Abstract
The DevS histidine kinase of Mycobacterium smegmatis contains tandem GAF domains (GAF-A and GAF-B) in its N-terminal sensory domain. The heme iron of DevS is in the ferrous state when purified and is resistant to autooxidation from a ferrous to a ferric state in the presence of O2. The redox property of the heme and the results of sequence comparison analysis indicate that DevS of M. smegmatis is more closely related to DosT of Mycobacterium tuberculosis than DevS of M. tuberculosis. The binding of O2 to the deoxyferrous heme led to a decrease in the autokinase activity of DevS, whereas NO binding did not. The regulation of DevS autokinase activity in response to O2 and NO was not observed in the DevS derivatives lacking its heme, indicating that the ligand-binding state of the heme plays an important role in the regulation of DevS kinase activity. The redox state of the quinone/quinol pool of the respiratory electron transport chain appears not to be implicated in the regulation of DevS activity. Neither cyclic GMP (cGMP) nor cAMP affected DevS autokinase activity, excluding the possibility that the cyclic nucleotides serve as the effector molecules to modulate DevS kinase activity. The three-dimensional structure of the putative GAF-B domain revealed that it has a GAF folding structure without cyclic nucleotide binding capacity.
Mycobacterium smegmatis is a nonpathogenic and fast-growing mycobacterium whose adaptive response to a gradual decrease in oxygen tension and exposure to NO is similar to that of Mycobacterium tuberculosis (7). This adaptive capability of mycobacteria has been suggested to allow them to persist in a latent state in the immune-competent host, especially in the case of M. tuberculosis (14, 24, 35, 52). The hypoxic conditions within granulomas, although this is controversial (1), and the NO synthesized by activated macrophages have been proposed to serve as possible signals for the transition of mycobacteria to the nonreplicating, latent state (35, 50, 52).
The DevSR (DosSR) two-component system plays a crucial role in the adaptation of mycobacteria to hypoxic and NO conditions. Approximately 48 genes of M. tuberculosis were reported to be induced under hypoxic conditions, as well as on exposure to NO. The upregulation of these genes is mediated by the DevSR system (35, 38, 39, 52). The DevSR two-component system consists of the DevS histidine kinase (HK) and its cognate response regulator (39, 40). In addition to DevS, the DosT HK was found to cross talk with DevR and to be functional in M. tuberculosis (39, 40). DevS and DosT show high sequence similarity to each other over the length of their primary structures. The N-terminal sensory domains of DevS and DosT contain two putative GAF domains. The first GAF domain (GAF-A) serves as a heme-binding domain, while the function of the second one (GAF-B) remains to be revealed (18, 22, 43, 46). It was recently demonstrated that either the binding of O2 to the ferrous form of hemes of both DevS and DosT or the oxidation of Fe2+ within the heme to Fe3+ led to a decrease in their autophosphorylation rate. DevS (DosT) appears to have an ability to discriminate between O2 and NO, as judged by the fact that the binding of NO to the hemes does not result in the reduction of their autophosphorylation rate, in contrast to the effect of O2 (22, 46). On exposure to O2, the deoxyferrous form of DevS is converted to the met form (Fe3+), while the heme iron of DosT is resistant to autooxidation (22). M. smegmatis possesses a single DevS HK that phosphorylates DevR. The DevSR two-component system has also been reported to be involved in the hypoxic adaptation of this bacterium (29).
For this study, we purified the full-length DevS and the truncated DevS lacking its N-terminal sensory domain (C-DevS), along with the H150A mutant form of DevS, and demonstrated that the heme accommodated by the GAF-A domain of DevS is directly responsible for its O2- and NO-sensory functions. The results of this study also show that neither the redox state of the quinone pool of the electron transport chain nor cyclic nucleotides affected the autophosphorylation rate of DevS. Finally, the three-dimensional structure of the GAF-B domain of DevS is presented in this paper.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. M. smegmatis strains were grown aerobically on Middlebrook 7H9 medium (Difco, Sparks, MD) supplemented with 0.2% (wt/vol) glucose at 37°C. When required, hygromycin (50 μg/ml) was added to the growth medium. To prevent clumping of bacterial cells, Tween 80 was added to 7H9 medium to a final concentration of 0.02% (vol/vol). For hypoxic culture, M. smegmatis strains were grown in a 250-ml flask filled with 150 ml of 7H9-glucose medium and tightly sealed with a rubber septum on a gyratory shaker (200 rpm) for 15 h following inoculation of the medium with aerobically grown preculture to an optical density at 600 nm (OD600) of 0.05. Escherichia coli strains were grown on Luria-Bertani (LB) medium at 37°C or 18°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | (φ80dlacZΔM15) ΔlacU169 recZ1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 | 19 |
| BL21(DE3) | F−ompT hsdSB(rB−mB−) dcm gal λ(DE3) | Promega |
| M. smegmatis mc2155 | High-transformation-efficiency mutant of M. smegmatis ATCC 607 | 44 |
| Plasmids | ||
| pBS II KS+ | Apr; lacPOZ′ | Stratagene |
| pNC | Hygr; promoterless lacZ | Y. M. Kim, unpublished data |
| pT7-7 | Apr; T7 promoter, ribosome binding site, and translation start codon overlapping with NdeI site | 48 |
| pT7SIHis | pT7-7 with an insertion of a 1,727-kb NdeI-EcoRI fragment containing devS with six His codons before its stop codon | This study |
| pT7STHis | pT7-T with an insertion of a 0.739-kb NdeI-EcoRI fragment containing the 3′ part of devS with six His codons before its stop codon | This study |
| pT7SIH150A | pT7SIHIS in which the codon for H150 is displaced with GCT | This study |
| pT7HIS9A | pT7-7 with an insertion of a 1.4-kb NdeI-PstI fragment containing prrB with nine His codons before its stop codon | 34 |
| pNChspX | pNC with an insertion of a 0.498-kb ClaI-XbaI fragment containing the hspX promoter region | This study |
| pGST-parallel1-GAFB | pGST-parallel1 containing an NcoI-EcoRI fragment encoding the GAF-B domain (D232 to S373) | B. S. Kang, unpublished data |
DNA manipulation and electroporation techniques.
Standard protocols or manufacturer's instructions were followed for recombinant DNA manipulations (42). The introduction of plasmids into M. smegmatis strains was carried out as described elsewhere (44).
Construction of plasmids. (i) pT7SIHIS and pT7STHIS.
A 1,727-bp fragment including the devS gene and the six histidine codons immediately before its stop codon was amplified with SIHIS+ (5′-ATGCCATATGGGGGGCGTGAACGGCCAC-3′) and SHIS− (5′-ATGCGGATCCTCAGTGGTGGTGATGGTGGTGGTCGGGGAGCGGCGCGGT-3′) using the M. smegmatis mc2155 genomic DNA as the template and Pfu DNA polymerase. The PCR product was restricted with NdeI and BamHI and cloned into pT7-7 digested with the same restriction enzymes, yielding pT7SIHIS. pT7STHIS was constructed in the same way as pT7SIHIS except for the primers used for PCR. The primer pair STHIS+ (5′-ATGCCATATGCCTTTCACCGCAGAACAGTTG-3′) and SHIS−, was employed for the construction of pT7STHIS. pT7STHIS carries the portion of devS encoding the C-DevS protein from which 342 N-terminal amino acids have been removed.
(ii) pNChspX.
A 498-bp fragment including the hspX gene promoter region was amplified with hspBamHI (5′-AATTGGATCCTCGGCGTCGGCGATCGTC-3′) and hspEcoRI (5′-CGAAGAATTCGGACATCTCCGGAAAGAG-3′), using M. smegmatis mc2155 genomic DNA as the template and Pfu DNA polymerase. The PCR product was restricted with EcoRI and BamHI and cloned into pBS II KS+ digested with the same restriction enzymes, yielding pBShspX. A 526-bp fragment from pBShspX was cloned into pNC digested with ClaI and XbaI, resulting in pNChspX.
Site-directed mutagenesis.
To mutate H150 of DevS to alanine, site-directed mutagenesis was performed using the plasmid pT7SIHIS as the template plasmid and a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Synthetic oligonucleotides 33 bases long, containing an alanine codon (GCT) in place of the histidine codon in the middle of their sequences, were used to mutagenize the histidine codon. The mutation was verified by DNA sequencing, and the resulting plasmid, pT7SIH150A, was introduced into E. coli strain BL21(DE3).
Protein purification. (i) Intact DevS.
The E. coli BL21(DE3) strain carrying either pT7SIHIS or pT7SIH150A was grown aerobically at 37°C in LB medium containing 100 μg/ml ampicillin to an OD600 of 0.4 to 0.5. The devS gene was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, and the cells were further grown for 4 h at 30°C. After 2 liter of culture was harvested, cells were resuspended in 30 ml of buffer A (20 mM Tris-HCl [pH 8.0] containing 100 mM NaCl) and disrupted by two passages through a French pressure cell. Following DNase I treatment (150 units) in the presence of 10 mM MgCl2 for 30 min on ice, cell-free crude extracts were obtained by centrifugation two times at 20,000 × g for 20 min. After the addition of imidazole to a final concentration of 5 mM, 1 ml of 80% (vol/vol) Ni-Sepharose high-performance resin (Amersham Biosciences, Piscataway, NJ) was added to the crude extracts and mixed gently by shaking for 2 h on ice. The protein-resin mixture was loaded into a column, and the column was washed with 40 volumes of buffer A containing 10 mM imidazole, followed by 20 volumes of buffer A containing 50 mM imidazole. The His6-tagged DevS protein was eluted with buffer A containing 200 mM imidazole. The fractions containing the DevS protein were collected and dialyzed against 2 liter of 20 mM Tris-HCl (pH 8.0) overnight at 4°C. The desalted DevS was concentrated by means of ultrafiltration (YM10 membrane; Millipore Co., Bedford, MA), and 0.1% (wt/vol) n-dodecyl β-d-maltoside (DM) was added to the concentrated protein.
(ii) C-DevS.
C-DevS was overexpressed in an E. coli BL21(DE3) strain carrying pT7STHIS. The strain was grown, and the devS gene was induced as described above for overexpression of the intact DevS. The purification procedure for C-DevS was the same as that for the intact DevS except for the washing and elution conditions during affinity chromatography; these consisted of 20 volumes of buffer B (20 mM Tris-HCl [pH 8.0] containing 500 mM NaCl) containing 5 mM imidazole in the first wash step, 40 volumes of buffer B containing 20 mM imidazole in the second wash step, and elution with buffer B containing 250 mM imidazole. The purified C-DevS was concentrated and stored at −70°C without the addition of DM. When DevS and C-DevS were purified to determine their autokinase activities, β-mercaptoethanol was added to all buffers used in the purification procedure to a final concentration of 20 mM.
(iii) The GAF-B domain.
Glutathione S-transferase-fused GAF-B (amino acids 231 to 379) was overexpressed in an E. coli BL21 strain carrying pGST-parallel1-GAFB. The proteins were purified by affinity chromatography using glutathione-Sepharose 4B (GE Healthcare, Piscataway, NJ). The recombinant protein was digested by using rTEV (Invitrogen, Carlsbad, CA) at 10°C in the presence of 0.5 mM EDTA and 1 mM dithiothreitol (DTT). After complete digestion, the glutathione S-transferase tag was removed by using a glutathione-Sepharose 4B column. Gel filtration was performed with a Superdex G75 column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5), and the fractions containing GAF-B were collected and concentrated by using Centriprep YM10 (Millipore) for crystallization. The purified protein contains an additional three amino acids (GAM) at the N terminus due to the cloning procedure.
In vitro autophosphorylation assay.
Protein phosphorylation was performed at either room temperature (RT) or 30°C in an assay buffer containing the appropriate protein components, 20 mM Tris-HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, 3 to 15 mM β-mercaptoethanol, 0.001 to 0.007% (wt/vol) DM. The autophosphorylation assay with deoxyferrous, oxyferrous, and NO-ferrous forms of DevS was performed inside an anaerobic chamber (Thermo 1025 Forma anaerobic system). The reaction was initiated by the addition of a mixture of [γ-32P]ATP and unlabeled ATP to a final concentration of 200 μM (1,000 Ci/mol). In the case of time course experiments, samples (10 μl) were removed at various time intervals, and reactions were stopped by the addition of 3 μl of 5× loading buffer (50 mM Tris-HCl, [pH 6.8], 4% [wt/vol] sodium dodecyl sulfate [SDS], 20% [wt/vol] glycerol, 20 mM DTT, 1% [vol/vol] β-mercaptoethanol, 0.1% [wt/vol] bromophenol blue, and 100 mM EDTA). To examine the effect of cyclic AMP (cAMP) and cGMP on DevS autophosphorylation, 80 pmol of DevS was incubated in an assay buffer in the presence or absence of cAMP and cGMP for 1 h on ice before the reaction was started. To test whether ubiquinone and menaquinone affect DevS autophosphorylation, the autophosphorylation rate of DevS was determined with the presence or absence of coenzyme Q1 (Sigma, St. Louis, MO) and vitamin K2 (Sigma) in the reaction mixture. They were dissolved in 95% (vol/vol) ethanol at a concentration of 12.5 mM as 50× stock solutions. The 50× stock solutions of coenzyme Q1 and vitamin K2 were added to the reaction mixtures to a final concentration of 250 μM (or the same volume of 95% ethanol was added as a negative control). Following the incubation of the reaction mixtures for 25 min at 30°C, the reaction was initiated by the addition of ATP.
Spectroscopic analyses. (i) Visible absorption spectrum.
The spectra of oxy-, deoxy-, and NO-ferrous forms of DevS were measured for the purified proteins on a spectrophotometer (Shimadzu UV-1650PC) in 20 mM Tris-HCl (pH 8.0) containing 10 mM DTT at RT. The spectra of the CN− form were measured in 20 mM Tris-HCl (pH 8.0) without DTT after the addition of KCN, to a final concentration of 10 mM. To test whether O2 and NO can act as ligands for DevS, protein samples were diluted with 20 mM Tris-HCl in a quartz cuvette stoppered with a rubber septum (Aldrich, St. Louis, MO). The protein samples were deoxygenated for 10 min with a bubbling flow of N2 gas, yielding deoxyferrous DevS. Oxyferrous DevS was prepared by a brief exposure of deoxyferrous DevS to air. NO-ferrous DevS was prepared by bubbling the purified NO gas through deoxyferrous DevS in the dark for 1 min. NO gas was purified by sequential passage through 0.5 M, 0.75 M, and 1 M KOH solutions.
(ii) Hemochrome assay.
To identify the heme type of DevS, 100 μl of DevS (5 mg/ml) was added to 3 ml of 50 mM NaOH containing 20% (wt/vol) pyridine and 6 μl of 0.1 M K3Fe(CN)6, and then an air-oxidized spectrum was recorded on the spectrophotometer. The same sample was reduced with 30 μl of 15% (wt/vol) sodium hydrosulfite (dithionite) for 2 min at RT, and then a reduced spectrum was recorded. The difference spectrum was obtained by subtraction of the oxidized spectrum from the reduced spectrum. Myoglobin (Sigma) was used as a reference protein containing the b-type heme.
β-Galactosidase assay and determination of protein concentration.
M. smegmatis cells were harvested, resuspended in the appropriate buffer, and disrupted by passage through a French pressure cell. Cell-free crude extracts were obtained by centrifugation at 20,000 × g. β-Galactosidase activity was assayed spectrophotometrically as described previously (33).
The protein concentration was determined by using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA), with bovine serum albumin as the standard protein.
Crystal structure determination.
The crystallization of GAF-B proteins was performed with a selenomethionine (SeMet)-labeled protein and a native GAF-B protein solution containing 1 M cAMP by using the sitting-drop vapor diffusion method at 294 K. The crystals were obtained under conditions of 1.25 M sodium citrate in 0.1 M Tris-HCl, pH 9.0, containing 0.6% (vol/vol) isopropanol. The data were collected at beamlines 4A and 6B at the Pohang Accelerator Laboratory (PAL, Korea), and all data were processed by using HKL-2000 (37). An initial GAF-B model was generated after subsequent improvements of the structure built automatically from Resolve (49) and used as templates for the molecular replacement procedures using AMoRe (32), with the data for the SeMet-labeled crystal at a 2.0-Å resolution and the data for a native crystal at a 2.1-Å resolution. Further model building was performed manually in the electron density map by using the programs O (20) and Coot (8), and the refinement with isotropic displacement parameters was performed with CCP4 refmac5 (31).
The Rwork and Rfree values of the structure with the SeMet substitution are 0.208 and 0.249, and the values of the native structure are 0.199 and 0.215, respectively. The statistics of the crystallographic data are summarized in Table 2.
TABLE 2.
Data collection and refinement statistics
| Parameter | X-ray sourcea
|
|
|---|---|---|
| PAL 4A | PAL 6C | |
| Experimental data | ||
| Wavelength (Å) | 1.0000 | 1.2398 |
| Space group | P61 | P61 |
| Unit cell parameters (Å) | 73.2 × 73.2 × 48.5 | 73.2 × 73.2 × 48.8 |
| Resolution limit (Å) | 40-2.0 (2.07-2.00) | 40-2.1 (2.18-2.10) |
| Total reflections | 72,234 | 92,286 |
| Unique reflections | 10,102 (968) | 8,716 (796) |
| Redundancy | 7.2 (6.5) | 10.6 (8.8) |
| Completeness (%) | 99.5 (97.1) | 99.1 (93.4) |
| Rsymb | 0.082 (0.376) | 0.084 (0.424) |
| Average I/σ (I) | 31.7 (5.3) | 31.2 (4.6) |
| Refinement details | ||
| Space group | P61 | P61 |
| Resolution (Å) | 30-2.0 | 30-2.1 |
| Reflection (working) | 9,590 | 8,282 |
| Reflection (test) | 485 | 409 |
| Rworkc | 0.208 | 0.199 |
| Rfreec | 0.249 | 0.215 |
| No. of water molecules | 41 | 44 |
| Root mean square deviation from ideal geometry | ||
| Bond length (Å) | 0.011 | 0.010 |
| Bond angle (°) | 1.273 | 1.283 |
| Avg B factors (Å) | ||
| Molecule A (main/side chain) | 32.9 (31.9/34.1) | 29.0 (28.3/29.9) |
| Waters | 37.7 | 33.4 |
The numbers in parentheses describe the relevant value for the last resolution shell.
Rsym = ΣIi − <I>/ΣI, where Ii is the intensity of the ith observation and <I> is the mean intensity of the reflections.
Rwork = Σ‖Fobs-Fcalc‖/ΣFobs, crystallographic R factor, and Rfree = Σ‖Fobs-Fcalc‖/ΣFobs when all reflections belong to a test set of randomly selected data.
Protein Data Bank accession numbers.
The final models of the SeMet-labeled and native structures have been deposited in the worldwide Protein Data Bank (3) under the PDB ID codes 2VJW and 2VKS, respectively.
RESULTS
Ligand-binding properties of purified DevS.
The soluble full-length DevS was purified, without using any detergent, by affinity chromatography. When purified under oxidizing conditions where reducing agents, such as β-mercaptoethanol and DTT, were not included in the buffers used during purification, the DevS protein showed a light brown color, indicating that DevS of M. smegmatis contains heme as a prosthetic group, like the DevS and DosT proteins of M. tuberculosis. The results of a hemochrome assay demonstrated that purified DevS displayed a major peak at 556 nm, like myoglobin containing b-type heme, indicating the presence of b-type heme in DevS (data not shown).
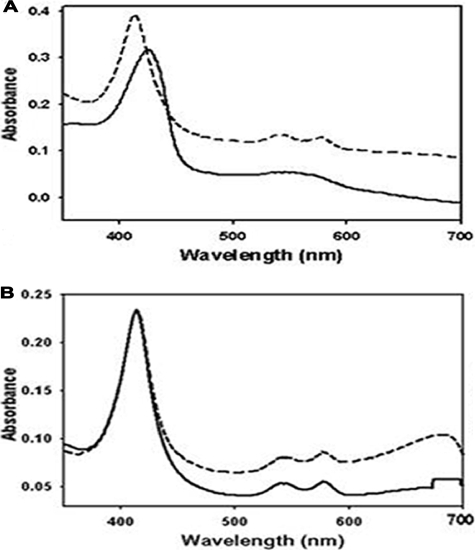
Using UV-visible spectroscopy, the redox state and O2 binding state of heme iron of DevS were determined. When the aerobically purified DevS was reduced to the ferrous form by treatment with DTT and deoxygenated by sparging with N2 gas, the deoxyferrous form of DevS had broad absorption in the visible region of 500 to 600 nm and a Soret band at 425 nm, implying that heme iron is in the pentacoordinate state (Fig. 1A). When the deoxyferrous form of DevS was reexposed to air, DevS showed well-resolved α and β bands at 576 and 541 nm, respectively, and the Soret band was blue-shifted from 425 to 413 nm and became sharper (Fig. 1A). This spectral characteristic indicates that air-exposed DevS has a hexacoordinate heme iron with O2 as the distal ligand (oxyferrous form). Interestingly, the results of absorption spectroscopy showed that aerobically purified DevS had distinct α and β bands, as well as the Soret band, at the same wavelengths as the oxyferrous form of DevS (Fig. 1B), indicating that aerobically purified DevS is in the ferrous state, even though it was not treated with any reducing agents. To confirm that heme iron of the aerobically purified DevS is in the ferrous state, the purified DevS was treated with KCN and the absorption spectrum was determined. The purified DevS treated with KCN displayed the same spectrum as the control that was not treated with KCN (Fig. 1B). When the purified DevS was oxidized with K3Fe(CN)6, its Soret band was shifted from 413 to 407 nm. The oxidized DevS treated with KCN showed a shift of its Soret peak from 407 to 417 nm (data not shown), indicating that CN− ion binds to the oxidized DevS. As judged by the fact that CN− ion binds to Fe3+ but not Fe2+, the aerobically purified DevS is in the ferrous state. When the deoxyferrous form of DevS was treated with NO gas, the Soret band was shifted from 413 to 425 nm (data not shown), indicating that the deoxyferrous form of DevS can bind NO.
FIG. 1.
Absorption spectra of purified DevS. The spectra were recorded using 300 μg of DevS in 20 mM Tris-HCl buffer at RT. The DevS protein was purified in the absence of any reducing reagent in the buffer used during the purification procedures. (A) DevS was reduced by the addition of DTT to a final concentration of 10 mM and sparged with N2 gas for 10 min to yield a deoxyferrous derivative of DevS (solid line). The deoxyferrous form of DevS was exposed to air to generate the oxyferrous form of DevS (dashed line). (B) The spectra of the purified DevS before (solid line) and after (dashed line) 10 mM KCN treatment.
The autokinase activity of DevS is controlled by the liganding state of the heme.
We next examined the effect of ligand binding on DevS autokinase activity. C-DevS, in which the heme-containing N-terminal sensory domain is removed, and the H150A mutant form of DevS were included as the controls. The H150 residue, which has been suggested to serve as the proximal axial ligand for heme iron (18, 43), was replaced with alanine in the H150A mutant form of DevS. The results of a hemochrome assay showed that the mutant form of DevS did not contain heme when purified from E. coli (data not shown).
As shown by the results in Fig. 2, the autophosphorylation rate of deoxyferrous DevS was higher than that of oxyferrous DevS. The NO-ferrous form of DevS was also autophosphorylated significantly faster than oxyferrous DevS. In contrast, when DTT-treated C-DevS and H150A DevS were sparged with N2 or NO, their autophosphorylation rates were not increased. Taken together, these results suggest that the sensory function of DevS for O2 and NO is mediated through their binding to the heme moiety located in the N-terminal sensory domain of DevS.
FIG. 2.
Kinase activities of the DTT-reduced ferrous DevS derivatives. Purified DevS was treated with 10 mM DTT and sparged with N2 gas for 10 min to generate the deoxyferrous form of DevS. Oxy- and NO-ferrous DevS derivatives were prepared as described in Materials and Methods. The autophosphorylation reactions were performed by using 80 pmol each of DevS, C-DevS, and H150A DevS and 200 mM (1,000 ci/mol) ATP in the anaerobic box at RT. At the time points indicated, samples (10 μl) were removed and added to 3 μl of loading buffer to stop the reaction. (A) The phosphorylation of the protein was assayed by SDS-polyacrylamide gel electrophoresis and subsequent autoradiography. (B) Quantitation of the band intensity of phosphorylated DevS was performed with a densitometer program, ImageJ (version 1.37), and the relative values are plotted as a function of the reaction time. Symbols: filled circle, oxyferrous DevS; open circle, deoxyferrous DevS; filled inverted triangle, NO-ferrous DevS.
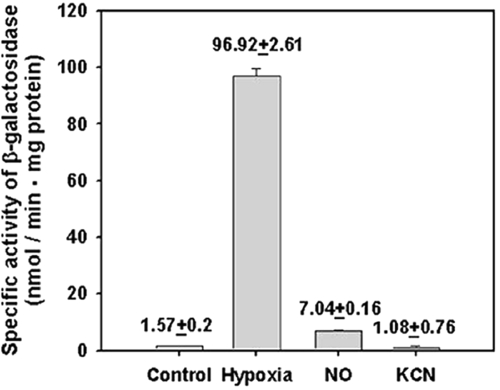
The heme iron of aerobically purified DevS is in the ferrous state, implying that DevS has a ferrous form of heme inside the cell and that the binding of the ligands, such as O2 and NO, rather than the redox state of the heme iron regulates DevS kinase activity in vivo. To test this assumption in vivo, the expression of the hspX gene belonging to the DevR regulon in M. smegmatis cells grown under various growth conditions was measured (Fig. 3). As expected, the expression of hspX in the cells grown under hypoxic stress was increased 62-fold in comparison to that in the cells grown aerobically. When aerobically grown M. smegmatis culture was treated with an NO generator, sodium nitroprusside, hspX expression was enhanced 4.5-fold over that in the aerobically grown culture. In contrast, the treatment of aerobically grown culture with KCN did not lead to an increase in hspX expression, which is in good agreement with the fact that CN− cannot bind to the ferrous form of heme iron.
FIG. 3.
Expression of hspX in M. smegmatis grown under various stress conditions: Control, the M. smegmatis strain carrying the hspX::lacZ transcriptional fusion plasmid pNChspX was grown aerobically in 7H9-glucose medium to an OD600 of 0.5; Hypoxia, the strain was grown in a 250-ml flask filled with 150 ml of 7H9-glucose medium and sealed with a rubber septum on a gyratory shaker (200 rpm) for 15 h following inoculation of the medium with aerobically grown preculture, to an OD600 of 0.05; NO and KCN, the strains were initially grown aerobically in 7H9-glucose medium to an OD600 of 0.3. The cultures were treated with either 500 μM sodium nitroprusside or 500 μM KCN and further grown aerobically for an additional 2 h. Cell-free crude extracts were used to determine β-galactosidase activity. The β-galactosidase activity is expressed as nanomoles per minute per milligram of protein. All values provided are the average of the results of two independent determinations. Error bars indicate standard deviations.
The GAF-B domain of DevS appears not to be involved in the sensing of cyclic nucleotides.
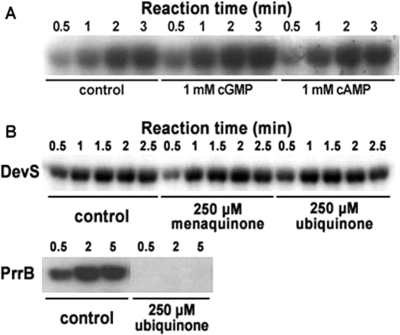
DevS has the tandem putative GAF domains in its N-terminal sensory domain, and the proximal GAF domain (GAF-A) from the N terminus is implicated in binding the heme moiety (43). The role of the second GAF domain (GAF-B) remains elusive. The GAF domains found in some cyclic-nucleotide phosphodiesterases (PDEs) and cyanobacterial adenylyl cyclases are known to serve as the binding domains for cyclic nucleotides, such as cGMP and cAMP (25). To assess whether cGMP and cAMP affect the DevS kinase activity as the regulatory ligands, the autophosphorylation rate of DevS in the presence or absence of the cyclic nucleotides was determined. As shown by the results in Fig. 4A, treatment with cGMP or cAMP did not lead to noticeable changes in the autophosphorylation rate of DevS.
FIG. 4.
Effects of cyclic nucleotides, menaquinone, and ubiquinone on DevS autokinase activity. (A) The autophosphorylation reactions were performed by using purified DevS (80 pmol) and 200 μM (1,000 ci/mol) ATP at 30°C. The reaction mixtures with either 1 mM cGMP or 1 mM cAMP were incubated on ice for 1 h before the addition of ATP. As a control, the reaction mixture without a cyclic nucleotide was included in the experiment. At the time points indicated, samples (10 μl) were removed and added to 3 μl of loading buffer to stop the reaction. The phosphorylation of the protein was assayed by SDS-polyacrylamide gel electrophoresis and subsequent autoradiography. (B) The autophosphorylation reactions were performed by using 80 pmol each of DevS or PrrB and 100 μM (1,000 Ci/mol) ATP at 30°C. The reaction mixtures with either 250 mM menaquinone or 250 μM ubiquinone were incubated at 30°C for 25 min before the addition of ATP. As a control, the reaction mixture without a quinone compound was included in the experiment.
The redox state of quinone/quinol does not affect the autokinase activity of DevS.
The autophosphorylation of the ArcB and RegB HKs, which are involved in redox sensing in E. coli and Rhodobacter capsulatus, respectively, has been suggested to be controlled by the redox state of the quinone/quinol pool of the electron transport chain (9, 23, 47). Ubiquinone was shown to inhibit the autophosphorylation of ArcB and RegB. Since DevS also pertains to redox sensing, the effects of ubiquinone and menaquinone on the autophosphorylation rate of DevS were investigated. As shown in Fig. 4B, when purified DevS was incubated in the presence of ubiquinone or menaquinone, its autokinase activity was not affected. As a control, PrrB HK (RegB homolog in Rhodobacter sphaeroides) was included in this experiment. The autophosphorylation of PrrB was completely abolished when purified PrrB was incubated in the presence of 250 μM ubiquinone before the autophosphorylation reaction. This result clearly demonstrates that the kinase activity of DevS is not controlled by the redox state of the quinone/quinol pool of the electron transport chain, which closely reflects the cellular redox state.
The three-dimensional structure of the GAF-B domain.
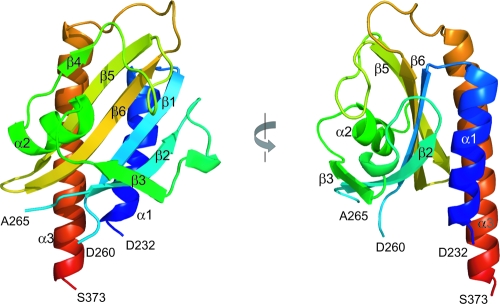
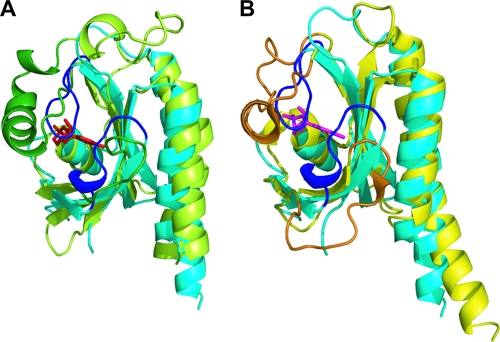
The three-dimensional structure of GAF-B (D232 to S373) from the DevS protein was determined by X-ray crystallography using single-wavelength anomalous dispersion and refined to a crystallographic R factor of 0.208 at a 2.0-Å resolution (Table 2). The structure of GAF-B is composed of three α-helices, six β-strands, and long connecting loops. The structure is built around a central antiparallel six-stranded β-sheet with the strand order 321654 (Fig. 5). The strongly curved β-sheet forms a half barrel and divides the three α-helices into two and one at either side. The N- and C-terminal portions of the sequence are the α1- and α3-helices, respectively, and form an outer layer of the barrel structure. The loop between the β3- and β4-strands, along with the α2-helix, fills the inside of the half barrel. There are missing residues (P261 to S264) in the loop region connecting the β1- and β2-strands due to a poor electron density at the region, suggesting flexibility of the loop. The structural comparison of DevS GAF-B with other known GAF-B domains revealed that the overall folding of the domain is similar to that of a typical GAF domain, except for the absence of an α-helix in the loop between the β4- and β5-strands (Fig. 6). The structure is also similar to those of PAS (Per-Arnt-Sim) domains, which contain a five-stranded β-sheet and α-helices on the sheet. Previously, sequence analysis of M. tuberculosis DevS suggested two transmembrane motifs (5), A289 to L295 and L325 to V341, within the GAF-B domain. These sequences correspond to the regions V278 to I294 and L319 to A335, respectively, of DevS from M. smegmatis (Fig. 7). In the crystal structure, the first transmembrane motif is around the β3-strand and the second one forms a β-hairpin structure with the β5- and β6-strands.
FIG. 5.
Ribbon diagrams of the GAF-B domain of DevS. The domain consists of a six-stranded antiparallel β-sheet and three α-helices. Residues P261 to S264 are missing in the structure. The left diagram is rotated 90° about the vertical axis.
FIG. 6.
Structural comparison of the DevS GAF-B domain with GAF domains containing cyclic nucleotides. The structure of the GAF-B domain of DevS (cyan) is superimposed on the structures of the GAF-A domain (green) of adenylyl cyclase CyaB2 (A) and the GAF-B domain (yellow) of PDE2A (B). For the binding of cAMP (red) or cGMP (magenta), the α3-helix and the loop between the β2- and β3-strands of the GAF domains of PDE2A (orange) and CyaB2 (dark green) form the binding pocket. In GAF-B of DevS, two loops corresponding to the orange and dark green regions are presented in blue.
FIG. 7.
The multiple sequence alignment of DevS of M. smegmatis with its homologs in M. tuberculosis. Amino acid sequences of DevS from M. smegmatis (Msm_DevS) and DosT (Mtb_DosT) and DevS (Mtb_DevS) from M. tuberculosis were aligned. Numbering was done using DevS of M. smegmatis. The arrows and coils above the aligned sequences indicate the elements of the secondary structure of the DevS GAF-B domain. Conserved residues are shown by the gray background. The putative transmembrane motifs, which were predicted with M. tuberculosis DevS, are denoted by thick lines below the alignment. The amino acid residues corresponding to those hydrogen bonded to a cyclic nucleotide in PDE2A and CyaB2 are indicated by the inverted triangles. Multiple alignment was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/) and visualized using ESPript (http://espript.ibcp.fr/ESPript/ESPript/).
A search with the DALI server (16) revealed the proteins that are structurally similar to the GAF-B domain of DevS. Although the sequence similarity is low, there were a few structurally similar proteins. The server yielded seven structures with Z-scores above 10 (data not shown). The sequence identities between GAF-B and the structurally similar proteins range between 7 and 18%. The root mean square differences of the structures from the GAF-B structure were between 2.1 and 3.0 Å. The GAF-B domain of mouse PDE2A (1LQ9) shows the highest Z-score, 15.7. The GAF domains of cyanobacterial adenylyl cyclase CyaB2 (1YKD) and the chromophore-binding domains from the phytochromes DrBphP (1ZTU) and RpBphP3 (2OOL) are also similar to the GAF-B domain of DevS. The GAF-B domain of PDE2A contains cGMP (27), and both the GAF-A and -B domain of CyaB2 bind cAMP in the cyclic-nucleotide binding pockets (26). The chromophore-binding domains of the phytochromes are also considered GAF structures, and they accommodate biliverdin molecules in their binding pockets (51, 53).
Since the PDE2A and CyaB2 GAF domains bind cyclic nucleotides, the structure of the DevS GAF-B domain was superimposed on the GAF domains bound by the cyclic nucleotides (Fig. 6). GAF-B of PDE2A and GAF-A of CyaB2 hold cGMP and cAMP, respectively, in their binding pockets. The binding pocket for a cyclic nucleotide consists of the half-barrel β-sheet structure and the α3-helix between the β4- and β5-strands covering the half-barrel structure. Unlike these GAF domains, a short loop of DevS GAF-B connecting strands β4 and β5 does not contain the α-helix structure corresponding to the α3-helix forming the binding cavity in the other GAF domains. The absence of the α3-helix in DevS GAF-B leaves a hydrophobic groove exposed on the surface of GAF-B (data not shown). The curved β-sheet and two outer helices of DevS GAF-B, as well as the α2-helix, at the bottom of the barrel structure fit well to the structure of the GAF domains of PDE2A and CyaB2. However, there is no space for cyclic-nucleotide binding in the structure of DevS GAF-B. Two loops connecting the β2- and β3-strands and the β4- and β5-strands are located close to the inside of the half-barrel structure and fill the putative cyclic-nucleotide binding pocket. The positioning of these loops close to the β-sheet allows no room for the binding of small molecules, such as cyclic nucleotides. We obtained the crystals with the native GAF-B protein from the drop containing cAMP and determined the structure of the native GAF-B domain (Table 2). However, this trial of cocrystallization of the GAF-B domain with cAMP failed to get the complex. This native crystal structure was almost identical to the structure of the SeMet-labeled crystal (data not shown), suggesting that the absence of space for a cyclic nucleotide was not due to the lack of ligands when it was crystallized.
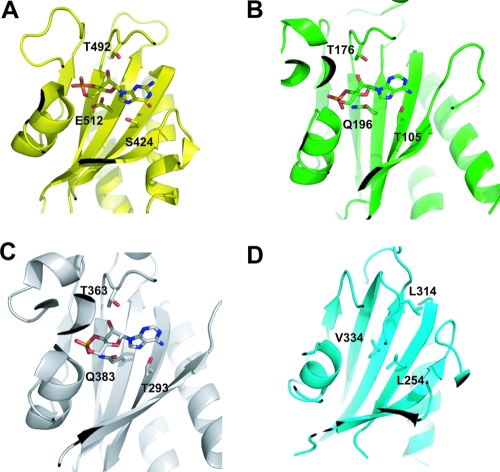
Several hydrophilic residues are involved in the binding of cAMP or cGMP in the GAF domains known to bind the cyclic nucleotide (Fig. 8). Among them, E512 at the β6-strand and S424 at the β1-strand are hydrogen bonded to cGMP in the GAF-B of PDE2A. The corresponding residues, Q196 and T105 in GAF-A of CyaB2 and Q383 and T293 in GAF-B of CyaB2, participate in the binding of cAMP. The threonine residues in the loop connecting to the β5-strand of PDE2A GAF-B (T492), as well as CyaB2 GAF-A (T176) and GAF-B (T363), are also implicated in the binding of the cyclic nucleotides. These three hydrophilic residues are replaced by the hydrophobic residues V334, L254, and L314 in GAF-B of DevS, implying that DevS GAF-B is unlikely to bind a cyclic nucleotide.
FIG. 8.
Hydrophilic residues involved in the binding of cyclic nucleotides in the GAF domains. The residues presented on the β-sheets and the loop linked to the β5 strand are hydrogen bonded directly to the cyclic nucleotides in the GAF-B domain of PDE2A (S424, T492, and E512) (A) and the GAF-A domain (T105, T176, and Q196) (B) and GAF-B domain (T293, T363, and Q383) (C) of adenylyl cyclase CyaB2. These hydrophilic residues are replaced by hydrophobic ones in the GAF-B domain of DevS (L254, V334, and L314) (D).
DISCUSSION
The heme iron within DevS is in the petacoordinate state in the absence of exogenous ligands.
The b-type heme within DevS was suggested to be coordinated by the GAF-A domain with H150 providing a proximal axial ligand (18, 43). The GAF domain is distantly related to the PAS domain with regard to the basic fold structure (17, 25). The PAS domains are implicated in heme binding in FixL HKs from bacterial origins and in DOS (direct oxygen sensor) from E. coli (13). In the case of E. coli DOS, the heme iron is coordinated to H77 on the proximal side and a displaceable M95 residue on the distal side, thereby exhibiting the absorption spectra typical of the hexacoordinate heme in the absence of any exogenous ligands, such as O2, NO, and CO (6). In contrast, the heme iron in FixL is coordinated by a proximal histidine residue and its distal axial position is not occupied in the absence of the exogenous ligands (11, 12, 30). When the purified DevS protein was deoxygenated by N2 sparging following DTT treatment, it showed an absorption spectrum characteristic of the pentacoordinate heme. The spectrum of deoxyferrous DevS of M. smegmatis was similar to those of deoxyferrous DevS and DosT of M. tuberculosis, as well as of FixL proteins of rhizobia, i.e., with no distinct α and β peaks in the region of 500 to 600 nm and a red-shifted Soret peak (11, 18, 22, 46). These results indicate that the distal axial position of the heme iron is not occupied by an endogenous ligand derived from the DevS polypeptide chain.
The DevS protein purified under oxidizing conditions showed the same absorption spectrum as the oxyferrous form of DevS and did not form a complex with CN−. Despite prolonged desalting by means of dialysis under atmospheric conditions, the purified DevS was in the ferrous state, indicating that the heme iron in DevS is resistant to autooxidation from an oxyferrous to a ferric state in the presence of O2 and that DevS inside the cell is in the ferrous state. It has been reported that there are two DevS homologs (DevS and DosT) in M. tuberculosis that cross talk with the DevR response regulator (39, 41). DevS of M. tuberculosis was shown to be rapidly autooxidized from an oxyferrous to a ferric form when exposed to air, whereas an oxyferrous DosT was relatively stable in air and was not easily converted to the ferric form (22). Regarding the autooxidation property, DevS of M. smegmatis is similar to DosT rather than DevS of M. tuberculosis. Multiple-alignment analysis of the GAF-A domains (amino acids 68 to 210 of M. smegmatis DevS) of the DevS homologs showed that DevS of M. smegmatis is more closely related to the corresponding domains of DosT (82% identity) than of DevS (69% identity) of M. tuberculosis (Fig. 7), which is consistent with the similarity of its autooxidation property to that of M. tuberculosis DosT. This finding indicates that DevS of M. smegmatis is an ortholog of M. tuberculosis DosT. Therefore, it is reasonable to rename DevS of M. smegmatis DosT. Phylogenetic analysis using the GAF-A domain revealed that the DosT homologs occur in Mycobacterium vanbaalenii and Mycobacterium gilvum, as well as M. tuberculosis, Mycobacterium bovis, and M. smegmatis, while the DevS homologs are found in M. tuberculosis, M. bovis, Mycobacterium marinum, and Mycobacterium ulcerans. M. vanbaalenii, M. marinum, and Mycobacterium avium contain DevS (DosT) homologs that form a phylogenetically distinct branch from both DosT and DevS of M. tuberculosis (data not shown).
DevS kinase activity is controlled by ligand binding rather than the redox state of its heme iron.
The heme iron of M. smegmatis DevS HK appears to be in the ferrous state inside the cell. Therefore, it is not the redox state (Fe2+ or Fe3+) of the heme iron that controls the kinase activity of DevS in vivo in response to changes in oxygen tension. The fact that the oxyferrous form of DevS has a lower autokinase activity than the deoxy- and NO-ferrous forms suggests that the kinase activity of DevS is inhibited by O2 binding. NO binding appears to lock the conformation of DevS into an active state which resembles that of the deoxyferrous form of DevS. Thus, NO might mimic hypoxia even in the presence of O2. The regulation of DevS autokinase activity by O2, NO, and CO was recently reported for DevS and DosT of M. tuberculosis (22, 46). In this study, we included the DevS derivatives lacking the heme group as the negative controls and provided direct evidence that DevS directly senses the diatomic gases, such as O2 and NO, by means of its heme group. The mechanism by which DevS discriminates O2 from NO can be revealed when the three-dimensional structure of the ligand-bound sensory domain of DevS is determined, although the basis of the discriminating mechanism has been suggested for FixL HKs (10, 15).
An increase in DevS kinase activity under hypoxic conditions is not the result of inhibition of aerobic respiration.
One of the mechanisms underlying redox sensing by HKs is that the HKs perceive the redox state of the quinone/quinol pool of the electron transport chain, since it reflects the availability of O2, the terminal electron acceptor of aerobic respiration, as well as the redox state of the environment. The equilibrium of ubiquinone and ubiquinol is shifted to the ubiquinone-dominant state when aerobic respiration takes place efficiently under aerobic conditions. The kinase activities of the ArcB and RegB HKs from E. coli and R. capsulatus, respectively, were shown to be inhibited by ubiquinone (9, 21, 23, 47).
It was demonstrated in this study that neither ubiquinone nor menaquinone had an inhibitory effect on DevS kinase activity. Furthermore, the addition of 500 μM KCN to an aerobically growing culture of M. smegmatis did not affect the expression of the hspX gene, which is under the control of the DevSR system (29, 36), although 500 μM KCN inhibited the O2 consumption rate (aerobic respiration) by ∼70% (data not shown). It was also recently reported that inactivation of the bc1-aa3 pathway of the M. smegmatis electron transport chain by mutations did not induce the DevR regulon (28). These results clearly indicate that an increase in DevS kinase activity under hypoxic conditions or in the presence of NO is not the result of inhibition of aerobic respiration and that the redox-sensing mechanism of DevS is not related to the functional state of the respiratory electron transport chain.
Implication of the GAF-B domain in the function of DevS.
The GAF-A domain of DevS has been known to be implicated in heme binding, while the role of the GAF-B domain remains to be determined (18, 22, 43, 46). The GAF domains have been found in a number of proteins which are involved in many signal transduction pathways and the regulation of gene expression, as exemplified by cGMP-regulated cyclic-nucleotide PDEs, some adenylyl cyclases, phytochromes, and the transcriptional regulator FhlA (25). The GAF domains in the cGMP-regulated cyclic-nucleotide PDEs and adenylyl cyclases serve as the binding domains for the cyclic nucleotides, which play an important role in signal transduction as secondary messengers (2, 4, 45). For this reason, the possibility that the cyclic nucleotides modulate the DevS kinase activity as effector molecules by their binding to the GAF-B domain was examined. Neither cGMP nor cAMP affected the DevS autophosphorylation rate, indicating that the cyclic nucleotides do not function as the regulatory molecules controlling the DevS activity and that the GAF-B domain of DevS might not be involved in the binding of the cyclic nucleotides. In good agreement with these results, structural analysis of DevS GAF-B revealed that it appears not to be suitable to bind a cyclic nucleotide, since its putative nucleotide binding pocket is collapsed and the hydrophilic residues involved in the binding of a cyclic nucleotide in the pocket are not conserved in the DevS homologs (Fig. 7).
The presence of transmembrane domains in the N-terminal sensory domain of DevS has been controversial (29). In silico analysis of the primary structure of M. tuberculosis DevS has suggested that the protein contains three transmembrane segments (5), although the DevS (DosT) proteins have been reported to be purified in the soluble form without any detergent (18, 22, 43, 46). The overlap of the predicted transmembrane segments and the regions in the GAF domains in DevS implies that DevS is not a membrane protein (Fig. 7).
The GAF-B domain of M. tuberculosis DevS was suggested to help in the formation of a better-defined distal pocket of the heme-binding domain (GAF-A) through interdomain interactions within DevS (54). Since GAF-B has a hydrophobic groove on the β-sheet, it might interact with a hydrophobic ridge of its binding partner.
Acknowledgments
This work was supported by grants (KRF-2005-041-C00399 and R01-2006-000-10846-0) from the Korea Research Foundation and Korea Science and Engineering Foundation, respectively.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Aly, S., K. Wagner, C. Keller, S. Malm, A. Malzan, S. Brandau, F. C. Bange, and S. Ehlers. 2006. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210298-305. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22458-459. [DOI] [PubMed] [Google Scholar]
- 3.Berman, H., K. Henrick, and H. Nakamura. 2003. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10980. [DOI] [PubMed] [Google Scholar]
- 4.Charbonneau, H., R. K. Prusti, H. LeTrong, W. K. Sonnenburg, P. J. Mullaney, K. A. Walsh, and J. A. Beavo. 1990. Identification of a noncatalytic cGMP-binding domain conserved in both the cGMP-stimulated and photoreceptor cyclic nucleotide phosphodiesterases. Proc. Natl. Acad. Sci. USA 87288-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuberc. Lung Dis. 80141-159. [DOI] [PubMed] [Google Scholar]
- 6.Delgado-Nixon, V. M., G. Gonzalez, and M. A. Gilles-Gonzalez. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 392685-2691. [DOI] [PubMed] [Google Scholar]
- 7.Dick, T., B. H. Lee, and B. Murugasu-Oei. 1998. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 163159-164. [DOI] [PubMed] [Google Scholar]
- 8.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 602126-2132. [DOI] [PubMed] [Google Scholar]
- 9.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 2922314-2316. [DOI] [PubMed] [Google Scholar]
- 10.Gilles-Gonzalez, M. A., and G. Gonzalez. 2005. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 991-22. [DOI] [PubMed] [Google Scholar]
- 11.Gilles-Gonzalez, M. A., G. Gonzalez, M. F. Perutz, L. Kiger, M. C. Marden, and C. Poyart. 1994. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry 338067-8073. [DOI] [PubMed] [Google Scholar]
- 12.Gong, W., B. Hao, S. S. Mansy, G. Gonzalez, M. A. Gilles-Gonzalez, and M. K. Chan. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 9515177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, J., and M. S. Paget. 2004. Bacterial redox sensors. Nat. Rev. Microbiol. 2954-966. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, S., and D. Chatterji. 2005. Stress responses in mycobacteria. IUBMB Life 57149-159. [DOI] [PubMed] [Google Scholar]
- 15.Hao, B., C. Isaza, J. Arndt, M. Soltis, and M. K. Chan. 2002. Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 4112952-12958. [DOI] [PubMed] [Google Scholar]
- 16.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233123-138. [DOI] [PubMed] [Google Scholar]
- 17.Huang, Z. J., I. Edery, and M. Rosbash. 1993. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364259-262. [DOI] [PubMed] [Google Scholar]
- 18.Ioanoviciu, A., E. T. Yukl, P. Moenne-Loccoz, and P. R. de Montellano. 2007. DevS, a heme-containing two-component oxygen sensor of Mycobacterium tuberculosis. Biochemistry 464250-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jessee, J. 1986. New subcloning efficiency competent cells: >1 × 106 transformants/μg. Focus 89. [Google Scholar]
- 20.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 47110-119. [DOI] [PubMed] [Google Scholar]
- 21.Kim, Y. J., I. J. Ko, J. M. Lee, H. Y. Kang, Y. M. Kim, S. Kaplan, and J. I. Oh. 2007. Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1895617-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 10411568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malpica, R., B. Franco, C. Rodriguez, O. Kwon, and D. Georgellis. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. USA 10113318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manabe, Y. C., and W. R. Bishai. 2000. Latent Mycobacterium tuberculosis: persistence, patience, and winning by waiting. Nat. Med. 61327-1329. [DOI] [PubMed] [Google Scholar]
- 25.Martinez, S. E., J. A. Beavo, and W. G. Hol. 2002. GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol. Interv. 2317-323. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, S. E., S. Bruder, A. Schultz, N. Zheng, J. E. Schultz, J. A. Beavo, and J. U. Linder. 2005. Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: modes of ligand binding and dimerization. Proc. Natl. Acad. Sci. USA 1023082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, S. E., A. Y. Wu, N. A. Glavas, X. B. Tang, S. Turley, W. G. Hol, and J. A. Beavo. 2002. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc. Natl. Acad. Sci. USA 9913260-13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsoso, L. G., B. D. Kana, P. K. Crellin, D. J. Lea-Smith, A. Pelosi, D. Powell, S. S. Dawes, H. Rubin, R. L. Coppel, and V. Mizrahi. 2005. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J. Bacteriol. 1876300-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayuri, G. Bagchi, T. K. Das, and J. S. Tyagi. 2002. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol. Lett. 211231-237. [DOI] [PubMed] [Google Scholar]
- 30.Miyatake, H., M. Mukai, S. Adachi, H. Nakamura, K. Tamura, T. Iizuka, Y. Shiro, R. W. Strange, and S. S. Hasnain. 1999. Iron coordination structures of oxygen sensor FixL characterized by Fe K-edge extended x-ray absorption fine structure and resonance raman spectroscopy. J. Biol. Chem. 27423176-23184. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 53240-255. [DOI] [PubMed] [Google Scholar]
- 32.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 50162-182. [Google Scholar]
- 33.Oh, J. I., and S. Kaplan. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 382688-2696. [DOI] [PubMed] [Google Scholar]
- 34.Oh, J. I., I. J. Ko, and S. Kaplan. 2004. Reconstitution of the Rhodobacter sphaeroides cbb3-PrrBA signal transduction pathway in vitro. Biochemistry 437915-7923. [DOI] [PubMed] [Google Scholar]
- 35.Ohno, H., G. Zhu, V. P. Mohan, D. Chu, S. Kohno, W. R. Jacobs, Jr., and J. Chan. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell. Microbiol. 5637-648. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, R., M. J. Smeulders, M. C. Blokpoel, E. J. Kay, K. Lougheed, and H. D. Williams. 2003. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 1851543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 38.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 27923082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saini, D. K., V. Malhotra, D. Dey, N. Pant, T. K. Das, and J. S. Tyagi. 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150865-875. [DOI] [PubMed] [Google Scholar]
- 41.Saini, D. K., V. Malhotra, and J. S. Tyagi. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 56575-80. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Sardiwal, S., S. L. Kendall, F. Movahedzadeh, S. C. Rison, N. G. Stoker, and S. Djordjevic. 2005. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J. Mol. Biol. 353929-936. [DOI] [PubMed] [Google Scholar]
- 44.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 45.Soderling, S. H., and J. A. Beavo. 2000. Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr. Opin. Cell Biol. 12174-179. [DOI] [PubMed] [Google Scholar]
- 46.Sousa, E. H., J. R. Tuckerman, G. Gonzalez, and M. A. Gilles-Gonzalez. 2007. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 161708-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swem, L. R., X. Gong, C. A. Yu, and C. E. Bauer. 2006. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 2816768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 821074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D. 56965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, J. R., J. S. Brunzelle, K. T. Forest, and R. D. Vierstra. 2005. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438325-331. [DOI] [PubMed] [Google Scholar]
- 52.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55139-163. [DOI] [PubMed] [Google Scholar]
- 53.Yang, X., E. A. Stojkovic, J. Kuk, and K. Moffat. 2007. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc. Natl. Acad. Sci. USA 10412571-12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yukl, E. T., A. Ioanoviciu, P. R. de Montellano, and P. Moenne-Loccoz. 2007. Interdomain interactions within the two-component heme-based sensor DevS from Mycobacterium tuberculosis. Biochemistry 469728-9736. [DOI] [PubMed] [Google Scholar]