Abstract
Among other functions, ATP-dependent proteases degrade misfolded proteins and remove several key regulatory proteins necessary to activate stress responses. In Bacillus subtilis, ClpX, ClpE, and ClpC form homohexameric ATPases that couple to the ClpP peptidase. To understand where these peptidases and ATPases localize in living cells, each protein was fused to a fluorescent moiety. We found that ClpX-GFP (green fluorescent protein) and ClpP-GFP localized as focal assemblies in areas that were not occupied by the nucleoid. We found that the percentage of cells with ClpP-GFP foci increased following heat shock independently of protein synthesis. We determined that ClpE-YFP (yellow fluorescent protein) and ClpC-YFP formed foci coincident with nucleoid edges, usually near cell poles. Furthermore, we found that ClpQ-YFP (HslV) localized as small foci, usually positioned near the cell membrane. We found that ClpQ-YFP foci were dependent on the presence of the cognate hexameric ATPase ClpY (HslU). Moreover, we found that LonA-GFP is coincident with the nucleoid during normal growth and that LonA-GFP also localized to the forespore during development. We also investigated LonB-GFP and found that this protein localized to the forespore membrane early in development, followed by localization throughout the forespore later in development. Our comprehensive study has shown that in B. subtilis several ATP-fueled proteases occupy distinct subcellular locations. With these data, we suggest that substrate specificity could be determined, in part, by the spatial and temporal organization of proteases in vivo.
Regulated proteolysis is important for the degradation of misfolded proteins in organisms ranging from bacteria to mammals (for a review, see references 16 and 50). In eukaryotes, the proteasome has roles in the degradation of regulatory proteins, providing an added level of control for several pathways, including cell cycle and programmed cell death (for a review, see references 16 and 76). Similarly, in bacteria, proteolysis regulates several stress responses, including DNA damage, heat shock, and oxidative stress (for a review, see reference 16).
Several ATP-dependent proteases have been characterized in bacteria, including Clp, Lon, and FtsH (HflB). The Clp proteases are two-component degradation machines. These proteases are composed of a hexameric AAA+ component that is required for both substrate recognition and ATP-dependent substrate unfolding (for a review, see reference 16). The hexameric ATPase associates with a multisubunit peptidase that is required for substrate destruction. ClpE, ClpC, ClpY (HslU), and ClpX all function in substrate recognition and ATP hydrolysis, while ClpP and ClpQ (HslV) are the peptidases in Bacillus subtilis (for a review, see reference 42). ClpP pairs with ClpX, ClpC, and ClpE, while ClpQ (HslV) pairs with ClpY (HslU) (12, 13).
In B. subtilis, Clp proteases carry out important roles during normal growth and during stress responses (for a review, see reference 11). During normal growth, ClpP is also important for protein turnover (22). ClpXP function is required for spore development during nutritional deprivation, DNA damage checkpoint release, oxidative stress, and alkaline stress and for the development of genetic competence for the uptake of exogenous DNA (13, 22, 23, 29, 36, 39, 40, 71).
In Escherichia coli, Lon is a 94-kDa ATP-dependent protease that forms a ring-shaped hexamer with a central cavity for degradation (44). The Lon protease in E. coli has been characterized extensively (for a review, see reference 16). Disruption of E. coli lon results in striking phenotypes, including cell filamentation, mucoid colony morphology, and sensitivity to DNA-damaging agents (17-19, 30, 55, 66, 75). B. subtilis encodes two lon homologs, lonA+ and lonB+ (49, 58). LonA has been shown to prevent σG-dependent transcription during vegetative growth (57), and lonB+ gene expression is controlled by σF in the forespore (58). In contrast to the drastic phenotypes associated with E. coli lon strains, deletion of lonA, lonB, or both in B. subtilis has no observable phenotype (54, 57).
The striking number of ATP-dependent proteases in B. subtilis and other bacteria raises questions about how cells target specific substrates to these abundant proteases in vivo. In E. coli, ClpXP recognizes specific N- and C-terminal motifs located within its substrates (10). Furthermore, experiments by many groups have shown that substrate targeting to several Clp proteases is based on peptide sequence (for a review, see reference 56). Although Lon-dependent degradation is less specific, critical amino acids within the DNA polymerase V accessory subunit UmuD′ have been identified (14). For many of the Clp proteases, adaptor proteins have been well characterized for specific targeting of substrates to their cognate hexameric ATPase (6, 9, 27, 36-38, 43, 72). These examples highlight the biochemical mechanisms used to specify substrate delivery to the abundant energy-dependent proteases found in bacteria (for a review, see reference 56).
In eukaryotic cells, the proteasome shows distinct subcellular localization (53) and this localization contributes to the degradation of specific substrates in vivo (for a review, see references 15, 50, 51, and 73). Less is known about the subcellular localization of proteases in bacteria. In B. subtilis, localization of ectopically expressed FtsH-GFP (green fluorescent protein) has been shown to be near midcell in vegetative cells and near the asymmetric septa of sporulating cells (70). Cells deficient for ftsH show defects in cell division, suggesting that FtsH subcellular localization contributes to the biological function of this protease (7). Furthermore, LonB-GFP has been shown to localize to the forespore in B. subtilis (58). Although lonB strains show accumulation of a forespore-specific sigma factor, defects in sporulation have not been observed (58).
The function of the subcellular localization of the Clp proteases in B. subtilis or E. coli is not well understood. E. coli ClpX-GFP and ClpP-GFP have been shown to form foci in a small percentage of cells when overexpressed from a plasmid (http://ecoli.aist-nara.ac.jp/GB5/search.jsp). For B. subtilis, electron microscopy imaging of ClpC, ClpX, and ClpP has shown that these proteins aggregate in heat-treated cells that were fixed (23). The best-understood example is from Caulobacter crescentus. In Caulobacter, ClpX and ClpP have been shown to localize at the cell pole, and polar localization has been shown to depend on a specific protein, CpdR (32). These studies suggest that at least a subset of the ATP-dependent proteases may localize to distinct subcellular positions.
In this study, we report the subcellular localization of the Clp and Lon proteases in live B. subtilis cells. Each of the functional fluorescent protein fusions examined was expressed from its native promoter at its native chromosomal locus. We found that these proteases and the associated ATPases occupy multiple subcellular positions in live cells and that ClpX and ClpP foci are dynamically regulated by temperature. We also show that LonA-GFP localizes to the nucleoid during normal growth and to the forespore during development. Furthermore, we find that LonB-GFP undergoes a redistribution that is developmentally regulated. LonB-GFP associates with the forespore membrane early during development, before redistributing to the entire forespore later in development. Taken together, we demonstrate that ATP-dependent proteases are spatially and temporally regulated in B. subtilis. We suggest that protease localization may help direct the degradation of specific substrates in vivo.
MATERIALS AND METHODS
Bacteriological methods.
All B. subtilis strains use in this study are described in Table 1. We constructed C-terminal translational fusions of clpX, clpP, clpE, clpC, and clpQ with gfp or yfp by using the EcoRI and XhoI sites in plasmids pKL147 for gfp and pBS243 for yfp (25, 62). For construction of lonA-gfp and lonB-gfp, we used a monomeric version of gfp with an extended flexible linker that has been described previously (2). We used site-directed mutagenesis to insert the A206K missense mutation into cfp(w7) in plasmid pKL189 (26). pLS24A was then digested with XhoI and SphI to release the mcfp(w7) fragment for replacement of gfp in plasmid pLS13 (clpP-gfp), yielding plasmid pLS24 [clpP-mcfp(w7)]. For construction of the lonA::spc allele, a 500-bp internal fragment of lonA was cloned into pJL74 by use of EcoR1 and XhoI sites encoded in the primers (24). All plasmids were used to transform the wild-type strain PY79. For subsequent transformation, chromosomal DNA was harvested from the established strain and used to transform subsequent strains, all of which were derivatives of PY79. Sporulation was induced by adding 1 mg/ml decoyinine to S7 medium after the cultures reached an optical density at 600 nm of 0.5 (61). All primer sequences used in this study are available upon request.
TABLE 1.
List of strains
| Straina | Relevant genotype | Reference or source |
|---|---|---|
| PY79 | Prototroph SPβ° | 74 |
| LAS31 | clpX::clpX-gfpmut2 (spc) | This work |
| LAS34 | clpP::clpP-gfpmut2 (spc) | This work |
| LAS56 | clpP::clpP-mcfp(w7)(A206K) (spc) | This work |
| LAS57 | clpE::clpE-yfpmut2 (cat) | This work |
| LAS58 | clpX::clpX-yfpmut2 (cat) | This work |
| LAS59 | clpC::clpC-yfpmut2 (cat) | This work |
| LAS62 | clpQ::clpQ-yfpmut2 (cat) clpY | This work |
| GJK485 | clpP::clpP-mcfp(w7)(A206K) (spc) clpX::clpX-yfpmut2 (cat) | R. Losick |
| LAS106 | lonA::lonA-mgfpmut2(A206K) (spc) | This work |
| LAS107 | lonB::lonB-mgfpmut2(A206K) (spc) | This work |
| LAS125 | lonA::spc | This work |
| LAS175 | clpX::spc | 28 |
| LAS176 | clpP::mls | 41 |
| LAS370 | clpE::neo | 54 |
| LAS371 | clpC::tet | 54 |
| LAS372 | clpQY::spcb | 54 |
| LAS373 | lonB::neo | 54 |
Medium and growth conditions.
Unless otherwise indicated, all strains were incubated at 30°C in S750 minimal medium (2) supplemented with 1% glucose, 0.1% glutamate, 40 μg/ml tryptophan, and phenylalanine prior to microscopy. When necessary, the following antibiotics were used, with the final concentration indicated: spectinomycin, 100 μg/ml; chloramphenicol, 5 μg/ml; erythromycin, 0.5 μg/ml; lincomycin, 12.5 μg/ml; and neomycin, 2.5 μg/ml. For nucleoid condensation, either 5 μg/ml chloramphenicol or 50 μg/ml kanamycin was used following addition to the growth medium for 60 min. For heat treatment, each strain was assembled onto slides, and the slide was incubated at 42°C for 20 min. We found that ClpP-CFP (cyan fluorescent protein) or ClpP-GFP focus numbers increased substantially even after a 10-min incubation at 42°C (data not shown). Cells were examined immediately following incubation, and all images presented in figures or used for scoring were acquired within 5 min following heat shock treatment. We found that the increase in focus intensity of ClpP-GFP following heat shock was stable at room temperature for at least 60 min (data not shown). For inhibiting protein translation, cells were incubated with 40 μg/ml chloramphenicol until growth arrest (∼2 h). Cells were then heat treated at 42°C for 20 min prior to microscopy.
Microscopy.
Live-cell microscopy was performed as described previously (59-62). Briefly, 300-μl aliquots of cells were stained with the vital membrane dye FM4-64 (Molecular Probes) and the DNA dye 4′,6′-diamidino-2-phenylindole (DAPI). We used Chroma filter sets 41001 for GFP, 31000 for DAPI, 41002C for FM4-64, 31044v2 for CFP, and 41029 for YFP (yellow fluorescent protein). Exposure time for FM4-64 was 150 ms, that for DAPI was 43 ms, and that for GFP ranged from 43 ms to 1 s depending on the GFP fusion protein (exposure times are indicated in each figure legend). For other images, YFP exposure was 1 s, and CFP exposure was 90 ms. All images were colored and merged using OpenLab software. Images were transferred to Adobe Photoshop and assembled using Adobe Illustrator. For colocalization experiments, ClpX-YFP (90 ms) and ClpP-CFP were captured after a 240-ms exposure for untreated samples and a 90-ms exposure for the heat-treated sample. The membrane was stained with FM4-64, and images were captured after 300-ms exposures. In all cases, heat treatment experiments were performed by placing the slide assembled with cells and coverslip at 42°C for 20 min. Images presented in figures or quantitation in tables were from at least two independent experiments. Single foci were scored using a random-number generator to remove systematic bias from the cell measurements, as described previously (2, 68).
Homology modeling.
The model of B. subtilis ClpQ was generated using SWISS-MODEL (http://swissmodel.expasy.org/SWISS-MODEL.html). The template used to generate the B. subtilis model was from the E. coli ClpQ crystal structure coordinates (3).
Protein structure accession number.
The Protein Data Bank accession number used was 1ned.
RESULTS
ClpP-GFP and ClpX-GFP foci form in the majority of cells.
To investigate the subcellular location of the ClpP peptidase, the ClpX ATPase, and all of the proteins examined in this work, we constructed a translational fusion of each gene to gfp (or other fluorescent moiety) under the control of its native promoter at its normal chromosomal locus (see Materials and Methods). Also, for all fluorescent protein fusions described in this study, the fusion represents the only source of each protein in the cell (see Materials and Methods).
The ClpX-GFP and ClpP-GFP fusion proteins that we constructed are functional because the resultant strains do not have any of the pleiotropic phenotypes associated with loss of clpX or clpP function. For example, loss-of-function mutations in clpX and clpP result in a defect in genetic competence. The transformation efficiencies of the clpX-gfp and clpP-gfp strains were similar to those of the wild-type strain, indicating that the fusions are functional (data not shown).
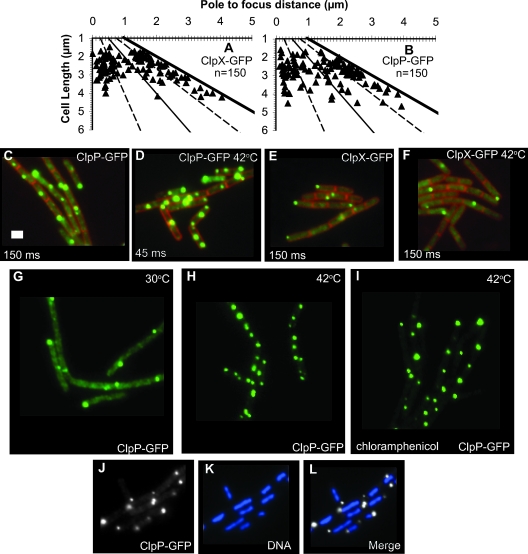
We found that ClpX-GFP and ClpP-GFP showed two types of localization patterns. ClpX-GFP and ClpP-GFP were diffuse and present throughout the cell (Fig. 1C to F). These proteins also formed discrete foci with fluorescence well above the diffuse background (Fig. 1C to F). We scored the positions of ClpP-GFP and ClpX-GFP foci and found that the majority of foci were located near cell poles (Fig. 1A to F; also see below and Fig. 6). We determined that the percentage of cells with one and two ClpP-GFP foci is higher than the percentage of cells with one and two ClpX-GFP foci (Table 2). During exponential growth, ∼67% (n = 372) of cells have a single ClpP-GFP focus and ∼10% (n = 372) of cells have two ClpP-GFP foci. In contrast, ∼52% (n = 730) of cells have a single ClpX-GFP focus, and we did not observe any cells with two ClpX-GFP foci (n = 730) (Table 2). Our finding that a higher percentage of cells have single and double ClpP-GFP foci than ClpX-GFP foci is statistically significant (P < 0.001) and is not surprising because ClpP pairs with other hexameric ATPases (12).
FIG. 1.
ClpX-GFP and ClpP-GFP form foci in nucleoid-excluded areas. (A) Positions of single ClpX-GFP foci scored from cell pole to focus and plotted relative to cell length. (B) Positions of single ClpP-GFP foci scored from cell pole to focus and plotted relative to cell length. For each plot, the left vertical axis denotes the near cell pole. The solid lines mark the midcell and the other cell pole, respectively. The dashed lines mark the cell quarter positions. The number of cells scored is indicated, and the focus scoring was performed as described previously (2, 68). (C) ClpP-GFP (green) with vital membrane stain FM4-64 (red); (D) ClpP-GFP with membrane stain following temperature shift for 20 min to 42°C. (E) ClpX-GFP with membrane; (F) ClpX-GFP with membrane following temperature shift for 20 min to 42°C. (G) ClpP-GFP at 30°C after 20 min; (H) ClpP-GFP at 42°C for 20 min; (I) ClpP-GFP pretreated with chloramphenicol prior to a shift to 42°C for 20 min. (J) ClpP-GFP foci (white) and (K) the corresponding nucleoid stained with DAPI (blue) are shown after treatment with chloramphenicol and heat shock at 42°C for 20 min; (L) merge of ClpP-GFP (white) and the nucleoid stained with DAPI (blue). Bar, 2 μm.
FIG. 6.
Distributions of ClpP-GFP, ClpX-GFP, ClpE-YFP, and ClpC-YFP foci relative to the nucleoid. We scored the positions of the ClpP-GFP peptidase and each ClpP-associated ATPase in single cells relative to the nucleoid during normal growth in S750 defined minimal medium. (A) Description of the position of the focus. (B) Schematic representation of the subcellular localization described. The boundary of the cell is represented by a solid black line, the nucleoid is dashed, and the focus is represented by the shaded oval. (C) Percentages of cells with foci localized to the indicated subcellular positions. The number of cells scored is indicated below each column. It should be noted that when foci were scored to be located at “midcell” the foci that qualified were located between nucleoids and did not appear to be located on the nucleoid edge. In the case of ClpE-YFP and ClpC-YFP, a considerable number of foci were located at “midcell” but were on the edge of the nucleoid and were thus scored as “coincident with the nucleoid edge.” The experiments used to generate the data presented were independent from those used to generate the data presented in Table 2.
TABLE 2.
Percentages of cells with Clp foci during normal growth and following heat shocka
| Fusion | Heat treatment | No. of cells | % of cells with:
|
|||
|---|---|---|---|---|---|---|
| 0 foci | 1 focus | 2 foci | >3 foci | |||
| ClpP-GFP | − | 372 | 20 | 67 | 10 | 2 |
| + | 376 | 2 | 52 | 38 | 7 | |
| ClpX-GFP | − | 730 | 48 | 52 | 0 | 0 |
| + | 549 | 72 | 27 | <1 | 0 | |
| ClpE-YFP | − | 900 | 88 | 11 | <1 | 0 |
| + | 494 | 81 | 19 | <1 | 0 | |
| ClpC-YFP | − | 717 | 82 | 17 | 1 | 0 |
| + | 482 | 86 | 13 | 1 | 0 | |
For each experiment, cultures were grown at 30°C in S750 minimal medium. Cells were stained with the vital membrane stain FM4-64 and DAPI, followed by assembly onto microscope slides containing agarose pads. For each strain, one slide was heat treated at 42°C for 20 min followed by microscopy (+, with heat treatment; −, without heat treatment). In cells without foci, ∼100% had diffusely distributed fluorescence for ClpP-GFP, ClpX-GFP, and ClpQ-YFP. For ClpE-YFP and ClpC-YFP, 100% of cells without foci showed nucleoid-associated fluorescence. ClpQ-YFP does form punctate foci in most cells (Fig. 4). These foci were too faint to score accurately to include in this table. ClpQ-YFP foci were also unchanged following heat shock at 42°C for 20 min. Statistical significance for the change in number of foci between normal growth and growth following heat shock was calculated using the chi-square test for homogeneity. A P value of <0.001 was obtained for ClpP-GFP, ClpX-GFP, and ClpE-YFP following heat shock.
Heat shock induces ClpP-GFP focus formation.
ClpP expression is induced following heat shock (12). We asked whether heat shock also affects the number or appearance of ClpP-GFP foci. Cells bearing the clpP-gfp allele were placed on slides with agarose pads at room temperature to allow the cells to settle. Slides were then incubated at either 42°C for 20 min to induce heat shock or 30°C for 20 min as a control for the effect of temperature. A temperature of 42°C instead of 50°C was used, because we did not want to risk desiccating our cells on the slides. We found that both the size and the number of ClpP-GFP foci increased after incubation at 42°C (Table 2 and Fig. 1C and D). It should be noted that the camera exposure time for image capture of cells with ClpP-GFP foci at 30°C was 150 ms and that the exposure time for our heat-treated cells was reduced to 43 ms to image the intense fluorescent foci present at 42°C. When we examined our 30°C control, we did not observe the striking increase in the number of foci that was observed following incubation at 42°C (Fig. 1C).
During normal growth, ClpP-GFP foci were not visible in ∼20% (n = 372) of cells. Following heat shock, only ∼2% (n = 376) of cells were devoid of ClpP-GFP foci (Table 2). In B. subtilis, the expression of ClpP, but not ClpX, has been shown to increase following heat treatment (12). Examination of ClpX-GFP foci following incubation at 42°C showed that the percentage of cells with foci decreased (Table 2). During normal growth, a single ClpX-GFP focus was observed in ∼52% (n = 730) of cells, and we observed nearly a twofold decrease (∼27%, n = 549) in single-focus cells following heat treatment (Table 2 and Fig. 1F). We determined that, for both ClpX-GFP and ClpP-GFP, the alterations in the percentage of cells with foci following heat shock that we observed were statistically significant (P < 0.001). It has been shown that ClpC expression is induced by heat shock (12). We suggest that the decrease in ClpX-GFP foci may coincide with an increase in assembly of the ClpCP protease in vivo (12). Taken together, we find that the number of ClpP-GFP foci increased following heat shock and the number of ClpX-GFP foci decreased. We conclude that ClpP-GFP and ClpX-GFP foci are dynamically regulated by temperature (Fig. 1C to F).
In B. subtilis, alterations in protein levels can be detected within 3 min of a temperature shift to 46°C or 50°C (1). To determine if the increase in the percentage of cells with ClpP-GFP foci was due to increased protein synthesis or a redistribution of existing ClpP-GFP protein, we inhibited protein synthesis with the addition of chloramphenicol prior to heat shock. Following transfer to 42°C, we still observed the striking increase in the percentage of cells with ClpP-GFP foci in cells arrested for protein synthesis (Fig. 1G to I). Scoring of these cells showed that only ∼4% of cells (n = 329) were devoid of at least a single ClpP-GFP focus. We conclude that heat shock induces a dynamic redistribution of ClpP-GFP into foci in vivo independently of protein synthesis. Because we were virtually unable to detect a fluorescence signal from the diffuse pool of ClpP-GFP following heat treatment (Fig. 1H and I), we speculate that the increase in ClpP-GFP foci is formed from the redistribution of diffusely localized ClpP-GFP into foci.
As mentioned above, it is established that ClpP-GFP expression does increase following heat shock (12), and we have shown that the increase in the percentage of cells with ClpP-GFP foci is independent of protein synthesis. We have found that when cells were heat treated for prolonged time periods (60 min) the size of the foci increased and the fluorescence signal from the diffuse fluorescence also increased in cells that were not arrested for protein synthesis (data not shown). Taken together, ClpP-GFP foci can redistribute following heat treatment in the absence of protein synthesis. If protein synthesis is not inhibited, then ClpP-GFP expression increases, and this is observed at the single-cell level as an increase in the fluorescence signal of the foci and diffusely localized protein.
ClpX and ClpP form discrete foci in nucleoid-free areas.
Under normal growth conditions, we observed ClpP-GFP foci coincident with the nucleoid in only ∼5% of cells (n = 267) (Fig. 6). It is difficult to discern if ClpP-GFP foci are actually coincident with the nucleoid or positioned closely adjacent to the nucleoid because the nucleoid is spread throughout most of the cell (61). In an effort to better determine nucleoid coincidence, we treated cells with chloramphenicol to condense the nucleoid away from the cell membrane to more easily image the position of ClpP-GFP foci relative to the DNA (61). After incubating cells with chloramphenicol for 1 h, we also heat treated these cells to increase the number of ClpP-GFP foci to determine the percentage of cells that showed ClpP-GFP foci coincident with the nucleoid. We found that only ∼3% of cells had ClpP-GFP foci coincident with the nucleoid (n = 237) (Fig. 1J and L). We found that ClpP-GFP foci form in mostly nucleoid-free areas usually near cell poles and that ClpP-GFP foci are rarely coincident with the nucleoid during normal growth conditions (Fig. 6) or following heat shock (Fig. 1J to L).
We also examined the frequency at which ClpX-GFP foci were coincident with the nucleoid. We found that virtually all of the ClpX-GFP foci were located near cell poles (Fig. 6). We observed ClpX-GFP foci coincident with the DNA in only ∼0.5% (n = 425) of cells. We conclude that the vast majority of ClpP-GFP and ClpX-GFP foci are not coincident with the DNA.
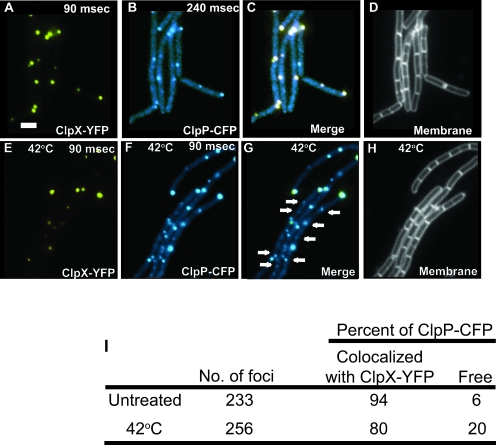
ClpP-CFP colocalizes with ClpX-YFP.
ClpX and ClpP have been shown to form a protease machine in vitro with the interacting interface well characterized for the E. coli proteins (for a review, see reference 56). In both B. subtilis and Caulobacter, ClpX and ClpP coimmunoprecipitate from whole-cell extracts, demonstrating an interaction between these proteins in vivo (12, 32). These biochemical observations motivated us to determine if ClpX-YFP and ClpP-CFP foci colocalize in single cells. To explore this, we constructed a strain expressing ClpX-YFP and ClpP-CFP fusions for simultaneous visualization. We found that strains bearing both the clpP-cfp and clpX-yfp alleles show some of the phenotypes associated with a clpX or clpP null strain, indicating that the combination caused a minor synthetic defect (data not shown). Overall, however, strains bearing both of these fusion alleles behaved more like the wild type than like either of the clpX or clpP disrupted strains. When we examined the numbers of ClpP-CFP and ClpX-YFP foci, we did observe a higher percentage of cells with ClpX-YFP foci (∼77%, n = 259) than with ClpP-CFP foci (∼65%, n = 232), which is not what we observed with the single ClpX-GFP fusion strain (Table 2). We hypothesize that the increase in the percentage of cells with ClpX-YFP foci is a consequence of having both fusion proteins present in the same cell and that this may contribute to the phenotype of the resultant strain. With this limitation, we examined the frequency of colocalization between the ClpP-CFP and ClpX-YFP foci.
We found that ∼94% (n = 233) of ClpP-CFP foci captured were coincident with ClpX-YFP foci, suggesting that the foci may represent a functional proteasome in vivo (Fig. 2A to D). Because the percentage of cells with ClpP-GFP foci increases following heat shock, we examined ClpP-CFP and ClpX-YFP colocalization following incubation at 42°C for 20 min. We found that ∼20% of ClpP-CFP foci (n = 256) were unpaired with ClpX-YFP following incubation at 42°C, compared with only ∼6% of ClpP-CFP foci (n = 233) during normal growth (Fig. 2I). This result was of interest. As mentioned above, ClpP peptidase has the capability to pair with several hexameric ATPases. We speculate that the increase in number of ClpP-CFP foci following heat shock coincides with an increase in ClpP peptidase pairing with ClpE and ClpC hexameric ATPases. Because both ClpE and ClpC provide significant roles in protein degradation during heat shock (38, 41), we hypothesize that, during heat shock, new ClpP foci will pair with existing ClpE or ClpC to degrade unfolded proteins caused by increased temperature. We conclude that ClpP-CFP and ClpX-YFP foci colocalize in vivo, and we suggest that the foci may represent sites of ClpXP-dependent protein degradation.
FIG. 2.
ClpX-YFP and ClpP-CFP foci colocalize during normal growth and following heat shock. (A) ClpX-YFP; (B) ClpP-CFP; (C) merge of ClpX-YFP and ClpP-CFP; (D) membrane stained with FM4-64. (E) ClpX-YFP at 42°C; (F) ClpP-CFP at 42°C; (G) merge of ClpX-YFP and ClpP-CFP; (H) corresponding membrane stain. (I) Quantitation of the percentage of ClpP-CFP foci paired with ClpX-YFP or free (unpaired) with ClpX. Arrows in panel G highlight free ClpP-CFP foci. The percentage of cells with ClpP-CFP was ∼98% (n = 414) with foci following heat treatment. For the ClpX-YFP images shown in panels A and E, the 90-ms exposure does not capture the diffuse fluorescence that ClpX-YFP forms. Bar, 2 μm.
ClpE-YFP and ClpC-YFP form foci on the nucleoid edge near the cell pole.
After examining the localization of ClpX and ClpP foci, we asked whether the hexameric ClpE and ClpC ATPases displayed similar or different subcellular localization properties. ClpE is important for CtsR-dependent derepression of heat shock genes (33), while ClpC contributes to the control of competence development and the heat shock response (38).
We found that ClpE-YFP fluorescence and ClpC-YFP fluorescence (not foci; for foci, see below) were nucleoid associated in ∼100% (n = 200) and ∼99% (n = 272) of cells, respectively (Fig. 3C to F). We define nucleoid association as protein fluorescence that is coincident with the DAPI-stained DNA (Fig. 3C to F). This pattern is in striking contrast to that of ClpX-GFP, which is not nucleoid associated (Fig. 6). For ClpE-YFP and ClpC-YFP, we can show nucleoid-associated fluorescence by condensing the nucleoid through inhibition of protein translation following addition of kanamycin to the growth medium (data not shown) (69). For ClpC-YFP, all of the fluorescence condenses with the nucleoid, demonstrating nucleoid association. For ClpE-YFP, some fluorescence observed is not DNA associated, indicating that ClpE-YFP is both DNA associated and free in the cytoplasm (Fig. 3A).
FIG. 3.
ClpE-YFP and ClpC-YFP form foci on nucleoid edges near cell poles. (A) Positions of single ClpE-YFP foci scored from cell pole to focus and plotted relative to cell length. (B) Positions of single ClpC-YFP foci scored from cell pole to focus and plotted relative to cell length. For each plot, the left vertical axis denotes the near cell pole. The solid lines mark the midcell and other cell pole, respectively. The dashed lines mark the cell quarter positions. The number of cells scored is indicated in the graph. (C) ClpE-YFP; (D) corresponding DAPI. (E) ClpC-YFP; (F) corresponding DAPI. (G) ClpE-YFP (green) and membrane (red). (H) ClpC-YFP (green) and membrane (red). The exposure time for ClpE-YFP and ClpC-YFP was 2 s. The cell membrane was stained with FM4-64. Bar, 2 μm.
In addition to nucleoid-associated fluorescence, ClpE-YFP and ClpC-YFP form foci in a subpopulation of cells (Fig. 3 and Table 2). During normal growth in defined S750 minimal medium, ClpE-YFP and ClpC-YFP formed foci in ∼11% and ∼17% of cells, respectively (n = 900 for ClpE-YFP and n = 717 for ClpC-YFP). ClpC-YFP and ClpE-YFP foci are coincident with the nucleoid edge closest to the cell pole in the majority of cells that have foci (Fig. 3 and 6). Following heat shock, the percentage of cells with ClpE-YFP foci increases (P < 0.001) and the percentage of cells with ClpC-YFP foci is almost unchanged (Table 2). Qualitatively, however, ClpC-YFP foci are more intense following heat shock (data not shown). We conclude that most of the ClpE fluorescence and all of the ClpC fluorescence that we detect are coincident with the nucleoid and that ClpE-YFP and ClpC-YFP foci are coincident with the nucleoid edge near the cell pole.
ClpQ-YFP (HslV) localizes as foci and diffuse fluorescence throughout the cell.
We were interested in understanding whether the ClpQY (HslVU) protease organized into focal assemblies as we have observed with the other Clp proteases. The function of the ClpQY protease in B. subtilis is poorly understood, although the ClpQY protease has been shown to degrade misfolded proteins in E. coli (35) and the structure of these proteins is known for E. coli and Haemophilus influenzae (3, 4, 64, 67).
Because we lacked an in vivo assay examining the functionality of these fusions, we used the available structural information to guide our analysis and predict if the fusion proteins were likely to be functional. For two reasons, we hypothesized that a C-terminal YFP fusion to ClpY was likely to disrupt interaction with ClpQ (HslV) and inactivate the protease in vivo. First, C-terminal mutations in E. coli ClpY inactivate peptide hydrolysis by the ClpQY protease machine in vitro (63). Second, the H. influenzae ClpQY protease structure shows that the C-terminal ClpY helices bind to sites in between the ClpQ monomer interfaces (47, 63), suggesting that a C-terminal YFP fusion to ClpY would inhibit docking of ClpY to ClpQ. We also considered investigating an N-terminal YFP-ClpY fusion but did not because the N terminus of Clp ATPases has a conserved role in substrate recognition. Because of these limitations, we did not pursue the subcellular localization of B. subtilis ClpY.
In contrast, the structures of both E. coli and H. influenzae show that the C terminus of ClpQ (HslV) is exposed at the surface of each monomer in the assembled ClpQY protease machine (3, 4, 64, 67). We modeled the B. subtilis ClpQ (HslV) monomer to determine if the C terminus of the B. subtilis protein was predicted to be surface exposed on the outside of the proteolytic chamber. Indeed, the C-terminal β-strand is modeled on the exterior of the ClpQ monomer (Fig. 4A). Based on the B. subtilis ClpQ homology model (Fig. 4A) and the crystal structure of the intact ClpQY protease machine (3, 4, 64), we predicted that the ClpQ-YFP fusion was unlikely to disrupt activity of the ClpQY protease in vivo (Fig. 4A) (63). With this information, we moved forward with characterization of the ClpQ-YFP fusion protein.
FIG. 4.
ClpQ-YFP is distributed diffusely and forms small foci. (A) Homology model of the B. subtilis ClpQ (HslV) monomer. The residues colored in red represent the C-terminal 8 amino acids of ClpQ (DQIILEEL). Shown is a side view. The “surface” represents the outside and the solvent-exposed exterior of the proteolytic chamber, and the “chamber” represents the site of peptide proteolysis when assembled into a dodecamer. (B) ClpQ-YFP and membrane in a clpY mutant background; (C) ClpQ-YFP in a clpY+ background; (D) merge of ClpQ-YFP and membrane (red). (E) Corresponding DNA (blue) and membrane (red) for ClpQ-YFP; (F) merge of ClpQ-YFP with DNA and membrane. Arrows highlight the positions of several ClpQ-YFP foci. The membrane was stained with FM4-64. Bar, 2 μm.
We examined the subcellular localization of ClpQ-YFP in a strain background that was deficient for clpY because integration of the clpQ-yfp fusion would disrupt expression of clpY directly downstream. We found that ClpQ-YFP was localized diffusely throughout most cells and formed very large single foci in ∼1% of cells (n = 521) (Fig. 4B). Strikingly, when we examined ClpQ-YFP localization following expression of clpY by using an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, we found that ClpQ-YFP formed very small foci in ∼100% of cells (n = 325). Because the small ClpQ-YFP foci are difficult to image, we did not score the number or position of these focal assemblies. It should be noted that ClpQ-YFP foci appear to be localized mostly to nucleoid-free areas near the cell membrane (Fig. 4C and D). The localization of ClpQ-YFP protein was unchanged following heat shock at 42°C for 20 min (data not shown). We conclude that ClpQ-YFP forms foci and that focus formation is dependent on clpY+, suggesting that these focal assemblies require formation of a protease machine in vivo.
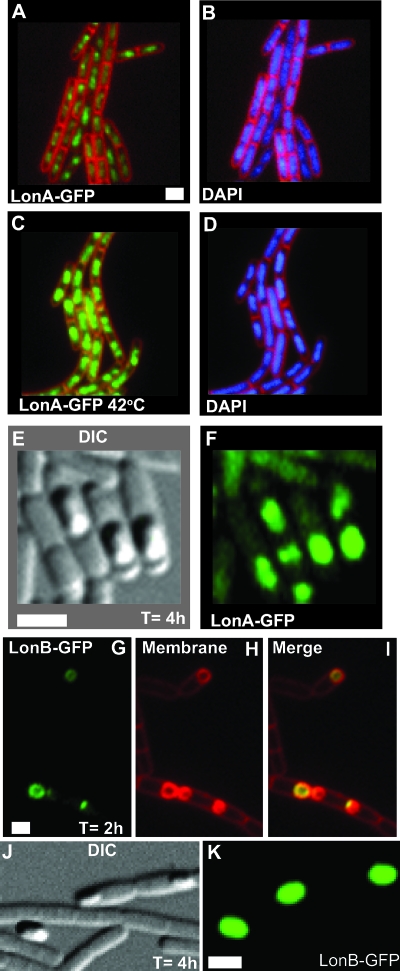
LonA-GFP localizes to the nucleoid and the forespore.
LonA and LonB of B. subtilis are homologs of the ATP-dependent E. coli Lon protease (49). E. coli Lon has been shown to regulate several pathways, including DNA damage response, heat shock, and colanic acid (capsular polysaccharide) production (16). We found that LonA-GFP is nucleoid associated and that LonA-GFP nucleoid fluorescence is enhanced by heat shock (Fig. 5A and C). We find this localization pattern particularly interesting because E. coli Lon has been shown to have DNA binding activity and because both single- and double-stranded DNA stimulate ATP-dependent proteolysis (5). Also, in E. coli, Lon has been shown to degrade many proteins that are involved in DNA metabolism. Thus, the nucleoid association of B. subtilis LonA may contribute to the function or selection of substrates for this protease in vivo.
FIG. 5.
LonA-GFP localizes to the nucleoid and the forespore. (A) LonA-GFP and membrane; (B) corresponding DNA (DAPI) and membrane. (C) LonA-GFP at 42°C and membrane; (D) corresponding DNA (DAPI) and membrane. (E) LonA-GFP differential interference contrast (DIC); (F) corresponding LonA-GFP. (G) LonB-GFP; (H) corresponding membrane; (I) merge of LonB-GFP with membrane. (J) DIC corresponding to LonB-GFP; (K) LonB-GFP. The time following induction of sporulation is indicated in the appropriate panels. The membrane was stained with FM4-64. Bar, 2 μm.
During sporulation, one copy of the chromosome is translocated into the developing forespore, while the other copy remains in the mother cell (for a review, see reference 52). Because we observed LonA-GFP to be coincident with the nucleoid, we asked if this protein also localized to the forespore DNA during development. Strikingly, we did not observe considerable enrichment of the forespore DNA with LonA-GFP (data not shown) early during sporulation. As the spore developed, LonA-GFP (t > 2 h) fluorescence became more intense and we observed considerable enrichment of LonA-GFP fluorescence in the forespore later in development (Fig. 5E and F). We conclude that LonA-GFP is produced in the forespore.
LonB-GFP localizes to the forespore membrane early in development.
LonB-GFP subcellular localization is temporally distinct from LonA-GFP. The lonB gene is transcribed from a σF promoter, and LonB-GFP localizes exclusively to the forespore during sporulation (58). We asked whether LonB-GFP has a distinct localization pattern during log-phase growth. LonB-GFP had very little if any fluorescence above that of a wild-type strain lacking a GFP fusion, and the fluorescence was distributed diffusely throughout the cell (data not shown). Heat shock of LonB-GFP did not alter this localization pattern (data not shown). We induced sporulation and confirmed previously published data that LonB-GFP fluorescence is forespore associated (Fig. 5J and K) (58). Strikingly, sporulation time course experiments revealed that LonB-GFP localized to the forespore membrane early in sporulation (t < 2 h) (Fig. 5G to I). As sporulation progressed, LonB-GFP was no longer localized exclusively to the forespore membrane but instead was present throughout the forespore (Fig. 5J and K) (58). We conclude that the subcellular localization of LonA and LonB is temporally regulated in B. subtilis during sporulation, with LonB-GFP localizing to the forespore membrane early during development. Later in development, both LonB-GFP and LonA-GFP are present throughout the forespore later during development (Fig. 5).
DISCUSSION
In the present study, we have examined the subcellular localizations of the Clp peptidases, Clp hexameric ATPases (except ClpY), and Lon proteases of B. subtilis. Our study examined each of these proteases or ATPases fused to a fluorescent moiety and expressed from their native loci in living cells. We also examined the subcellular localization of each of these proteins during normal growth and following heat shock. Our results show that several of the Clp and Lon ATP-dependent proteases show distinct spatial and/or temporal subcellular localization patterns in B. subtilis.
Examination of the ClpP peptidase showed that this protein forms single foci in most cells (∼67%) and multiple foci in a subset of cells (∼12%) during normal growth (Table 2). ClpX forms foci in nucleoid-free areas and is rarely coincident with the nucleoid. In contrast, ClpE-YFP and ClpC-YFP show fluorescence that is coincident with the nucleoid and foci that form on nucleoid edges near cell poles (Fig. 6). Although the Clp ATPases form foci in locations distinct from each other, the ClpP peptidase localizes to all of these subcellular locations but is more prevalent in areas occupied by ClpX (Fig. 6). Consistent with this observation, the majority of ClpP-CFP foci are colocalized with ClpX-YFP during normal growth (Fig. 2). Upon heat shock, the number of ClpP-CFP foci increases, and we propose a model where a limited number of ClpP peptidases are complexed with ClpE and ClpC during normal growth; however, following heat shock, the new ClpP foci that are formed pair with ClpE and ClpC. We also showed that ClpP-GFP foci are dynamically regulated by temperature and that the percentage of cells with foci increases following heat treatment even in the absence of protein synthesis.
We investigated the subcellular localization of ClpQ (HslV) and found that ClpQ-YFP forms small punctate foci in areas near the cell membrane. We also determined that ClpQ-YFP focus formation was dependent on clpY+. We pursued similar experiments with ClpX and ClpP and found that ClpX-YFP focus formation was not dependent on clpP and that ClpP-CFP focus formation was not dependent on clpX (data not shown). Furthermore, in an accompanying work, Kain et al. (21) show that the abilities of ClpX-GFP, ClpP-GFP, and ClpC-GFP to form foci are independent from one another. Taken together, focus formations by ClpP, ClpC, and ClpX are not interdependent, but focus formation by ClpQ-YFP does depend on clpY.
In B. subtilis, the subcellular localization of Lon proteases shows spatial and temporal separation. LonA-GFP is localized to the nucleoid, possibly for degradation of proteins involved in DNA metabolism, while LonB-GFP is localized to the forespore (58) (Fig. 5) and is not observed in the mother cell. We investigated LonB-GFP localization further and found that this protein localizes to the forespore membrane early during development (Fig. 5G to I) but that later in development LonB-GFP is localized to the entire forespore (Fig. 5J and K), confirming previous observations (58). These data suggest a developmentally regulated redistribution in the localization of LonB-GFP. As mentioned above, lonB-deficient strains lack a sporulation phenotype; however, we find the colocalization of LonB-GFP with the forespore membrane interesting. Several proteins that localize preferentially to the forespore membrane early during development have been identified (for a review, see reference 48). It is tempting to consider a model where LonB contributes to the degradation of forespore-specific membrane proteins.
In Caulobacter crescentus, ClpXP has been shown to form polar foci (32). Polar ClpXP degrades the essential cell cycle regulator CtrA, only at the cell pole (32). ClpXP localization is dynamically regulated in Caulobacter. The unphosphorylated form of CpdR is required for polar localization of ClpXP (20). When Caulobacter cells are deficient for cpdR, ClpXP polar localization is disturbed and two ClpXP substrates, CtrA and McpA, are not degraded (20). These data indicate that in Caulobacter proper subcellular positioning of ClpXP is important for the role of ClpXP in cell cycle regulation. CpdR does not have homologs in B. subtilis or E. coli, and thus the mechanism of polar localization in these organisms remains unknown.
These data raise questions about how ClpX and ClpP are targeted near cell poles in B. subtilis. In an accompanying paper, Kain et al. (21) show that the polar localization of ClpX and ClpP is not dependent on the presence of the polar protein DivIVA (31, 45, 46, 65). The anionic phospholipid cardiolipin also accumulates at cell poles (8, 34), but ClpP and ClpX still form foci in cells deficient for cardiolipin production, demonstrating that this phospholipid is not the sole targeting signal. Furthermore, their work shows that B. subtilis ClpP and ClpX still localize near cell poles when expressed in E. coli. Their results demonstrate that these proteins intrinsically recognize cell poles. Taken together, our data and the Caulobacter results show that ClpXP polar localization is conserved between B. subtilis, Caulobacter, and possibly other bacteria. A future challenge will be to determine the protein(s) or signals that direct localization of the Clp peptidases and ATPases to localize near cell poles in B. subtilis.
Acknowledgments
We thank Bryan Davies, Adam Breier, Brad Weart, Hajime Kobayashi, C. Lee, and Mary Ellen Wiltrout for comments on the manuscript. We also thank Robert Sauer, Peter Chien, and Saskia Neher for helpful discussions. We are indebted to Rich Losick and James Kain (Harvard University) for sharing information, strains, and helpful discussions prior to publication.
L.A.S. was supported by a postdoctoral fellowship from the National Cancer Institute (CA113124). G.C.W. was supported by an American Cancer Society research professorship, the Massachusetts Institute of Technology Center for Environmental Health Sciences (P30 ES002109), and a National Cancer Institute grant (CA21615-27). This work was also supported by NIH grant GM50895 to A.D.G.
Footnotes
Published ahead of print on 8 August 2008.
REFERENCES
- 1.Arnosti, D. N., V. L. Singer, and M. J. Chamberlin. 1986. Characterization of heat shock in Bacillus subtilis. J. Bacteriol. 1681243-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkmen, M. B., and A. D. Grossman. 2006. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol. Microbiol. 6257-71. [DOI] [PubMed] [Google Scholar]
- 3.Bochtler, M., L. Ditzel, M. Groll, and R. Huber. 1997. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl. Acad. Sci. USA 946070-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochtler, M., C. Hartmann, H. K. Song, G. P. Bourenkov, H. D. Bartunik, and R. Huber. 2000. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403800-805. [DOI] [PubMed] [Google Scholar]
- 5.Bolhuis, A., A. Matzen, H. L. Hyyrylainen, V. P. Kontinen, R. Meima, J. Chapuis, G. Venema, S. Bron, R. Freudl, and J. M. van Dijl. 1999. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J. Biol. Chem. 27424585-24592. [DOI] [PubMed] [Google Scholar]
- 6.Bolon, D. N., D. A. Wah, G. L. Hersch, T. A. Baker, and R. T. Sauer. 2004. Bivalent tethering of SspB to ClpXP is required for efficient substrate delivery: a protein-design study. Mol. Cell 13443-449. [DOI] [PubMed] [Google Scholar]
- 7.Deuerling, E., A. Mogk, C. Richter, M. Purucker, and W. Schumann. 1997. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol. Microbiol. 23921-933. [DOI] [PubMed] [Google Scholar]
- 8.Dowhan, W., E. Mileykovskaya, and M. Bogdanov. 2004. Diversity and versatility of lipid-protein interactions revealed by molecular genetic approaches. Biochim. Biophys. Acta 166619-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn, J. M., I. Levchenko, R. T. Sauer, and T. A. Baker. 2004. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 182292-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn, J. M., S. B. Neher, Y. I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11671-683. [DOI] [PubMed] [Google Scholar]
- 11.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, gram-positive bacteria. Mol. Microbiol. 631285-1295. [DOI] [PubMed] [Google Scholar]
- 12.Gerth, U., J. Kirstein, J. Mostertz, T. Waldminghaus, M. Miethke, H. Kock, and M. Hecker. 2004. Fine-tuning in regulation of Clp protein content in Bacillus subtilis. J. Bacteriol. 186179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28787-802. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, M., E. G. Frank, A. S. Levine, and R. Woodgate. 1998. Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: in vitro degradation and identification of residues required for proteolysis. Genes Dev. 123889-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon, C. 2002. The intracellular localization of the proteasome. Curr. Top. Microbiol. Immunol. 268175-184. [DOI] [PubMed] [Google Scholar]
- 16.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30465-506. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman, S., and D. Zipser. 1978. Deg phenotype of Escherichia coli lon mutants. J. Bacteriol. 133844-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossman, A. D., R. R. Burgess, W. Walter, and C. A. Gross. 1983. Mutations in the lon gene of E. coli K12 phenotypically suppress a mutation in the sigma subunit of RNA polymerase. Cell 32151-159. [DOI] [PubMed] [Google Scholar]
- 19.Hamelin, C., and Y. S. Chung. 1975. Characterization of mucoid mutants of Escherichia coli K-12 isolated after exposure to ozone. J. Bacteriol. 12219-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iniesta, A. A., P. T. McGrath, A. Reisenauer, H. H. McAdams, and L. Shapiro. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. USA 10310935-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kain, J., G. He, and R. Losick. 2008. Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J. Bacteriol. 1906749-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kock, H., U. Gerth, and M. Hecker. 2004. The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis. J. Bacteriol. 1865856-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krüger, E., E. Witt, S. Ohlmeier, R. Hanschke, and M. Hecker. 2000. The Clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 1823259-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 2821516-1519. [DOI] [PubMed] [Google Scholar]
- 26.Lemon, K. P., and A. D. Grossman. 2000. Movement of replicating DNA through a stationary replisome. Mol. Cell 61321-1330. [DOI] [PubMed] [Google Scholar]
- 27.Levchenko, I., M. Seidel, R. T. Sauer, and T. A. Baker. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 2892354-2356. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol. Microbiol. 33415-428. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J., and P. Zuber. 2000. The ClpX protein of Bacillus subtilis indirectly influences RNA polymerase holoenzyme composition and directly stimulates sigma-dependent transcription. Mol. Microbiol. 37885-897. [DOI] [PubMed] [Google Scholar]
- 30.Long, W. S., C. L. Slayman, and K. B. Low. 1978. Production of giant cells of Escherichia coli. J. Bacteriol. 133995-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 123419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGrath, P. T., A. A. Iniesta, K. R. Ryan, L. Shapiro, and H. H. McAdams. 2006. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124535-547. [DOI] [PubMed] [Google Scholar]
- 33.Miethke, M., M. Hecker, and U. Gerth. 2006. Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J. Bacteriol. 1884610-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mileykovskaya, E., and W. Dowhan. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 1821172-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Missiakas, D., F. Schwager, J. M. Betton, C. Georgopoulos, and S. Raina. 1996. Identification and characterization of HsIV HsIU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 156899-6909. [PMC free article] [PubMed] [Google Scholar]
- 36.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27899-914. [DOI] [PubMed] [Google Scholar]
- 37.Msadek, T., F. Kunst, D. Henner, A. Klier, G. Rapoport, and R. Dedonder. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172824-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Msadek, T., F. Kunst, and G. Rapoport. 1994. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc. Natl. Acad. Sci. USA 915788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano, S., G. Zheng, M. M. Nakano, and P. Zuber. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 1843664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nanamiya, H., E. Shiomi, M. Ogura, T. Tanaka, K. Asai, and F. Kawamura. 2003. Involvement of ClpX protein in the post-transcriptional regulation of a competence specific transcription factor, ComK protein, of Bacillus subtilis. J. Biochem. (Tokyo) 133295-302. [DOI] [PubMed] [Google Scholar]
- 41.Ogura, M., L. Liu, M. Lacelle, M. M. Nakano, and P. Zuber. 1999. Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis. Mol. Microbiol. 32799-812. [DOI] [PubMed] [Google Scholar]
- 42.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure-diverse function. Genes Cells 6575-597. [DOI] [PubMed] [Google Scholar]
- 43.Park, E. Y., B. G. Lee, S. B. Hong, H. W. Kim, H. Jeon, and H. K. Song. 2007. Structural basis of SspB-tail recognition by the zinc binding domain of ClpX. J. Mol. Biol. 367514-526. [DOI] [PubMed] [Google Scholar]
- 44.Park, S. C., B. Jia, J. K. Yang, D. L. Van, Y. G. Shao, S. W. Han, Y. J. Jeon, C. H. Chung, and G. W. Cheong. 2006. Oligomeric structure of the ATP-dependent protease La (Lon) of Escherichia coli. Mol. Cells 21129-134. [PubMed] [Google Scholar]
- 45.Perry, S. E., and D. H. Edwards. 2004. Identification of a polar targeting determinant for Bacillus subtilis DivIVA. Mol. Microbiol. 541237-1249. [DOI] [PubMed] [Google Scholar]
- 46.Perry, S. E., and D. H. Edwards. 2006. The Bacillus subtilis DivIVA protein has a sporulation-specific proximity to Spo0J. J. Bacteriol. 1886039-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramachandran, R., C. Hartmann, H. K. Song, R. Huber, and M. Bochtler. 2002. Functional interactions of HslV (ClpQ) with the ATPase HslU (ClpY). Proc. Natl. Acad. Sci. USA 997396-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramamurthi, K. S., and R. M. Losick. 2005. Protein localization: reach out and touch the forespore. Curr. Biol. 15R165-R167. [DOI] [PubMed] [Google Scholar]
- 49.Riethdorf, S., U. Volker, U. Gerth, A. Winkler, S. Engelmann, and M. Hecker. 1994. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J. Bacteriol. 1766518-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivett, A. J. 1998. Intracellular distribution of proteasomes. Curr. Opin. Immunol. 10110-114. [DOI] [PubMed] [Google Scholar]
- 51.Rivett, A. J., G. G. Mason, R. Z. Murray, and J. Reidlinger. 1997. Regulation of proteasome structure and function. Mol. Biol. Rep. 2499-102. [DOI] [PubMed] [Google Scholar]
- 52.Rudner, D. Z., and R. Losick. 2001. Morphological coupling in development: lessons from prokaryotes. Dev. Cell 1733-742. [DOI] [PubMed] [Google Scholar]
- 53.Russell, S. J., K. A. Steger, and S. A. Johnston. 1999. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 27421943-21952. [DOI] [PubMed] [Google Scholar]
- 54.Ruvolo, M. V., K. E. Mach, and W. F. Burkholder. 2006. Proteolysis of the replication checkpoint protein Sda is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis. Mol. Microbiol. 601490-1508. [DOI] [PubMed] [Google Scholar]
- 55.Sasakawa, C., Y. Uno, and M. Yoshikawa. 1987. lon-sulA regulatory function affects the efficiency of transposition of Tn5 from lambda b221 cI857 Pam Oam to the chromosome. Biochem. Biophys. Res. Commun. 142879-884. [DOI] [PubMed] [Google Scholar]
- 56.Sauer, R. T., D. N. Bolon, B. M. Burton, R. E. Burton, J. M. Flynn, R. A. Grant, G. L. Hersch, S. A. Joshi, J. A. Kenniston, I. Levchenko, S. B. Neher, E. S. Oakes, S. M. Siddiqui, D. A. Wah, and T. A. Baker. 2004. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 1199-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt, R., A. L. Decatur, P. N. Rather, C. P. Moran, Jr., and R. Losick. 1994. Bacillus subtilis lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor σG. J. Bacteriol. 1766528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serrano, M., S. Hövel, C. P. Moran, Jr., A. O. Henriques, and U. Völker. 2001. Forespore-specific transcription of the lonB gene during sporulation in Bacillus subtilis. J. Bacteriol. 1832995-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simmons, L. A., B. W. Davies, A. D. Grossman, and G. C. Walker. 2008. Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol. Cell 29291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmons, L. A., A. D. Grossman, and G. C. Walker. 2007. Replication is required for the RecA localization response to DNA damage in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1041360-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, B. T., A. D. Grossman, and G. C. Walker. 2002. Localization of UvrA and effect of DNA damage on the chromosome of Bacillus subtilis. J. Bacteriol. 184488-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, B. T., A. D. Grossman, and G. C. Walker. 2001. Visualization of mismatch repair in bacterial cells. Mol. Cell 81197-1206. [DOI] [PubMed] [Google Scholar]
- 63.Song, H. K., C. Hartmann, R. Ramachandran, M. Bochtler, R. Behrendt, L. Moroder, and R. Huber. 2000. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. USA 9714103-14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sousa, M. C., C. B. Trame, H. Tsuruta, S. M. Wilbanks, V. S. Reddy, and D. B. McKay. 2000. Crystal and solution structures of an HslUV protease-chaperone complex. Cell 103633-643. [DOI] [PubMed] [Google Scholar]
- 65.Thomaides, H. B., M. Freeman, M. El Karoui, and J. Errington. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 151662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waksman, G., G. Thomas, and A. Favre. 1984. The lon gene and photoprotection in Escherichia coli K-12. Photochem. Photobiol. 39337-342. [DOI] [PubMed] [Google Scholar]
- 67.Wang, J. 2001. A corrected quaternary arrangement of the peptidase HslV and ATPase HslU in a cocrystal structure. J. Struct. Biol. 13415-24. [DOI] [PubMed] [Google Scholar]
- 68.Wang, J. D., M. E. Rokop, M. M. Barker, N. R. Hanson, and A. D. Grossman. 2004. Multicopy plasmids affect replisome positioning in Bacillus subtilis. J. Bacteriol. 1867084-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber, M. H., A. V. Volkov, I. Fricke, M. A. Marahiel, and P. L. Graumann. 2001. Localization of cold shock proteins to cytosolic spaces surrounding nucleoids in Bacillus subtilis depends on active transcription. J. Bacteriol. 1836435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wehrl, W., M. Niederweis, and W. Schumann. 2000. The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J. Bacteriol. 1823870-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiegert, T., and W. Schumann. 2001. SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol. 1833885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams, M. D., T. X. Ouyang, and M. C. Flickinger. 1994. Starvation-induced expression of SspA and SspB: the effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol. Microbiol. 111029-1043. [DOI] [PubMed] [Google Scholar]
- 73.Wojcik, C., and G. N. DeMartino. 2003. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35579-589. [DOI] [PubMed] [Google Scholar]
- 74.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 121-9. [DOI] [PubMed] [Google Scholar]
- 75.Zhestianikov, V. D. 1971. Post-radiation cell repair. II. Survival of Escherichia coli B following exposure to UV, - and -rays in relation to the phenotypic expression of the lon- mutation. Tsitologiia 131148-1156. (In Russian.) [PubMed] [Google Scholar]
- 76.Zolkiewski, M. 2006. A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol. Microbiol. 611094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]