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate (Sigma).
DNA sequencing.
All DNA sequencing was carried out at the Rhode Island Genomics and Sequencing Center (University of Rhode Island, Kingston, RI) using the ABI3130XL genetic analysis system (Applied Biosystems, Inc.). Fluorescently labeled dideoxynucleotides and Taq DNA polymerase in a thermal cycling program were used in the sequence reactions. DNA samples were mixed with the appropriate primer prior to sequencing.
RESULTS
Mini-Tn10Km mutagenesis.
We hypothesized that since Staroscik et al. (25) had demonstrated that EmpA activity was dependent upon processing the secreted 46-kDa pro-EmpA into mature 36-kDa EmpA, a specific protease was responsible for the processing. Mini-Tn10Km mutagenesis (13) was used to create mutants of V. anguillarum M93Sm that exhibited altered protease activity. Approximately 3,000 mini-Tn10Km-containing colonies created by a single round of mutagenesis were screened for altered protease activity on NSSM-plus-1% skim milk agar plates by measuring the zones of proteolysis around the colonies after 24 h at 27°C. Several reduced-protease and protease-negative mutants were observed. A clone bank was created and screened by PCR with EmpA forward (EmpA-F1) and reverse (EmpA-R1) primers (Table 2) to identify any potential empA insertion mutants. One clone (MSD1; Table 1) that exhibited reduced protease activity compared to wild-type strain M93Sm and did not contain an insertion in empA was identified (data not shown). No protease-overproducing mutants were observed during the mini-Tn10Km mutagenesis screening.
Cloning and identification of a V. anguillarum putative protease gene.
In order to identify and characterize the gene interrupted by the mini-Tn10Km insertion, the region surrounding the mini-Tn10Km insertion was cloned into the SacI site of pBluescript SKII+. The resulting plasmid was designated pMV01 (Table 1). Restriction digestion of pMV01 using SacI yielded a 13-kbp insertion.
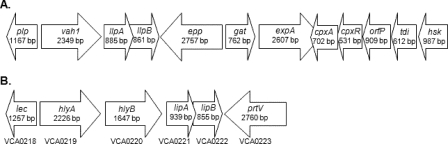
Forward and reverse primers from the mini-Tn10Km (KanDS1 and KanDS4) and modified T7 and T3 pBluescript-specific primers (Table 2) were used to initiate sequencing of pMV01. The sequence of the inserted DNA was determined by primer walking. DNA sequence analysis (GenBank accession number EU650390) by BLASTn and BLASTx (1) resulted in the identification of an open reading frame (containing the mini-Tn10Km insertion) encoding a 918-amino-acid putative protease previously undescribed for V. anguillarum. Additionally, several other previously unidentified genes, including those encoding a putative glutamine amidotransferase, a sensor protein, a transcriptional regulator, a periplasmic protein, a thiol:disulfide interchange protein, and a homoserine kinase as well as an additional putative extracellular protease were also found (Fig. 1A). The area containing the insertion was also found to contain the previously described lactonizing lipase activator gene (llpB), which is part of the V. anguillarum M93Sm hemolysin cluster 1 (vah1 hemolysin gene cluster; GenBank accession number DQ008059) (23) (Fig. 1A). BLASTx analysis of the cloned region revealed that the amino acid sequence of epp is highly conserved in both V. cholerae and V. splendidus, as well as in several non-Vibrio species (Table 3). The epp amino acid sequence was found to have 78% identity and 88% similarity to the prtV protease of V. cholerae. It also shares sequence homology with immune inhibitor A (inhA) of Aeromonas hydrophila and peptidase M6 of Shewanella sp. strain MR-7 (Table 3). Additionally, SignalP 3.0 (4) analysis predicts that the epp gene product (Epp) contains a signal peptide sequence with a predicted cleavage site between amino acids 23 and 24, suggesting that Epp is an extracellular protease (data not shown).
FIG. 1.
(A) Maps of pMV01. V. anguillarum DNA, containing the EmpA-processing protease (epp) gene and flanking sequences, that was cloned into pBluescript SKII+ (pMV07) and the adjacent hemolysin cluster 1 are shown. Genes coding for the following products are represented: phospholipase/lecithinase (plp), hemolysin (vah1), lactonizing lipase (llpA), lactonizing lipase activator (llpB), EmpA-processing protease (epp), putative glutamine amidotransferase (gat), putative exoprotease (expA), sensor protein (cpxA), transcriptional regulator (cpxR), putative periplasmic protein (orfP), thiol:disulfide interchange protein (tdi), and a putative homoserine kinase (hsk). Arrows indicate the direction of transcription. (B) Map of hemolysin cluster 1, containing a putative protease gene from V. cholerae O1 biovar El Tor strain N16961. Genes and the corresponding tag numbers coding for the following products are represented: lecithinase (lec), hemolysin (hlyA), chemotaxis transducer (hlyB), lipase accessory (lipAB), and putative metalloprotease (prtV). Arrows indicate the direction of transcription. Sizes are approximate.
TABLE 3.
Epp sequence similarity with proteins from other Vibrio and non-Vibrio species
| Epp or predicted protein showing homology to Epp | GenBank accession no. | Predicted protein size (amino acid residues) | % Amino acid:
|
|
|---|---|---|---|---|
| Identity | Similarity | |||
| V. anguillarum M93Sm Epp protease | EU650390 | 918 | 100 | 100 |
| V. cholerae O1 biovar El Tor N16961 protease | NP_232622 | 918 | 78 | 88 |
| V. splendidus protease | ZP_00988167 | 918 | 77 | 88 |
| Moritella sp. strain PE36 protease | ZP_01897859 | 919 | 68 | 82 |
| Marinomonas sp. strain MED121 protease | ZP_01074777 | 840 | 63 | 76 |
| Aeromonas hydrophila immune inhibitor A | YP_857947 | 758 | 50 | 65 |
| Shewanella sp. strain MR-7 peptidase M6, immune inhibitor A | YP_736182 | 936 | 39 | 56 |
Effect of an epp mutation on protease activity.
An epp mutant of V. anguillarum M93Sm, M101, was created by single-crossover insertional mutagenesis (see Materials and Methods) to determine its effect on protease activity compared to what was seen for other V. anguillarum strains. The pNQ705-1-derived suicide vector pMV07 (Table 1) was introduced into M93Sm by conjugation and inserted into epp via a single-crossover event. PCR and sequence analysis confirmed that the insertion is located at bp 914 of the epp gene (data not shown). Experiments were carried out in LB20 and NSSM, and these experiments demonstrated that M101 grows at approximately the same rate (g = 0.5 h) and to the same cell density as the wild-type strain M93Sm in LB20 and NSSM, with maximum cell densities of 3.5 × 109 CFU/ml and 4.1 × 109 CFU/ml, respectively.
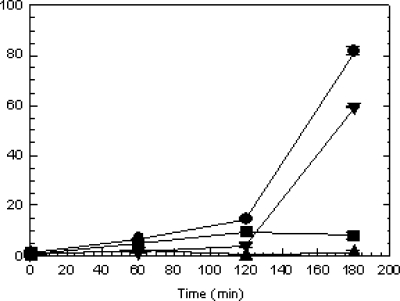
In order to demonstrate the effect of epp on secreted protease activity cultures of V. anguillarum strains M93Sm, M99 (empA mutant; Table 1), and M101, the strains were grown 16 h in LB20, washed twice in NSS, resuspended in NSSM at ∼2 × 109 CFU/ml, and incubated at 27°C. Samples (1 ml) were taken at 0, 60, 120, and 180 min, cell-free culture supernatants were assayed for protease activity, and their values were normalized to CFU (28). The data presented in Fig. 2 show that protease activity was induced in M93Sm culture supernatants by 120 min, reaching a maximum activity level of ∼82 U at 180 min. A low level of activity (8.1 U) was detected for M99 culture supernatant. No protease activity was detected for M101 culture supernatant (Fig. 2). In order to determine whether the mutation in epp was directly responsible for the loss of protease activity shown in Fig. 2, the wild-type epp gene was cloned into pSUP202, and the resulting plasmid, pSUP202-epp (Table 1), was introduced into M101 by conjugation. The complemented strain, M102, was able to restore the protease activity to ∼73% of that of the M93Sm wild-type strain (Fig. 2). These results demonstrate that the loss of a functional epp product results in a loss of protease activity, as observed for the M101 strain.
FIG. 2.
Induction of protease activity in the wild-type strain (M93Sm [•]) and mutant strains (M99 [empA mutant] [▪], M101 [epp mutant] [▴], and M102 mutant [pSUPepp] [▾]) of V. anguillarum at high cell densities. Cultures were grown with shaking for 16 h in LB20 at 27°C. Cells were harvested by centrifugation, washed two times with NSS, and then resuspended in NSSM (∼2 × 109 CFU/ml) at 27°C. Samples (1 ml) were taken at 0, 60, 120, and 180 min, and the supernatants were sterile filtered through a 0.22-μm filter. Protease activity was determined and then normalized to CFU as described by Denkin and Nelson (6). The data presented are from a representative experiment. Experiments were performed in triplicate and repeated at least three times. Error bars indicate one standard deviation.
These data also suggest that Epp protease activity can be detected in the absence of EmpA, when cells are incubated in mucus. No measurable protease activity was detected in the absence of Epp, which strongly suggests that Epp is the protease responsible for the activity observed for the empA mutant strain. Results also suggest that epp is directly involved in the processing of the V. anguillarum EmpA metalloprotease, and thus its product was designated as the EmpA-processing protease.
Effect of an epp mutation on EmpA processing.
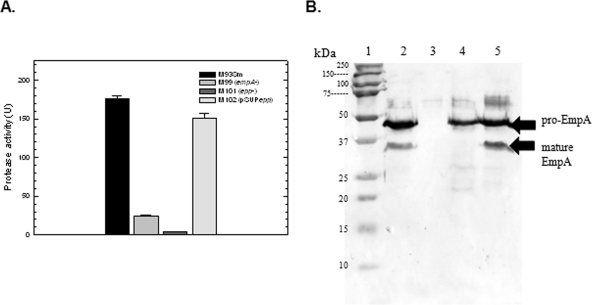
Since a mutation in epp resulted in a loss of protease activity when cells were incubated in mucus (Fig. 2), Western blot analysis was used to determine the role of epp in EmpA processing under the same conditions. Cultures of M93Sm, M99, and M101 were grown (18 h) to ∼2 × 109 CFU/ml in NSSM. Culture supernatants were sterile filtered and used for determining protease activity and for TCA protein precipitation. The wild-type M93Sm had a protease activity of 176.6 U. No protease activity was detected for M101. M99 had a protease activity of 24.3 U, which is approximately 13% of the wild-type activity level (Fig. 3A). Complemented strain M102 restored protease activity to ∼85% of wild-type levels (Fig. 3A), further suggesting that a functional epp product is necessary for protease activity in NSSM-grown cells. Note that unlike the previous experiment, which showed protease activity by LB20-grown stationary-phase cells induced with mucus for 3 h, this experiment used cells grown continuously in mucus to stationary phase (see Materials and Methods).
FIG. 3.
(A) Protease activity of V. anguillarum strains M93Sm, M99 (empA mutant), M101 (epp mutant), and M102 (pSUPepp) grown to stationary phase in NSSM. Cultures were grown with shaking for 18 h in NSSM at 27°C (∼2 × 109 CFU/ml). Cells were harvested by centrifugation and the supernatants were filtered through a 0.22-μm filter. Protease activity was determined and normalized to CFU. Samples from left to right: M93Sm, M99, M101, and M102. The data presented are from a representative experiment. Experiments were performed in triplicate and repeated at least three times. Error bars indicate 1 standard deviation. (B) Western blot analysis of EmpA secretion and activation. TCA-precipitated proteins from 1.5 ml of 18-h cell-free culture supernatants (∼80 μg protein) from strains M93Sm, M99 (empA mutant), M101 (epp mutant), and M102 (pSUPepp) grown in NSSM were separated by SDS-PAGE, transferred to nitrocellulose, probed with rabbit anti-LasB antibody followed by immunoglobulin G-labeled goat anti-rabbit antibody, and visualized using TMB as described in Materials and Methods. Lanes: 1, molecular size protein ladder; 2, M93Sm; 3, M99 (empA mutant); 4, M101 (epp mutant); 5, M102 (pSUPepp). The positions of secreted pro-EmpA (∼48 kDa), and mature EmpA (∼36 kDa) are indicated with arrows on the right. The data presented are from a representative experiment. Experiments were repeated at least three times.
TCA-precipitated proteins from cell-free culture supernatants were separated by SDS-PAGE and transferred to a nitrocellulose membrane for Western blot analysis. The amount of protein loaded in each lane was equivalent to 1.5 ml of the original NSSM culture (∼80 μg). Western blot analysis revealed two anti-LasB reactive bands in M93Sm culture supernatants with estimated molecular masses of ∼46 kDa and ∼36 kDa, representing pro-EmpA and mature EmpA, respectively (Fig. 3B, lane 2). Only the 46-kDa pro-EmpA could be detected in M101 supernatant (Fig. 3B, lane 4). M99 supernatant was used as a control and did not react with the anti-LasB antibodies (Fig. 3B, lane 3). Analysis of the complemented strain M102 culture supernatant revealed both pro-EmpA and mature EmpA bands equivalent in intensity to those observed for M93Sm supernatant (Fig. 3B, lane 5). These data show that Epp is responsible for the processing of the 46-kDa pro-EmpA form to the 36-kDa mature active enzyme.
Mixing protease-negative culture supernatants results in protease activation.
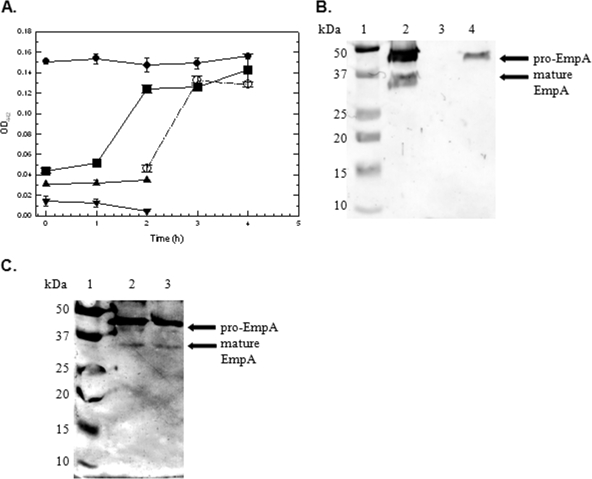
Based on the results shown in Fig. 3B, we hypothesized that if Epp was not dependent on empA transcription, then it would be secreted into the supernatant. We have shown that M101 secretes pro-EmpA when cells are incubated in mucus, as seen in Fig. 3B. We speculated that M99 cultures produce and secrete Epp under the same conditions. Moreover, we also hypothesized that Epp produced and secreted by M99 can process pro-EmpA in M101 culture supernatants. To test this, cultures were grown 16 h in LB20, washed twice in NSS, resuspended in NSSM at ∼2 × 109 CFU/ml, and incubated at 27°C for 180 min. Earlier results (Fig. 2) show that protease activity is fully induced by 180 min in wild-type strain M93Sm. At 180 min, cells from the M93, M99, and M101 NSSM cultures were harvested and supernatants were sterile filtered as described in Materials and Methods. Sterile-filtered cell-free supernatants from M99 and M101 were mixed in a 1:1 ratio (mix 1) and incubated at 27°C for 4 h. Aliquots of M93Sm and M99/M101 (mix 1) cell-free supernatants were taken at 0, 1, 2, 3, and 4 h, and protease activities were determined by OD readings at 442 nm. The remaining M99 and M101 cell-free supernatants were incubated at 27°C for 2 h and were then mixed in a 1:1 ratio and incubated for an additional 2 h (mix 2). Aliquots of M99/M101 (mix 2) cell-free supernatant were taken at 0, 1, and 2 h. The protease activities of cell-free supernatants were determined by OD readings at 442 nm. Only trace amounts of protease activity were detected in unmixed M99 and M101 cell-free supernatants (Fig. 4). When M99 and M101 cell-free supernatants were mixed 1:1 in both M99/M101 (mix 1) and M99/M101 (mix 2), protease activity was detected by 1 h and then reached an activity level close to that of the wild type by 2 h. Activity did not increase with further incubation. Protease activity levels remained constant over a 24-h period (data not shown).
FIG. 4.
(A) Protease activity of V. anguillarum strains (M93Sm [•], M99 [empA mutant] [▴], M101 [epp mutant] [▾], M99 plus M101 after first mixing [▪], and M99 plus M101 after second mixing [○]). Cultures (10 ml) were grown with shaking for 16 h in LB20 at 27°C. Cells were harvested by centrifugation, washed two times with NSS, and then resuspended in NSSM (∼2 × 109 CFU/ml) at 27°C. Cells from all cultures were harvested at 180 min and the supernatants were sterile filtered through a 0.22-μm filter. Aliquots (5 ml) of M99 and M101 culture supernatant were mixed 1:1 (mix 1) and incubated at 27°C with shaking. Remaining M99 and M101 culture supernatants (4 ml) were incubated separately at 27°C with shaking. Samples (750 μl) of M99/M101 mix 1 supernatant were taken at 0, 1, 2, 3, and 4 h. Samples (750 μl) of individual M99 and M101 supernatants were taken at 0, 1, and 2 h. At 2 h, M99 and M101 supernatants were mixed 1:1 (mix 2) and incubated at 27°C. Samples (600 μl) of mix 2 were taken at 0, 1, and 2 h after mixing (2, 3, and 4 h after the start of the experiment). M93Sm was used as a control. Protease activity was determined by OD readings at 442 nm (OD442). The data presented are from a representative experiment. Experiments were performed in triplicate and repeated at least three times. Error bars indicate 1 standard deviation. (B) Western blot of individual supernatants. TCA-precipitated proteins of cell-free supernatants from NSSM-grown cultures of M93Sm, M99 (empA mutant), and M101 (epp mutant) were separated by SDS-PAGE, transferred to nitrocellulose, probed with rabbit anti-LasB antibody followed by immunoglobulin G-labeled goat anti-rabbit antibody, and visualized using TMB liquid substrate as described in Materials and Methods. Lanes: 1, molecular size protein ladder; 2, M93Sm; 3, M99 (empA mutant); 4, M101 (epp mutant). (C) Western blot of M99/M101 mix 1 and M99/M101 mix 2. Lanes: 1, molecular size protein ladder; 2, M99/M101 mix 1 supernatant; 3, M99/M101 mix 2 supernatant. Positions of the secreted pro-EmpA (∼48 kDa) and mature EmpA enzyme (∼36 kDa) are indicated with arrows on the right. The data presented are from a representative experiment. Experiments were repeated at least three times.
To show that the protease activity detected as shown in Fig. 4A was a result of pro-EmpA activation by Epp, M99/M101 (mix 1) and M99/M101 (mix 2) supernatants were analyzed by Western blotting. TCA-precipitated proteins from cell-free culture supernatants from the control strains (M93Sm, M99, and M101) as well as the supernatants from M99/M101 (mix 1) and M99/M101 (mix 2) were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Western blotting revealed the presence of both pro-EmpA (∼46 kDa) and mature EmpA (∼36 kDa) in the M93Sm supernatant (Fig. 4B, lane 2) as well as for M99/M101 (mix 1) and M99/M101 (mix 2) (Fig. 4C, lanes 2 and 3). Only pro-EmpA (∼46 kDa) could be detected in the M101 supernatant (Fig. 4B, lane 4). M99 did not react with anti-LasB antibodies (Fig. 4B, lane 3). It should be noted that the processed EmpA bands in the Western blot were fainter than those previously observed. The most likely explanation is the fact that the protein obtained from the experiment depicted in Fig. 4C was from cultures induced with mucus for 180 min rather than from cultures grown for 18 h in NSSM, as was the case in the experiment whose results are shown in Fig. 3B.
In all, these results support our hypothesis that the epp gene product is responsible for the processing of EmpA, and without a functional epp gene, EmpA is not able to be converted from the immature pro-EmpA form to the mature active enzyme.
DISCUSSION
Previous studies have shown that incubation of V. anguillarum in salmon GI mucus specifically induces a number of proteins (9), including the EmpA metalloprotease, during stationary phase (6). Western blot analysis using V. anguillarum M93Sm has shown that the cytoplasmic 66.7-kDa pre-pro-EmpA protein is secreted as an ∼46-kDa proenzyme that is activated extracellularly by the removal of a 10-kDa peptide, resulting in the mature ∼36-kDa enzyme (25). However, the gene(s) involved in the processing of the EmpA protein from a nascent protein to an active mature metalloprotease was unknown. Staroscik et al. (25) hypothesized that the most likely mechanism by which this occurs is via the production and secretion of an additional protease that acts to remove the propeptide from EmpA and activate the enzyme.
In this study, a single gene of V. anguillarum (epp) was identified and characterized with regard to its ability to promote the processing of extracellular pro-EmpA to mature EmpA. Initial mini-Tn10Km mutagenesis created a mutant of V. anguillarum (MSD1) that lost all extracellular protease activity found in the wild-type strain M93Sm (data not shown). The region surrounding the insertion mutation was cloned and sequenced, revealing the previously described hemolysin gene cluster 1 (vah1 hemolysin gene cluster) (Fig. 1A) (23), as well as epp, which encodes a 918-amino-acid putative protease previously undescribed for V. anguillarum. A second epp mutation, M101, created by site-specific insertional mutagenesis, exhibited a phenotype identical to that of MSD1 and was used in all subsequent experiments.
The deduced amino acid sequences of the V. anguillarum epp gene and the previously reported Vibrio cholerae El Tor prtV gene show high degrees of identity and similarity, with an identity of 78% and a similarity of 88%. The Epp amino acid sequence also shares high degrees of homology with those of other putative proteases found in other Vibrio and non-Vibrio species (Table 3). As illustrated in Fig. 1A, the epp gene lies downstream from the vah1 hemolysin gene cluster. The gene order of this region is identical to that found for the V. cholerae hlyA hemolysin (Fig. 1B) region, with the exception that V. anguillarum does not have a hlyB homologue (20). This gene arrangement is highly conserved in several other Vibrio species (8). It has been proposed that the genetic organization of this region of V. cholerae, which includes prtV, is part of a pathogenicity island encoding products capable of damaging host cells (20). Moreover, it suggests a possible role for PrtV in V. cholerae, as a processing protease for the HA/protease (a homologue of EmpA). This may also be the case for other epp homologues which have yet to be fully characterized.
Protease activity was lost in M101 NSSM-grown cultures, and Western blot analysis revealed only the ∼46-kDa pro-EmpA present in the cell-free supernatant. Taken together, these data strongly suggest that Epp is the protease responsible for the cleavage of the 10-kDa EmpA propeptide from pro-EmpA to yield mature active EmpA. The presence of stable pro-EmpA in M101 supernatant supports the findings reported by Staroscik et al. (25), suggesting that EmpA activation is not autolytic. The epp mutation was complemented in the M102 strain, restoring protease activity to wild-type levels. Western blots of M102 NSSM-grown supernatants revealed mature EmpA bands comparable to those observed for wild-type strain M93Sm, thereby providing further evidence to support Epp as being the EmpA-processing protease. The ability of M102 to fully complement the epp mutation shows that epp is directly responsible for the differences observed between M93Sm and M101 with regard to protease activity and EmpA processing. Further, the gene orientation of the region containing epp strongly suggests that our findings are not the result of a cis polar effect. We have shown that the epp gene product, Epp, is a protease that is responsible for the processing and activation of pro-EmpA to the active mature metalloprotease.
Recent studies using recombinant EmpA synthesized in E. coli (29-31) have suggested that the presence of a stable ∼36-kDa EmpA form is the result of a thermoinduced self-proteolysis of a 44.6-kDa precursor (30). The 36-kDa peptide was detected only if protein samples were boiled prior to SDS-PAGE and Western blot analysis (29-31). Similar results were obtained using V. anguillarum W-1 supernatant (31). Our results, however, revealed no difference in EmpA processing between boiled and nonboiled samples (data not shown). Western blot analysis showed that M101 pro-EmpA could not be converted to the 36-kDa form by boiling. Also, the amount of mature ∼36-kDa EmpA did not increase if samples were boiled when equal amounts of protein were analyzed in either the wild type or the complemented strain. Perhaps the heat-induced proteolysis of recombinant EmpA was an artifact of cloning into E. coli, or it may have been specific to the EmpA of V. anguillarum W-1 (31); however, we were unable to observe heat-induced proteolysis. In contrast, our data clearly demonstrate that in V. anguillarum M93Sm, the conversion of ∼46-kDa pro-EmpA to mature ∼36-kDa EmpA requires epp.
When M99 and M101 supernatants were mixed in a 1:1 ratio, protease activity quickly approached wild-type levels. This protease activity was stable and remained constant over a 24-h period (data not shown). Western blots reveal that Epp is secreted by M99 (empA null mutant) into the culture supernatant, where it is stable and able to process pro-EmpA secreted by M101 (epp null mutant) to the mature EmpA protease. Thus, our data demonstrate that pro-EmpA processing occurs extracellularly and is mediated by the secreted protease, Epp. This provides insight into the regulation of empA and epp. EmpA secretion is independent of epp transcription and Epp secretion is independent of empA transcription. Both of these proteins are secreted into the culture supernatant independent of one another when cells are grown in salmon GI mucus. We have consistently detected low levels of protease activity in M99 EmpA-free supernatants by use of assays with azocasein. The protease activity levels of these cultures are always low, 5 to 10% of the activity seen for wild-type cultures. This seems to suggest that Epp has some general protease activity and is able to react with azocasein. It also suggests that Epp is secreted at low levels compared to those for EmpA secretion in NSSM culture conditions.
Previous studies by Denkin and Nelson (7) have demonstrated that the regulation of empA transcription is complex, requiring RpoS and VanT (a LuxR homologue), and is also positively affected by salmon GI mucus. Preliminary observations suggest that epp transcription is also positively regulated by salmon GI mucus.
Extracellular proteases are virulence factors for many pathogenic bacteria and are highly regulated proteins that are activated in response to different environmental factors. For example, the V. cholerae HA/protease (Hap) has been shown to nick and activate the A subunit of cholera enterotoxin (5), and the V. vulnificus Vvp metalloprotease causes a hemorrhagic reaction by degrading type IV collagen in basement membranes (17). The LasB elastase of P. aeruginosa is required for tissue destruction during opportunistic infections (18). These proteases are highly homologous to EmpA metalloprotease of V. anguillarum (6, 7, 16).
How secreted proteases are processed extracellularly to become active virulence factors is not fully understood. Multiple processing steps have been proposed for several other EmpA homologues, such as the Hap protease of V. cholerae (12), the Vvp protease of V. vulnificus (17), and the LasB elastase of P. aeruginosa (16). The Hap protease undergoes several steps of processing, including cleavage of the signal peptide of the 69.3-kDa propeptide to generate a 45-kDa form. It undergoes further proteolytic processing of the C-terminal region to form the mature 32-kDa enzyme (12). In in vivo systems, the 45-kDa form can be processed to the 35-kDa form autolytically as well as by other general proteases found in the intestine. Additional studies have shown that HA/protease has bifunctional properties; the 45-kDa form is responsible for the enterotoxic response and the fully processed 35-kDa protease causes the HA activity responsible for the cytotoxic effects caused by this organism (10). We note that the processing of the pro-LasA to mature LasA protease in P. aeruginosa has been shown to involve the action of other secreted proteases, including elastase, lysine-specific protease (protease IV or PrpL), and alkaline proteinase (14). Kessler et al. (14) showed that purified preparations of each protease were able to convert the secreted 42-kDa pro-LasA into the mature 20-kDa LasA, with the transient accumulation of a 28-kDa intermediate. In contrast, pro-EmpA processing by V. anguillarum involves only one additional protease, Epp, and we did not detect any transient intermediate.
Finally, Western blot analysis reveals that pro-EmpA is always detected in culture supernatants, regardless of culture conditions or length of incubation, suggesting that not all pro-EmpA is converted to mature EmpA. This raises the possibility that pro-EmpA has a role during infection by V. anguillarum that differs from that for mature EmpA metalloprotease. Perhaps EmpA, similar to Hap protease of V. cholerae, has bifunctional properties (10). How pro-EmpA may contribute to virulence of V. anguillarum is currently under investigation.
Acknowledgments
We thank Joan Olson (West Virginia University) for providing the LasB antisera used in this study.
This research was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service, grant no. 2005-35204-16294, and by a USDA Cooperative State Research, Education and Extension Service award, grant no. 2006-34438-17306, awarded to D.R.N. This research is based in part upon work conducted using the Rhode Island Genomics and Sequencing Center, which is supported in part by the National Science Foundation/EPSCoR (grant no. 0554548). This research was also made possible in part by use of the RI-INBRE Research Core Facility, supported by grant no. P20 RR16457 from NCRR, NIH.
Footnotes
Published ahead of print on 8 August 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids. Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: disease of farmed and wild fish, 3rd ed. Springer Praxis Books, Berlin, Germany.
- 3.Ausubel, F. M. 1988. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 5.Crowther, R. S., N. W. Roomi, R. E. Fahim, and J. F. Forstner. 1987. Vibrio cholerae metalloproteinase degrades intestinal mucin and facilitates enterotoxin-induced secretion from rat intestine. Biochim. Biophys. Acta 924393-402. [DOI] [PubMed] [Google Scholar]
- 6.Denkin, S. M., and D. R. Nelson. 1999. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl. Environ. Microbiol. 653555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denkin, S. M., and D. R. Nelson. 2004. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl. Environ. Microbiol. 704193-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiore, A. E., J. M. Michalski, R. G. Russell, C. L. Sears, and J. B. Kaper. 1997. Cloning, characterization, and chromosomal mapping of a phospholipase (lecithinase) produced by Vibrio cholerae. Infect. Immun. 653112-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia, T., K. Otto, S. Kjelleberg, and D. R. Nelson. 1997. Growth of Vibrio anguillarum in salmon intestinal mucus. Appl. Environ. Microbiol. 631034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh, A., D. R. Saha, K. M. Hoque, M. Asakuna, S. Yamasaki, H. Koley, S. S. Das, M. K. Chakrabarti, and A. Pal. 2006. Enterotoxigenicity of mature 45-kilodalton and processed 35-kilodalton forms of hemagglutinin protease purified from a cholera toxin gene-negative Vibrio cholerae non-O1, non-O139 strain. Infect. Immun. 742937-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girouard, L., D. C. Laux, S. Jindal, and D. R. Nelson. 1993. Immune recognition of human Hsp60 by Lyme disease patient sera. Microb. Pathog. 14287-297. [DOI] [PubMed] [Google Scholar]
- 12.Hase, C. C., and R. A. Finkelstein. 1991. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J. Bacteriol. 1733311-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessler, E., M. Safrin, J. K. Gustin, and D. E. Ohman. 1998. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J. Biol. Chem. 27330225-30231. [DOI] [PubMed] [Google Scholar]
- 15.Marden, P., A. Tunlid, K. Malmcrona-Friberg, G. Odham, and S. Kjelleberg. 1985. Physiological and morphological changes during short term starvation of marine bacteriological isolates. Arch. Microbiol. 142326-332. [Google Scholar]
- 16.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 1747235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyoshi, S., H. Nakazawa, K. Kawata, K. Tomochika, K. Tobe, and S. Shinoda. 1998. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect. Immun. 664851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicas, T. I., and B. H. Iglewski. 1985. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31387-392. [DOI] [PubMed] [Google Scholar]
- 19.Norqvist, A., B. Norrman, and H. Wolf-Watz. 1990. Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect. Immun. 583731-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogierman, M. A., A. Fallarino, T. Riess, S. G. Williams, S. R. Attridge, and P. A. Manning. 1997. Characterization of the Vibrio cholerae El Tor lipase operon lipAB and a protease gene downstream of the hly region. J. Bacteriol. 1797072-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsson, J. C., A. Joborn, A. Westerdhal, L. Blomberg, S. Kjelleberg, and P. L. Conway. 1996. Is the turbot, Scophthalums maximus (L.), intestine a portal of entry for the fish pathogen Vibrio anguillarum? J. Fish Dis. 19225-234. [Google Scholar]
- 22.O'Toole, R., J. Von Hofsten, R. Rosqvist, P. E. Olsson, and H. Wolf-Watz. 2004. Visualisation of zebrafish infection by GFP-labelled Vibrio anguillarum. Microb. Pathog. 3741-46. [DOI] [PubMed] [Google Scholar]
- 23.Rock, J. L., and D. R. Nelson. 2006. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect. Immun. 742777-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon, R., U. Prefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1787-796. [Google Scholar]
- 25.Staroscik, A. M., S. M. Denkin, and D. R. Nelson. 2005. Regulation of the Vibrio anguillarum metalloprotease EmpA by posttranslational modification. J. Bacteriol. 1872257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaantanen, P. 1976. Microbiological studies in coastal waters of the northern Baltic Sea. I. Distribution and abundance of bacteria and yeasts in the Tvarminne area. Walter Ander Nottbeck Found. Sci. Rep. 11-58. [Google Scholar]
- 28.Windle, H. J., and D. Kelleher. 1997. Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 653132-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, H., J. Chen, G. Yang, X. H. Zhang, and Y. Li. 2007. Mutational analysis of the zinc metalloprotease EmpA of Vibrio anguillarum. FEMS Microbiol. Lett. 26756-63. [DOI] [PubMed] [Google Scholar]
- 30.Yang, H., J. Chen, G. Yang, X. H. Zhang, Y. Li, and M. Wang. 2007. Characterization and pathogenicity of the zinc metalloprotease empA of Vibrio anguillarum expressed in Escherichia coli. Curr. Microbiol. 54244-248. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, F., J. Chen, Z. Chi, and L. F. Wu. 2006. Expression and processing of Vibrio anguillarum zinc-metalloprotease in Escherichia coli. Arch. Microbiol. 18611-20. [DOI] [PubMed] [Google Scholar]