Abstract
Thirty-five putative integrative conjugative elements and related elements were identified at 15 locations in the eight sequenced genomes of Streptococcus agalactiae. Twelve are composite, likely resulting from site-specific accretions. Circular forms were detected for five elements. Macroarray analysis confirmed their high plasticity and wide distribution in S. agalactiae.
Streptococcus agalactiae (group B streptococcus) is the predominant cause of invasive bacterial diseases in human neonates and is responsible for bovine mastitis in industrial countries. The genomes of eight S. agalactiae strains belonging to the major lineages infecting neonates have been sequenced (8, 19, 20). Whole-genome comparisons have pointed to the diversity of the flexible gene pool of S. agalactiae, suggesting that its pan-genome is open (19). They also revealed a composite organization of the chromosome, with a stable backbone and 11 to 14 large variable regions per genome, most of which are thought to derive from mobile genetic elements (8, 19, 20). In this work, we showed that 48 genomic islands (GIs) correspond to putative integrative conjugative elements (ICEs) or related elements.
ICEs, also called conjugative transposons, were defined as elements which encode their excision from a host replicon, their intercellular transfer by conjugation, and their integration into a replicon of a recipient cell, whatever the specificity and mechanism of integration and conjugation (4, 5). Most of the ICEs encode a tyrosine integrase which catalyzes recombination between identical sequences carried by the attL and attR recombination sites flanking the element (4, 5). This enables the excision of a circular form harboring an attI site and leads to a chromosome carrying an attB empty site. After transfer, the circular ICE integrates into a replicon, mainly at the 3′ end of a tRNA or protein-encoding gene (4, 5). Elements called integrative and mobilizable elements (IMEs), or mobilizable transposons, encode their own excision and integration but only for some of the functions needed for their conjugative transfer (5, 16). Most of them carry an origin of transfer (oriT) and genes encoding one to five mobilization proteins, including a relaxase, but use the mating apparatus of other conjugative elements (mobilization in trans). In ICEs and IMEs, genes involved in the same function, for example, conjugation or regulation, are grouped within modules. New genetic elements may appear by the acquisition or deletion of these modules. In Streptococcus thermophilus, GIs called cis mobilizable elements (CIMEs) were shown to derive from ICEs by the deletion of conjugation and integration modules, while the attL and attR recombination sites were retained (12). A related ICE may integrate by site-specific recombination between its attI site and one of the att sites flanking the CIME, a phenomenon called accretion (12). Such accretion would lead to a composite ICE corresponding to an attL-CIME-attI-ICE-attR structure.
We thoroughly investigated the eight sequenced genomes of S. agalactiae to identify putative ICEs and related elements. First, homologs of integrases found in ICEs of firmicutes were searched using the tBLASTn and BLASTn programs (expected value < 10−5). Second, the limits of the elements were determined by searching direct repeats of the end of the gene in which the element is inserted and by genomic comparison of the eight strains. The integrase gene was considered to be at the right end of the element. Recombination sites attR, attL, and internal attI, which include the direct repeats and adjacent sequences required for integrase attachment, were deduced from sequence alignments between related elements. Third, a putative oriT, similar to the previously described oriT of ICEBs1 (11) and Tn916 (15), was searched in the relaxase gene region. Finally, the putative nature of the elements (ICE, IME, or CIME) was inferred by identifying putative recombination and/or transfer functions using BLASTp and PSI-BLAST.
We identified 48 genetic elements corresponding to putative ICEs or related elements in the sequenced genomes (Fig. 1). This corresponds to two-thirds of the 69 regions of diversity defined by Tettelin et al. (19). Since some elements are present in different strains or exist as several copies, a total of 35 putative ICEs or related elements were distinguished: 12 putative ICEs, 6 putative IMEs, 13 putative CIMEs, and 4 other elements. Whereas the initial definition of a CIME did not include the presence of a complete recombination module (12), among the 13 putative CIMEs, 6 include an integrase gene and thus could be excisable. It will perhaps be worth creating a novel category of elements to reflect these new characteristics. Each strain carried a unique combination of four to eight elements. All these elements are thoroughly described in Table S1 in the supplemental material. The identified elements are inserted at 15 different loci. Most of them are integrated in a unique specific site which corresponds to the 3′ end of a tRNA gene (tRNALeu, tRNAThr, or tRNALys), a gene encoding a ribosomal protein (rpsI, rplL, or rpmG-3), or the guaA gene. The exceptions are Tn916 and TnGBSs (a family of ICEs encoding a DDE transposase [to be published elsewhere]), both of which were detected at four different sites.
FIG. 1.
Chromosome map indicating the positions of ICEs, IMEs, and CIMEs in the eight analyzed genomes of S. agalactiae. ICEs appear as circles, IMEs as triangles, CIMEs as squares (marked by an X when they include an integrase gene), and other GIs as crosses. The color of the symbol indicates the strain which harbors the element. Elements which are identical in several strains appear side by side on the same line. The origin of replication of the chromosome is indicated, as are the genes in which the site-specific integrated elements are inserted.
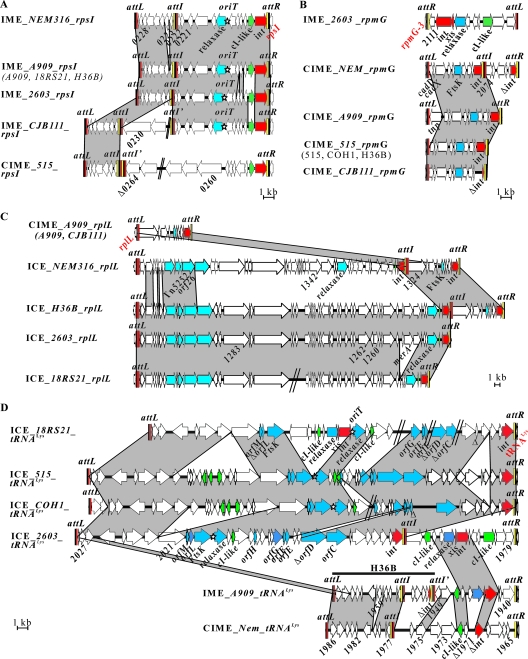
Elements integrated at the rpsI, rpmG-3, rplL, and tRNALys loci nicely illustrate the plasticity and diversity of the elements, so we chose to focus on these elements here. First, four different putative IMEs and one putative CIME were found at the rpsI locus (Fig. 2A). Their integrase exhibits 87% identity with one of the GIs integrated at the rpsI site in Streptococcus pyogenes MGAS9429. Second, seven strains harbor either a putative IME or four different putative CIMEs integrated into the 3′ end of the rpmG-3 gene (Fig. 2B). Their integrase exhibits 87% identity with one of the GIs inserted at the same locus in S. pyogenes MGAS10270. Third, four putative ICEs and one putative CIME were also identified at the 3′ end of the rplL gene (Fig. 2C). Many proteins encoded by these elements have closely related homologs in S. pyogenes, Streptococcus suis, Streptococcus pneumoniae, and Streptococcus dysgalactiae subsp. equisimilis (see Table S1 in the supplemental material). Finally, four putative ICEs, a putative IME, and two putative CIMEs were characterized at the 3′ end of a tRNALys gene (Fig. 1 and 2D). These ICEs are closely related to the putative ICE RD2 of S. pyogenes (9), to the putative ICE SmuE of Streptococcus mutans (5), and to ICESt3 of S. thermophilus (5, 12) (see Table S1 in the supplemental material).
FIG. 2.
Open reading frame (ORF) organization and comparison of the elements inserted into the 3′ end of the rpsI gene (A), the rpmG-3 gene (B), the rplL gene (C), and the tRNALys gene (D) in the eight analyzed genomes. To allow unambiguous denomination of the elements and to reflect their diversity, we chose to give them a name that includes the putative nature of the element (ICE, IME, CIME, or other GI), the host strain, and its insertion site. Elements differing by at least one ORF or displaying different putative mobility properties were given distinct names. When elements sharing identical gene contents (nucleic identity higher than 95%) were identified in more than one strain, we included in the name of the element only the name of a representative strain (usually one with a completely sequenced genome), and the other strains are cited in parentheses. CIME_H36B_tRNALys, which corresponds to the attL-attI′ part of IME_A909_tRNALys, is indicated by a horizontal black line above the latter. Due to a lack of information, a few elements do not appear on the figure: GI_COH1_rpsI, GI_COH1_rplL, and GI_CJB111_tRNALys. ORFs appear as arrows labeled with the number given in the annotated genomes (with a delta, if truncated) (8, 19, 20). For more clarity, only the names of the genes used as probes for the macroarray (Fig. 3) are indicated. Genes where the element is integrated are indicated in red. Integrase genes are indicated as “int” (red arrow) and excisionase genes as “xis.” Genes of the conjugation module (the gene encoding an FtsK-related protein, the relaxase gene, orfC, and orfD, etc., are indicated with blue arrows) and regulation module (cI-like repressors with green arrows) were named according to their putative function or similarity to the ORF of Tn5252 (2) or ICESt1/St3 of S. thermophilus (5). Putative oriT is indicated by a star. Recombination sites are drawn as vertical rectangles. Black rectangles indicate identical sequences found in attL, attR, and attI sites; yellow rectangles indicate the arm of attR sites and the related arm of attI sites; and red rectangles indicate the arm of attL sites and the related arms of attI sites. Protein identity higher than 80% is indicated in gray. Gaps in the genome due to missing contigs are indicated by a double slash. The scale of each island is indicated below each diagram.
Twelve of the elements integrated at the rpsI, rpmG-3, rplL, and tRNALys loci carry one or two putative attI internal sites. Therefore, these elements are composite (Fig. 2) and likely result from site-specific accretions. Accretion was proposed only for ICEs and related CIMEs in S. thermophilus (12). However, the ICEs SXT from Vibrio cholerae and pSAM2 from Streptomyces ambofaciens can transfer to a recipient strain and integrate into the att site of a closely related resident ICE, leading to the accretion of the two ICEs (10, 13). Reanalysis of published data also suggests the existence of accretion events for several other ICEs (12). Accretion by site-specific recombination could thus be a key mechanism of evolution of ICEs and related elements.
Excision of the elements carrying an integrase gene was tested by PCR. Genomic DNA was extracted as previously described (14). Fragments containing attI attachment sites resulting from the excision of circular forms were amplified and sequenced. A second PCR was done to amplify the empty chromosomal insertion site (attB). The sequence of oligonucleotides is provided in Table S2 in the supplemental material. Circular forms were detected for five elements, in particular for the elements integrated at the rpsI, rplL, and tRNALys loci. However, subsequent conjugal transfer of some putative ICEs (elements inserted at the rplL and tRNALys loci except in strain 515) after excision could be compromised due to truncated conjugation genes. A trans mobilization could occur for the six IMEs identified, in particular for the one which was shown to excise as a circular form.
DNA array analysis was conducted for 36 strains belonging to various multilocus sequence type groups (see Table S3 in the supplemental material) in order to study the prevalence and diversity of ICE-related elements in S. agalactiae. Probes correspond to the whole-genome set of the NEM316 strain (3) and to 383 genes located in variable regions and absent in at least one of the eight sequenced genomes. Oligonucleotides are listed in Table S4 in the supplemental material. Labeling, hybridization, and detection were performed as previously described (3). Elements related to those characterized in sequenced genomes were found in several other strains (see Table S5 in the supplemental material), in particular, elements identified at the rpsI, rpmG-3, rplL, and tRNALys loci, which were detected in most of the 36 strains (Fig. 3). Hybridization patterns obtained for elements integrated at the tRNALys locus appear highly mosaic, reflecting the diversity revealed by sequence comparison (Fig. 3). Distribution patterns of ICEs and related elements in S. agalactiae indicated that most of them were identified in unrelated strains, suggesting that they were laterally transferred.
FIG. 3.
DNA array hybridizations of genomic DNA of 36 strains (including the eight strains with sequenced genomes) belonging to different genetic lineages, using probes specific for elements inserted at the rpsI, rpmG-3, rplL, and tRNALys loci. Positive signals are indicated in gray. The positions of the probes are indicated in Fig. 2.
ICEs and related elements, thus, are major contributors to the overall diversity of the genome of S. agalactiae and probably participate in horizontal gene transfer with other species. Whereas only some ICEs have been characterized, previous in silico analyses revealed that numerous bacterial GIs could be ICEs (4, 7, 18). Only a few IMEs have been described (1, 6, 17), probably because they are difficult to detect, and examples of CIMEs are even scarcer (12). However, it can be anticipated that a meticulous examination of bacterial genomes would likely reveal ICEs, IMEs, and CIMEs in most of them. Through their plasticity and potential for gene acquisition and transfer, these elements probably play a crucial role in bacterial genome evolution and gene exchange with other species.
Supplementary Material
Acknowledgments
M.B. acknowledges support by the CNRSI (Caisse Nationale du Régime Social des Indépendants).
We thank Pascal Rainard for providing strains from the collection of the INRA-Nouzilly and Isabelle Rosinski Chupin for fruitful discussions.
Footnotes
Published ahead of print on 15 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, V., D. Lyras, K. A. Farrow, and J. I. Rood. 2002. The clostridial mobilisable transposons. Cell. Mol. Life Sci. 592033-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alarcon-Chaidez, F., J. Sampath, P. Srinivas, and M. N. Vijayakumar. 1997. Tn5252: a model for complex streptococcal conjugative transposons. Adv. Exp. Med. Biol. 4181029-1032. [DOI] [PubMed] [Google Scholar]
- 3.Brochet, M., E. Couvé, M. Zouine, T. Vallaeys, C. Rusniok, M. C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 81227-1243. [DOI] [PubMed] [Google Scholar]
- 4.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46601-610. [DOI] [PubMed] [Google Scholar]
- 5.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 4877-97. [DOI] [PubMed] [Google Scholar]
- 6.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 551911-1924. [DOI] [PubMed] [Google Scholar]
- 7.Gaillard, M., T. Vallaeys, F. J. Vorhölter, M. Minoia, C. Werlen, V. Sentchilo, A. Pühler, and J. R. van der Meer. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 1881999-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couvé, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 451499-1513. [DOI] [PubMed] [Google Scholar]
- 9.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. LeFebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192760-770. [DOI] [PubMed] [Google Scholar]
- 10.Hochhut, B., J. W. Beaber, R. Woodgate, and M. K. Waldor. 2001. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 1831124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, C. A., and A. D. Grossman. 2007. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 1897254-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guédon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150759-774. [DOI] [PubMed] [Google Scholar]
- 13.Possoz, C., C. Ribard, J. Gagnat, J. L. Pernodet, and M. Guerineau. 2001. The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol. Microbiol. 42159-166. [DOI] [PubMed] [Google Scholar]
- 14.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, A. MacGowan, J. McLauchlin, and P. Courvalin. 1992. Genetic basis of tetracycline resistance in clinical isolates of Listeria monocytogenes. Antimicrob. Agents Chemother. 36463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocco, J. M., and G. Churchward. 2006. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J. Bacteriol. 1882207-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salyers, A. A., N. B. Shoemaker, and L.-Y. Li. 1995. In the driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J. Bacteriol. 1775727-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoemaker, N. B., G.-R. Wang, and A. A. Salyers. 1996. The Bacteroides mobilizable insertion element, NBU1, integrates into the 3′ end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J. Bacteriol. 1783594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.te Poele, E. M., M. Samborskyy, M. Oliynyk, P. F. Leadlay, H. Bolhuis, and L. Dijkhuizen. 2008. Actinomycete integrative and conjugative pMEA-like elements of Amycolatopsis and Saccharopolyspora decoded. Plasmid 59202-216. [DOI] [PubMed] [Google Scholar]
- 19.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 10213950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 9912391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.