Abstract
The capacity to both ferment and oxidize l-ascorbate has been widely documented for a number of enteric bacteria. Here we present evidence that all the strains of Klebsiella pneumoniae tested in this study ferment l-ascorbate using the ula regulon-encoded proteins. Under aerobic conditions, several phenotypes were observed for the strains. Our results showed that the yiaK-S system is required for this aerobic metabolic process. Gel shift experiments performed with UlaR and YiaJ and probes corresponding to the specific promoters indicated that l-ascorbate-6-phosphate is the effector molecule recognized by both regulators, since binding of the repressors to their recognition sites was impaired by the presence of this compound. We demonstrated that in K. pneumoniae cells l-ascorbate-6-phosphate is formed only by the action of the UlaABC phosphotransferase system. This finding explains why strains that lack the ula genetic system and therefore are unable to form the inducer intracellularly cannot efficiently use this vitamin as a carbon source under either anaerobic or aerobic conditions. Thus, efficient aerobic metabolism of l-ascorbate in K. pneumoniae is dependent on the presence of both the yiaK-S and ula systems. The expression of the yiaK-S operon, but not the expression of the ula regulon, is controlled by oxygen availability. Both systems are regulated by the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex and by IHF.
Several studies have described the ability of a number of enteric bacteria to both ferment and oxidize l-ascorbate (7, 31, 33). Dissimilation of l-ascorbate by Escherichia coli under anaerobic conditions has been extensively documented (4, 5, 32, 34) and has been shown to be carried out by proteins encoded by the ula regulon (Fig. 1D). The ula system of E. coli is formed by two divergently transcribed operons (5): the ulaG operon, which is thought to encode the l-ascorbate-6-phosphate lactonase (32), and the ulaA-F operon, which encodes the three components of the l-ascorbate phosphotransferase transport system (UlaABC) (34), as well as three catabolic enzymes (UlaDEF) (32). The UlaA, UlaB, and UlaC gene products (formerly designated SgaT, SgaB, and SgaA, respectively) are involved in the uptake and phosphorylation of l-ascorbate (34). Intracellular l-ascorbate-6-phosphate may be transformed by l-ascorbate-6-phosphate lactonase to 3-keto-l-gulonate-6-phosphate. It has been proposed that this compound is decarboxylated by UlaD to l-xylulose-5-phosphate, which is then converted to d-xylulose-5-phosphate by the sequential action of UlaE (having 3-epimerase activity) and UlaF (having 4-epimerase activity) (32). Thus, the functions of the gene products of the ula system are transport of l-ascorbate and transformation of this compound into d-xylulose-5-phosphate (Fig. 1E) (32), which is subsequently metabolized by the pentose phosphate pathway. The ula regulon is under the control of the UlaR repressor (4, 5), which belongs to the DeoR repressor family (http://us.expasy.org/uniprot/P0A9W0; http://pfam.janelia.org/family?acc=PF00455).
FIG. 1.
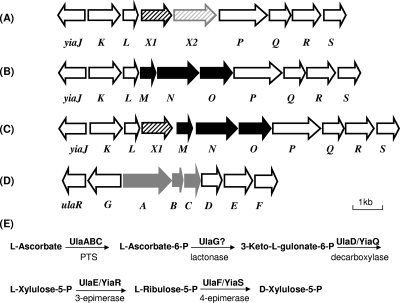
Gene organization of the yiaK-S operon and the ulaA-F regulon. (A) yiaK-S operon of K. pneumoniae strain ATCC 13822, which contains yiaX2 encoding a secondary transport protein (arrow with gray stripes) and yiaX1 encoding a chemotaxislike protein (arrow with black stripes). (B) yiaK-S operon of E. coli K-12, which contains the yiaMNO genes encoding a binding protein-dependent secondary transporter (24) (black arrows). (C) yiaK-S operon of S. enterica serovar Typhimurium, which encodes a chemotaxis-like protein (yiaX1 gene) (arrow with black stripes) and a binding protein-dependent secondary transporter (yiaMNO genes) (black arrows). (D) ulaA-G regulon of E. coli K-12, which encodes a phosphotransferase system (PTS) (ulaABC genes) (gray arrows). The arrows show the extent and direction of transcription of the genes. (E) Metabolic map of l-ascorbate utilization reported by Yew et al. (32).
The available information, also obtained using E. coli, indicates that the yiaK-S operon participates in the aerobic dissimilation of l-ascorbate. This large operon (nine genes), which has an unknown function, has been studied for several years in our laboratory (12, 13, 26), and the results have permitted us to define the participation of yiaK-S in l-ascorbate metabolism. We now know that the aerobic dissimilation of l-ascorbate involves three paralogous proteins, UlaD/YiaQ, UlaE/YiaR, and UlaF/YiaS, which are a decarboxylase, a 3-epimerase, and a 4-epimerase, respectively. In contrast, l-ascorbate enters the cells through the ula-encoded phosphotransferase transport system and not through the yiaMNO-encoded ABC transporter (6). Proteomic analysis showed that there was enhanced expression of the alkyl hydroperoxide reductase encoded by ahpC, indicating that there was a response to oxidative stress generated in the aerobic metabolism of l-ascorbate (2). Control of ahpC expression by the OxyR global regulator in response to the l-ascorbate concentration is consistent with the formation of hydrogen peroxide in the medium (6). The presence of certain amino acids, such as proline, threonine, or glutamine, in the culture medium allowed aerobic growth of E. coli cells on l-ascorbate. This effect could be explained by the ability of these amino acids to allow yiaK-S operon induction, thus increasing the metabolic flux of l-ascorbate dissimilation (6). Alternatively, these amino acids may decrease the rate of l-ascorbate oxidation. Transcriptional fusions and proteomic analysis indicated that both the ula regulon and the yiaK-S operon are required for the aerobic utilization of this compound in the presence of casein acid hydrolysate (CAA) in E. coli (6).
Although l-ascorbate metabolism in E. coli has been studied, to date there is no information concerning this metabolic process in the enterobacterium Klebsiella sp. Here we present evidence that only strains of K. pneumoniae carrying the yiaK-S and ula genetic systems have the capacity to grow on l-ascorbate under aerobic conditions. We also found that l-ascorbate-6-phosphate generated during the internalization of l-ascorbate by the UlaABC phosphotransferase system is the inducer of both the ula regulon and the yiaK-S operon.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The genotypes and sources of the bacterial strains and plasmids are shown in Table 1. Rifampin (Rf)- or streptomycin (Sm)-resistant derivatives of K. pneumoniae ATCC 13882 were obtained by spontaneous mutation.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tcr)] | Stratagene |
| JA134 | ECL1 Lyx+ | 26 |
| JA184 | JA134 himA::cat | 12 |
| S17-1(λ-pir) | Tpr SmrrecA thi pro hsdR−M+RP4:2-Tc:Mu:Km Tn7 λpir | Biomedal |
| Klebsiella strains | ||
| K. pneumoniae ATCC 13882 | K. pneumoniae subsp. pneumoniae deposited as K. aerogenes | ATCC |
| K. pneumoniae ATCC 13883 | K. pneumoniae subsp. pneumoniae | ATCC |
| K. pneumoniae KC2653 | hutC515 dad-1 Δ[bla]-2 str-6 | 16 |
| K. oxytoca | Wild type | ATCC |
| K. pneumoniae C3 | O1:K66 | M. Regué |
| EB6193 | RP4-2 tet:Mu-1 Kan::Tn7 integrant leu-63::IS10 recA1 creC510 hsdR17 endA1 zbf-5 uidA (ΔMuI):pir+thi Spr/Smr | R. A. Bender |
| Plasmids | ||
| pBluescript | AprlacZ | Stratagene |
| pGEM-T | AprlacZYA | Promega |
| pMAL-C2x | AprmalE α-lacZ lacIq | Biolabs |
| pMAL-C2x-yiaJ | yiaJ in pMAL-C2x | This study |
| pMAL-C2x-ulaR | ulaR in pMAL-C2x | This study |
| pRS415 | AprlacZYA | 27 |
| pCB1583 | Apr Kmr oriR6K mobRP4 rpsL rbsA′ lacZ rbsK′ | R. A. Bender |
| pUTmini-Tn5 Km | Apr oriR6K mobRP4 tnp* mini-Tn5 Km | Biomedal |
| pUTmini-Tn5 Tc | Apr oriR6K mobRP4 tnp* mini-Tn5 Tc | Biomedal |
| pCAT19 | Apr Tn9-CAT(Cmr) | 8 |
Growth conditions.
Cells were grown in Luria broth (LB) or minimal medium and harvested as described previously (3). l-Ascorbate or glucose was added to a basal inorganic medium (3) at a concentration of 10 mM for aerobic growth and at a concentration of 20 mM for anaerobic growth. CAA was routinely used at a concentration of 0.5%. Other carbon sources were used at final carbon concentrations of 60 mM under aerobic conditions and 120 mM under anaerobic conditions. To increase the cell yield under anaerobic conditions, 20 mM nitrate was added to the cultures (32). When required, antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 30 μg/ml; kanamycin (Km), 50 μg/ml; Rf, 25 μg/ml; Sm, 10 μg/ml; and tetracycline (Tc), 12 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) and isopropyl-β-d-thiogalactoside (IPTG) were used at concentrations of 30 and 10 μg/ml, respectively.
β-Galactosidase activity.
To determine β-galactosidase activity, cells were grown at 30°C to an optical density at 600 nm of 0.5, collected by centrifugation, washed in 1% KCl, and suspended to obtain a concentration of 1 to 1.5 mg of protein per ml. β-Galactosidase activity was assayed using detergent-treated whole cells and ο-nitrophenyl-β-d-galactopyranoside as the substrate and was expressed in U/mg of cell protein. One unit of β-galactosidase activity corresponded to the amount of enzyme that hydrolyzed 1 nmol of ο-nitrophenyl-β-d-galactopyranoside per min. The data reported below are averages of at least four separate experiments performed in triplicate and are expressed as means ± standard deviations. The protein concentration was determined by the method of Lowry et al. (17) with bovine serum albumin as a standard.
DNA manipulation and sequencing.
Bacterial genomic DNA was obtained using a Wizard genomic DNA purification kit (Promega), and plasmid DNA was prepared using the Wizard Plus SV Midipreps DNA purification system (Promega). DNA manipulations were performed essentially as described by Sambrook and Russell (25). DNA fragments were amplified by PCR using chromosomal DNA as the template. When necessary, specific restriction sites were incorporated at the 5′ ends of the primers to facilitate cloning of the fragments in the appropriate vector. PCRs were performed with PFU DNA polymerase under standard conditions. DNA was sequenced using an automated ABI 377 DNA sequencer and fluorescent dye termination methods.
For Southern blot analysis, DNA was digested with HindIII, SmaI, KpnI, BamHI, and EcoRI and (for double digestion) with EcoRI/SmaI, HindIII/SmaI, BamHI/SmaI, and KpnI/SmaI. Restriction fragments were separated by agarose gel electrophoresis, blotted onto nylon filters, and fixed by incubation at 80°C for 2 h. DNA hybridization was performed using digoxigenin (DIG)-labeled fragments.
Mini-Tn5 Km-1 random mutagenesis.
Random Tn5 insertion mutants of K. pneumoniae were obtained by conjugation using the pUTmini-Tn5 Km delivery vector (Biomedal, Spain). The recipient strain, K. pneumoniae ATCC 13882 Rfr, and the donor strain, E. coli S17-1(λpir) harboring pUTmini-Tn5 Km, were grown overnight at 30°C in LB. Samples (50 μl) of each culture were mixed in 1 ml of sterile 10 mM MgSO4, and cells were then collected by centrifugation and resuspended in 20 μl of 10 mM MgSO4. A drop containing the resuspended cells was placed on an LB plate and incubated overnight at 30°C. Cells grown on the plate were then suspended in 1 ml of 10 mM MgSO4, and different amounts of the suspension were plated on LB containing Rf and Km. This medium counterselected the donor strain and selected recipient cells carrying the Km transposon marker. l-Ascorbate mutants were subsequently selected by replica plating on medium containing 10 mM l-ascorbate.
Mini-Tn5 insertions were mapped by inverse PCR. The protocol involved isolation of genomic DNA, digestion with restriction enzyme HhaI, which cut within the transposon and numerous times within the genome, and ligation to circularize all linear genomic fragments, followed by PCR using two outward-facing, transposon-specific primers, Imini-Tn5A (5′-CTCGCTAGATTGTTAATGCG-3′) and Imini-Tn5B (5′-GCTTGCTCAATCAATCACCG-3′). Amplified products were sequenced, and homology to genes and open reading frames deposited in databases was determined using the BLAST search algorithm at the National Center for Biotechnology Information. Following the computational analysis, the sites of transposon insertion in 10 isolated mutants were identified.
Directed mutagenesis of ulaA and yiaX1.
To create a UlaA-defective mutant of K. pneumoniae ATCC 13882, a 1,340-bp fragment encompassing ulaA was amplified with primers ulaACm-Fw (5′-GCGGAATTCGCGGCCGCTCTACACCATGGGAGTGTGTTATGG-3′) and ulaACm-Rv (5′-GCGGGATCCGCGGCGCACGGCCATAACGCCAGCCCGATGAGCG-3′), which contained NotI (bold type) and EcoRI or BamHI (underlined) restriction sites at their 5′ ends. The product was digested with BamHI and EcoRI and cloned into pBluescript, yielding pBS-ulaA. Recombinant plasmid pBS-ulaA was then digested with PstI (with a single restriction site in ulaA) and ligated with a Cm cassette amplified from plasmid pCAT19 (8) using primers CmPstI-Fw (5′-AACTGCAGTGTGACGGAAGATCACTTCG-3′) and Cm-PstI-Rv (5′-AACTGCAGACCAGGCGTTTAAGGGCACC-3′) having PstI restriction sites (underlined) at their 5′ ends. Recombinant plasmid pBS-ulaA::cat was then digested with NotI to obtain the insert corresponding to mutated ulaA::cat. This fragment was cloned in the NotI restriction site of pUTmini-Tn5 Tc and introduced into E. coli S17-1(λpir) by electroporation. To introduce the ulaA::cat mutation into the K. pneumoniae chromosome, conjugation was performed with Rf-resistant derivatives of K. pneumoniae ATCC 13882 and ATCC 13882 yiaL::Tn5Km as recipient strains. Transconjugants having the Rfr Cmr Tcs and Kmr Cmr Tcs phenotypes were selected, respectively. Chromosomal insertions were confirmed by performing PCR.
To construct a YiaX1-deficient mutant, the yiaX1 gene was amplified using primers yiaX1-mut-Fw (5′-CGCGCGGCCGCAGCATTATTTTCAGGAGCACATTATGAAAA-3′) and yiaX1-Rv (5′-CGCGCGGCCGCTTGTTGTGCTTTATTTAAGGCAGCGATCCC-3′) and cloned in plasmid pGEM-T (Promega). yiaX1 was disrupted as described above for ulaA. In this case the Cm cassette was introduced into the HindIII restriction site present in the yiaX1 coding region. The recipient strain was K. pneumoniae ATCC 13882 Rfr, and mutants were selected by using the Rfr Cmr Tcs phenotype. Chromosomal insertions were confirmed by performing PCR.
Mapping of the 5′ end of the yiaK-S transcript.
The 5′ region of the yiaK-S transcript was determined by the rapid amplification of cDNA 5′ ends (RACE) method (25) using a commercial 5′-RACE kit (Roche Diagnostics, GmbH). Total RNA was isolated from K. pneumoniae ATCC 13882 cells grown aerobically to an optical density at 600 nm of 0.5 with l-ascorbate as the sole carbon source using a Qiagen RNeasy total RNA kit and then treated with RNase-free DNase. The cDNA was transcribed from the RNA with the specific yiaK antisense oligonucleotide 5′-GGATGATATCGCCATTGTCC-3′. A homopolymeric deoxyribosyladenine tail was added (via terminal transferase) to the 3′ terminus of the yiaK cDNA. Amplification of reverse transcription products was performed with nested yiaK-specific primer 5′-AAGCAGTACCTGATTGAAGG-3′ and an oligo(dT) anchor primer. The products obtained were cloned into a pGEM vector for sequencing and subsequent manipulation.
Construction of lacZ transcriptional fusions.
Transcriptional fusions were constructed by inserting the promoter fragments into plasmid pRS415 (27). This plasmid carries a cryptic lacZ operon and confers resistance to Ap. To construct Φ(yiaK-lacZ), the 350-bp fragment encompassing the yiaJ-yiaK intergenic region was amplified by PCR with primers YiaKprom-Fw (5′-CCGGAATTCAGCA TCAGTCCACGGAACAG-3′) and YiaK-prom-Rv (5′-GCGGGATCCAAGCAGTA CCTGATTGAAGG-3′), digested with BamHI and EcoRI (restriction sites underlined), and cloned into plasmid pRS415. Plasmid DNA was sequenced to ensure that the fragment was inserted in the correct orientation and that no mutations had been introduced during the amplification reaction. This promoter fusion comprised 299 bp upstream of the translational start site of yiaK. To transfer a single copy of Φ(yiaK-lacZ) to the K. pneumoniae chromosome, the recombinant plasmid was first digested with EcoRI and SacI, and the fragment containing the promoter fusion was subcloned into plasmid pCB1583 (16). pCB1583 is a λpir-dependent plasmid whose lacZ gene is flanked by genes of the K. pneumoniae d-ribose operon, thus allowing integration of the cloned fusion by homologous recombination into the d-ribose operon of the K. pneumoniae recipient strain. The recombinant plasmids containing Φ(yiaK-lacZ) were selected after transformation of strain EB6193 as blue colonies on LB plates containing X-Gal and Km and were then introduced into E. coli S17-1(λ pir) by electroporation. To transfer Φ(yiaK-lacZ) into the K. pneumoniae chromosome, conjugation was performed with the Rf- and Sm-resistant derivative of K. pneumoniae strain ATCC 13882. After several rounds of selection in several growth media, stable recombinants were isolated by selecting recombinants having a Kms Smr d-ribose-negative phenotype.
To construct Φ(yiaJ-lacZ), we used the same approach, except that the 350-bp fragment encompassing the yiaJ-yiaK intergenic region was cloned in pRS415 in the opposite direction.
Transcriptional fusions corresponding to ulaA and ulaG were obtained by the same procedure. In this case the 408-bp fragment encompassing the ulaG-ulaA intergenic region was amplified using the YJFR EcoRI primer (5′-GCGGAATTCAGGATTCATGATTCACGGG-3′) and the ULAA BamHI primer (5′-TGCTCAGGATCCAACACACTCCCGATGGTG-3′) to construct Φ(ulaG-lacZ) and with the YJFR BamHI primer (5′-TGCTCAGGATCCAGGATTCATGATTCACGGG-3′) and the ULAA EcoRI primer (5′-GCGGAATTCAACACACTCCCGATGGTG-3′) to construct Φ(ulaA-lacZ) (BamHI and EcoRI sites are underlined).
DNA binding studies.
For electrophoretic mobility shift assays, several PCR-amplified fragments were used as probes. The fragments used in the gel shift experiments were labeled with T4 polynucleotide kinase, [γ-32P]ATP (3,000 Ci/mmol; Amersham Pharmacia Biotech), or terminal transferase and DIG-ddUTP by following the manufacturer's instructions (Roche). A nonradioactive DIG gel shift kit for 3′ end labeling of DNA fragments (Roche Applied Science, Indianapolis, IN) was used for protein-DNA binding assays when crude extracts were used as the protein source. The radioactive probes were used in the gel shift experiments performed with purified YiaJ and UlaR proteins. Labeled DNA fragments were incubated with purified YiaJ or UlaR or with crude extracts, obtained as described by Nunoshiba et al. (20), in 10 mM Tris-HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 10% glycerol, 2 mM dithiothreitol in a 20-μl (total volume) mixture. Poly(dI-dC) was used as a nonspecific competitor. Where indicated, l-ascorbate intermediate metabolites were added to binding reaction mixtures at different concentrations up to 50 mM. The l-ascorbate-6-phosphate used in the gel shift experiments was chemically synthesized by reacting 6-bromo-6-deoxy-l-ascorbic acid with the hydrogen phosphate ion at 25°C, essentially as described by Liao et al. (15).
The binding mixtures were incubated for 20 min at 25°C and loaded onto a prerun 5% native polyacrylamide gel containing 10% glycerol in 1× Tris-borate-EDTA buffer. When probes were labeled with DIG, blotting was performed using a Bio-Rad electroblotting system (model Trans blot) by following the manufacturer's instructions. Chemiluminescence of DIG-labeled DNA-protein complexes on the nylon membranes was detected using Hyperfilm ECL (Amersham Pharmacia).
Expression and purification of YiaJ and UlaR.
YiaJ and UlaR were purified using the malE fusion system. For this, yiaJ was amplified by PCR with primers YiaJ-EcoRI-Fw (5′-CGCGAATTCATGGTGATGAAAGAGAGCGAGAC-3′) and YiaJ-BamHI (5′-CGCGGATCCTCATTTGCCTGACGCTTCACGCACCGAAACC-3′) and cloned into the BamHI and EcoRI restriction sites of plasmid pC2x, yielding plasmid pC2x-yiaJ. Primer YiaJ-EcoRI-Fw was designed to fuse the ATG start codon of yiaJ in frame with the malE gene. UlaR was obtained using the same procedure with primers UlaR-EcoRI-Fv (5′-CGCGAATTCATGACGGAAGCGCAAAGGCA-3′) and UlaR-BamHI-Rv (5′-CGCGGATCCTTAAACGCGGATTACGCTGA-3′). (BamHI and EcoRI sites are underlined.)
Overproduction of the MalE-YiaJ or MalE-UlaR fusion protein in strain XL1-Blue carrying recombinant plasmid pC2x-yiaJ or pC2x-ulaR was achieved after induction by IPTG (0.3 mM) in LB containing glucose and Ap for 3 h at 37°C. The fusion proteins were then purified by affinity chromatography with amylose resin (New England BioLabs, United States) by following the manufacturer's instructions. The fusion proteins were digested with factor Xa by incubating them at room temperature for 12 h, and the cleaved proteins were used in gel shift experiments.
Nucleotide sequence accession number.
We deposited the sequence corresponding to the yiaJ-S region of K. pneumoniae ATCC 13882 in the GenBank database under accession number DQ516067 (identification code GI:98989859).
RESULTS
Utilization of l-ascorbate by K. pneumoniae.
To test the abilities, of different Klebsiella strains to utilize l-ascorbate, cells were inoculated onto solid or liquid medium containing this compound as a sole carbon source and incubated under either aerobic or anaerobic conditions. All the strains tested were able to grow at similar rates and to produce similar yields on l-ascorbate under anaerobic conditions (not shown). However, under aerobic conditions, only the following three organisms showed growth on l-ascorbate: K. pneumoniae ATCC 13882, K. pneumoniae C3, and K. oxytoca (Table 2). Although these three organisms grew well on l-ascorbate, they did not grow on the intermediate dihydroascorbate or the intermediate 2,3-diketogulonate as a sole carbon source. This lack of growth may be attributed to the inability of the organisms to transport these compounds inside the cell.
TABLE 2.
Characterization of aerobic growth on l-ascorbate of different Klebsiella strains
| Strain | Optical density at 420 nm | Doubling time (min) |
|---|---|---|
| K. pneumoniae ATCC 13882 | 1.68 | 180 |
| K. pneumoniae C3 | 1.62 | 223 |
| K. oxytoca | 1.59 | 114 |
| K. pneumoniae ATCC 13883 | NGa | NG |
| K. pneumoniae KC2653 | NG | NG |
| K. pneumoniae ATCC 13882 ulaA::cat | <0.40 | 480 |
| K. pneumoniae ATCC 13882 ulaA::cat yiaL::mini-Tn5 Km | NG | NG |
NG, no growth.
Genetic system for l-ascorbate utilization.
To search for K. pneumoniae genes involved in l-ascorbate metabolism under aerobic conditions, we generated a set of mutants selected on the basis of their inability to utilize l-ascorbate in the presence of oxygen. Random Tn5 insertion mutants of strain ATCC 13882 were obtained after conjugation using the pUTmini-Tn5 Km delivery vector and selection on LB plates containing Rf and Km. The l-ascorbate-negative mutants were selected after replica plating on l-ascorbate, and the mutation was mapped and sequenced as described in Materials and Methods. Analysis of this sequence showed that the mini-Tn5 transposons were inserted into genes with unknown functions showing high levels of similarity to yiaL of E. coli or to yiaL, yiaX1, or yiaX2 of K. oxytoca.
The complete sequences of the yiaK-S genetic systems of E. coli (accession number NC_000913) and K. oxytoca (accession number AF282849) are available from GenBank. Comparison of these sequences revealed some differences in genome organization between the two species (Fig. 1). This genetic system was not present in the genome of K. pneumoniae subsp. pneumoniae MGH 78578, whose genome sequence is available from the K. pneumoniae Genome Sequencing Project released to the NCBI GenBank (accession number NC_009648). This observation led us to study the genetic organization of the yiaK-S operon in K. pneumoniae ATCC 13882. For this purpose, using a collection of primers 100% homologous to the E. coli or K. oxytoca genome, we amplified the yiaJ-yiaQ region of strain K. pneumoniae ATCC 13882, and the fragments obtained were subsequently sequenced. For unknown reasons, the region between yiaQ and yiaS was not amenable to amplification when this methodology was used. We therefore constructed a restriction map of the region encompassing the yiaJ-S region of K. pneumoniae ATCC 13882 by Southern blot analysis using different probes of the yiaK-S operon of E. coli. With this information, a BamHI genomic library of strain K. pneumoniae ATCC 13882 in plasmid pBluescript was constructed. Using this library, a plasmid containing sequences homologous to a specific yiaQ probe of E. coli was identified by the method of Grunstein and Hogness (10) and subsequently isolated and sequenced. Sequence analysis confirmed that the gene organization of the yiaJ-S system of K. pneumoniae ATCC 13882 was identical to that of K. oxytoca (accession number AF282849). We deposited the sequence corresponding to the yiaJ-S region of K. pneumoniae ATCC 13882 in the GenBank database. This genetic system is located between a gene encoding a 161-amino-acid protein with an unknown function and displaying similarity to kpn_03937 of K. pneumoniae MGH 78578 or to ysaA of E. coli (encoding a putative electron transport protein with an Fe-S center) and a gene with a high level of similarity to selB of K. pneumoniae MGH 78578.
Given the diversity of this genomic region in the strains of K. pneumoniae, we analyzed the presence of this region in the strains unable to grow on l-ascorbate. Southern blot analysis and PCR amplification of the yiaJ-S region with specific primers for strain K. pneumoniae ATCC 13882 showed that these genes were not present in K. pneumoniae strains ATCC 13883 and KC2653, which were unable to grow aerobically on l-ascorbate.
On the basis of the observation that the ula regulon is present in the genomes of K. oxytoca and K. pneumoniae and on the basis of the role of the ula regulon in the anaerobic metabolism of l-ascorbate in E. coli, we examined the possible role of the ula regulon in the metabolism of l-ascorbate in K. pneumoniae. To this end, we performed directed mutagenesis of ulaA in the genetic backgrounds of strain K. pneumoniae ATCC 13882 and its derivative yiaL mutant strain. The ulaA mutant strains were analyzed by determining their abilities to utilize l-ascorbate under aerobic and anaerobic conditions. Our results indicated that mutations in ulaA impaired the anaerobic metabolism of l-ascorbate in both genetic backgrounds and decreased the aerobic utilization of this compound in plates containing l-ascorbate as the sole carbon source. An analysis of the growth rate and culture yield in liquid medium was also performed. The results showed that the utilization of l-ascorbate under aerobic conditions by the ulaA mutant was slower than the utilization by the wild type and that the yield was very poor (Table 2). The ulaA yiaL double mutant failed to grow aerobically on l-ascorbate (Table 2).
YiaX1 is not required for l-ascorbate metabolism.
To analyze if the yiaX1 gene product is involved in l-ascorbate metabolism, a yiaX1::cat mutant strain was obtained as described in Materials and Methods. This mutant was able to grow aerobically on l-ascorbate with the same duplication time and yield as wild-type strain ATCC 13882, indicating that a lack of the yiaX1 product does not alter l-ascorbate metabolism. At present, the function of YiaX1 remains unknown.
Transcriptional regulation of the yiaK-S operon and the ula regulon.
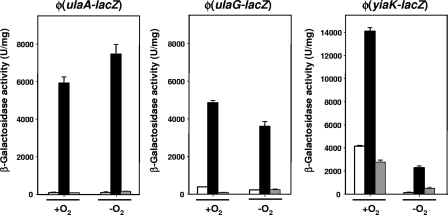
Expression of the yiaK-S system, which is involved in the aerobic utilization of l-ascorbate in K. pneumoniae, was analyzed by monitoring the growth and β-galactosidase activity of strain ATCC 13882 carrying the transcriptional fusions Φ(yiaK-lacZ) and Φ(yiaJ-lacZ) as reporters of the expression of structural and regulator genes, respectively. The results for Φ(yiaK-lacZ) expression indicated that under aerobic conditions, a high basal level of activity was observed under noninducing conditions (around 4,000 U/mg), which increased around fourfold when the organism was grown in the presence of l-ascorbate (Fig. 2). As expected for a genetic system that works only under aerobic conditions, the levels of induction of the structural genes under anaerobic conditions were very low (200 U/mg under noninducing conditions and 2,000 U/mg in the presence of l-ascorbate) and never reached the noninduced levels observed under aerobic conditions (Fig. 2). These results indicated that this operon is induced by growth on l-ascorbate and is positively regulated by oxygen availability. The presence of glucose lowered the β-galactosidase activity to levels below that obtained in the presence of CAA, indicating that there was total inhibition of the expression of the yiaK-S system (Fig. 2). This effect was not observed when the fusion of the regulator gene yiaJ was analyzed (not shown). Thus, the structural genes, but not the regulator gene, appear to be controlled by catabolite repression. Expression of the Φ(yiaJ-lacZ) fusion was not dependent on the presence of l-ascorbate, and the level of constitutive expression was 650 U/mg. This finding is consistent with the role of a transcriptional regulator assigned to the yiaJ gene product.
FIG. 2.
β-Galactosidase activities of Φ(ulaA-lacZ), Φ(ulaG-lacZ), and Φ(yiaK-lacZ) transcriptional fusions in the K. pneumoniae strain ATCC 13882 genetic background. Cells were grown under aerobic or anaerobic conditions with CAA (open bars), l-ascorbate (black bars), or l-ascorbate plus glucose (gray bars). The bars indicate means, and the error bars indicate standard deviations.
To analyze ula regulon expression, a fragment encompassing the ulaG and ulaA intergenic region was prepared, cloned, and introduced into the genetic background of K. pneumoniae strain ATCC 13882, as described in Materials and Methods. Expression οf Φ(ulaG-lacZ) and Φ(ulaA-lacZ) was measured after aerobic or anaerobic growth in medium containing CAA or l-ascorbate. Regardless of oxygen availability, growth in the presence of l-ascorbate clearly induced Φ(ulaG-lacZ) and Φ(ulaA-lacZ) and hence ula regulon expression (Fig. 2).
Mapping of the 5′ end of the yiaK-S transcript.
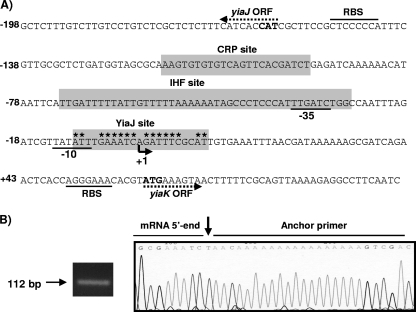
In this study the 5′ end of the yiaK-S mRNA was determined by the 5′-RACE method (25). Several clones were isolated through 5′-RACE of an oligo-nested cDNA pool derived from K. pneumoniae ATCC 13822 cells grown in the presence of l-ascorbate. In all cases analysis of the 5′-RACE products revealed only one transcriptional initiation site (Fig. 3B), which was located 61 nucleotides upstream of the predicted ATG start codon (Fig. 3A). Inspection of the DNA sequences upstream of nucleotide 1, the mRNA start site, revealed the presence of the putative −35 and −10 sequences (TTGATC-17 nucleotides-TATATT) (Fig. 3A).
FIG. 3.
Organization of transcriptional regulatory elements of the yiaK-S system. (A) The yiaK-S promoter sequence is numbered relative to the 5′ end determined in this study, which is indicated by an arrowhead labeled +1. The −10 and −35 promoter sequences of yiaK are underlined. The directions of the yiaJ and yiaK open reading frames (ORF) are indicated by dotted arrows. Predicted ATG start codons are indicated by bold type, and the corresponding ribosome binding sites (RBS) are also indicated. The IHF, cAMP-CRP, and YiaJ binding sites are indicated by shading. The inverted repeat of the YiaJ operator is indicated by asterisks. (B) Identification of yiaK-S 5′ end by sequencing across ligation sites of the 5′-RACE product, shown at the left site. The chromatogram shows the sequences at the ligation site of a typical cloned 5′-RACE product derived from a transcript obtained from K. pneumoniae ATCC 13882 grown on l-ascorbate. The arrow indicates the transcription initiation site.
Binding of YiaJ, CRP, and IHF to the yiaK promoter region.
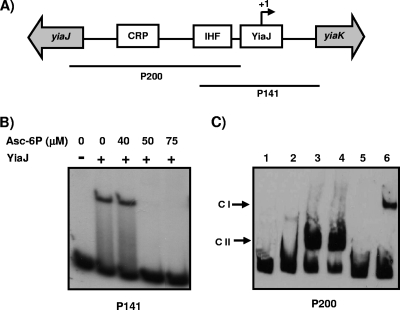
Identification of the 5′ end and analysis of the similarity between the yiaK promoter sequence of K. pneumoniae strain ATCC 13882 and the E. coli promoter sequence allowed us to identify the putative YiaJ binding site overlapping the RNA polymerase binding site. This location is consistent with the repressor role of YiaJ. Putative IHF (positions −72 to −28) and CRP (positions −115 to −100) recognition sites were also identified by computational analysis (Fig. 3A), suggesting that, as reported for E. coli (12), yiaK-S operon expression in K. pneumoniae is regulated by these proteins. The role of YiaJ in the regulation of yiaK-S operon expression was analyzed by performing an electrophoretic mobility shift assay using purified YiaJ and DNA probe P141 (Fig. 4A), which encompasses the predicted YiaJ operator site. A retarded complex was observed with the YiaJ protein, indicating that there was binding of this protein to the P141 fragment. To identify the effector molecule recognized by YiaJ, binding of YiaJ to P141 was also performed in the presence of several compounds, such as l-ascorbate, dihydroascorbate, 2,3-diketogulonate, l-ascorbate-6-phosphate, d-xylulose-5-phosphate, and l-ribulose-5-phosphate. Under our experimental conditions, only l-ascorbate-6-phosphate at concentrations higher than 50 μM impaired the formation of the YiaJ-DNA complex (Fig. 4B), indicating that l-ascorbate-6-phosphate is the effector molecule recognized by YiaJ.
FIG. 4.
Interactions of IHF, CRP, and YiaJ with different fragments of the yiaJ-yiaK intergenic region and role of l-ascorbate-6-phosphate as an inducer effector of YiaJ. (A) Diagram showing the locations of the putative IHF, CRP, and YiaJ binding sites (open boxes) between the yiaJ and yiaK genes (gray arrows) and the fragments used as probes in the gel shift experiments (lines). (B) Binding of YiaJ to probe P141 using 1.5 μg of purified protein in the presence of different amounts of l-ascorbate-6-phosphate (Asc-6P). (C) Binding to probe P200 of 1.5 μg of purified YiaJ (lane 2), 10 μg of crude extract of K. pneumoniae strain ATCC 13882 (lane 3), 10 μg of crude extract of E. coli strain JA134 (lane 4), 10 μg of crude extract of E. coli strain JA184 (lane 5), and 10 μg of crude extract of strain JA184 in the presence of 125 μM cAMP (lane 6). Lane 1 contained a control without protein. CI, cAMP-CRP complex; CII, IHF complex.
The high level of similarity between the IHF proteins of E. coli and K. pneumoniae led us to study the binding of IHF to probe P200 (Fig. 4A) using crude extracts of E. coli strain JA134 and its IHF mutant derivative strain JA184. A retarded complex with strain JA134, but not with the IHF mutant strain, was observed (Fig. 4C). When cAMP at a concentration of 125 μM was added to the binding reaction mixtures containing extracts of strain JA184, an additional complex was detected, indicating that CRP is involved in glucose-mediated catabolite repression (Fig. 4C). Binding of YiaJ was also analyzed with P200, which did not contain the putative YiaJ site. Consistently, no retarded complex was observed in this case (not shown).
Binding of UlaR, CRP, and IHF to the ulaG-ulaA intergenic region.
After PCR amplification and sequencing of the region from ulaG to ulaA, no significant differences were observed between K. pneumoniae ATCC 13882 and K. pneumoniae strain MGH 78578. Computational analysis of the sequence allowed us to identify by electrophoretic mobility similarity to the ulaG-ulaA intergenic region of E. coli the putative UlaR binding sites, two putative CRP sites, and an IHF binding site (Fig. 5A). To test whether UlaR was able to bind to the putative operator sites, we expressed and purified UlaR of K. pneumoniae, as described in Materials and Methods. Two fragments containing the putative UlaR binding sites were obtained and used as probes (P198 and P238) in gel shift experiments (Fig. 5B). In both cases, retarded complexes appeared with these probes, indicating that UlaR bound to the proposed operator sites (Fig. 5C). Since UlaABC of E. coli is a phosphotransferase transport system involved in the uptake of l-ascorbate, which is internalized as l-ascorbate-6-phosphate, we tested this phosphorylated compound as a possible effector molecule of UlaR. Binding experiments performed in the presence of different amounts of l-ascorbate-6-phosphate demonstrated that this compound impaired UlaR binding to the corresponding operators sites when it was used at a concentration higher than 5 mM (Fig. 5C). This observation indicates that this phosphorylated sugar is an effector molecule that is also recognized by UlaR. Other related compounds, such as l-ascorbate, dihydroascorbate, and l-xylulose-5-phosphate, were unable to dissociate the UlaR-DNA complexes (not shown).
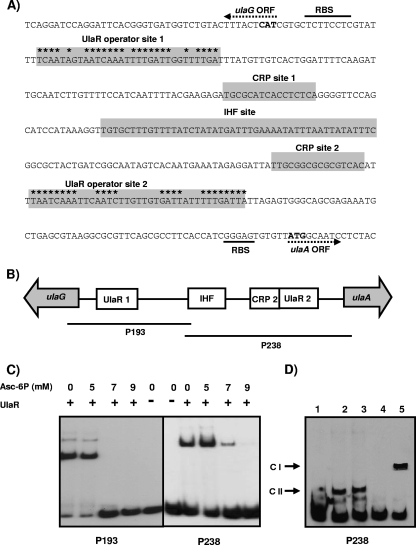
FIG. 5.
Regulatory elements of the ulaA-G regulon. (A) Sequence of the ulaA-G intergenic region. The directions of the ulaA and ulaG open reading frames (ORF) are indicated by dotted arrows. Predicted ATG start codons are indicated by bold type, and the corresponding ribosome binding sites (RBS) are also indicated. The IHF, cAMP-CRP, and UlaR binding sites are indicated by shading. The inverted repeat of the UlaR operator sites is indicated by asterisks. (B) Diagram showing the locations of the putative IHF, CRP, and UlaR binding sites (open boxes) between the ulaA and ulaG genes (gray arrows) and the fragments used as probes in the gel shift experiments (lines). (C) UlaR binding to probes P193 and P238 in the presence of different concentrations of l-ascorbate-6-phosphate (Asc-6P). A control lane with no protein was also included. (D) Complexes of P238 with no protein (lane 1), with 10 μg of crude extract of K. pneumoniae strain ATCC 13882 (lane 2), with 10 μg of crude extract of E. coli strain JA134 (lane 3), with 10 μg of crude extract of E. coli strain JA184 (lane 4), and with10 μg of crude extract of JA184 in the presence of 125 μM cAMP (lane 5).
To study the functionality of the IHF binding site identified (Fig. 5A), gel shift experiments were performed with probe P238 and crude extract of E. coli strain JA134 or its IHF mutant derivative strain JA184. A retarded complex with strain JA134, but not with the IHF mutant strain, was observed, indicating that IHF regulates ula regulon expression in K. pneumoniae (Fig. 5D). To analyze the functionality of the two putative CRP sites, additional experiments were performed using probes P193 and P238 and crude extracts of E. coli strain JA184. Addition of 125 μM cAMP to the binding reaction mixture resulted in a new complex when probe P238 was used (Fig. 5D) but not when P198 was used (not shown). This finding indicates that binding of CRP to CRP site 2 located near ulaA is the only binding that is functional and able to mediate the catabolite repression observed when bacteria are grown on glucose (Fig. 2).
Ascorbate-6-phosphate is generated by the action of the ula system but not by the action of yiaK-S.
Previous experiments performed with crude extracts obtained from K. pneumoniae ATCC 13882 cells grown aerobically on l-ascorbate did not reveal any phosphorylation activity on l-ascorbate in vitro. On the basis of the observation that a ulaA-deficient mutant of K. pneumoniae ATCC 13882 did not grow efficiently on l-ascorbate under aerobic conditions, we tested whether the ulaABC-encoded phosphotransferase system accounted for the intracellular l-ascorbate-6-phosphate. To this end, we analyzed the expression of Φ(yiaK-lacZ) in the genetic background of the ulaA-deficient mutant cells grown aerobically in medium containing CAA in the presence or absence of l-ascorbate. No induction of Φ(yiaK-lacZ) was obtained with the ulaA mutant genetic background since the same level of β-galactosidase activity (around 4,000 U/mg) was obtained under both conditions.
DISCUSSION
The results of the growth experiments with l-ascorbate described in this report reflect the great variety of phenotypes in the genus Klebsiella. Strain-to-strain differences in the capacity to use specific substrates are frequent in this taxonomic group (19). Rapid loss of genetic information when it is not required or acquisition via lateral transfer of genes is more likely in Klebsiella than in other enterobacteria, leading to the variety of phenotypic profiles in species of this genus. Given this genetic variability, the decision to select the K. pneumoniae ATCC 13882 strain was made on the basis of two considerations: (i) the availability of genetic manipulation procedures for this strain since it displayed resistance only to Ap and (ii) the presence of the yiaK-S and ulaA-G genetic systems, which allow good growth on l-ascorbate under aerobic and anaerobic conditions.
The inability of strains lacking the yiaK-S genetic system to grow on l-ascorbate reinforces the idea that this system is required for aerobic utilization of this carbon source. Consistently, K. oxytoca and K. pneumoniae C3, which are able to grow aerobically on l-ascorbate, displayed the same organization of the yiaK-S genes as K. pneumoniae ATCC 13882, whereas strains ATCC 13883 and KC2653, which were unable to grow on this compound, lacked this genetic system. In the case of impairment of ula regulon function, the slow growth observed in l-ascorbate-containing medium in the presence of oxygen indicated that the ula genetic system is necessary for rapid and complete utilization of this compound under aerobic conditions. Thus, it seems that K. pneumoniae requires the contributions of gene products of both systems for efficient utilization of l-ascorbate under aerobic conditions (Fig. 6).
FIG. 6.
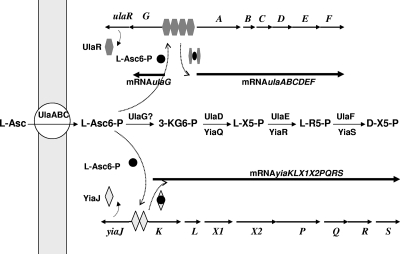
Coordinated regulation and roles of the yiaKLX1X2PQRS and ulaABCDEFG genetic systems in l-ascorbate metabolism. L-Asc, l-ascorbate; L-Asc6-P, l-ascorbate-6-phosphate; 3-KG6-P, 3-keto-gulonate 6 phosphate; LX5-P, l-xylulose-5-phosphate; L-R5-P, l-ribulose-5-phosphate.
The disappearance of the band corresponding to the formation of the YiaJ-DNA retarded complex in the presence of l-ascorbate-6-phosphate suggests that YiaJ acts as a repressor and that this compound is the effector molecule responsible for YiaJ dissociation. In this regard, the repressor character of YiaJ is consistent with the most common behavior of the regulators belonging to the ICLR family, to which YiaJ belongs on the basis of its amino acid sequence. Likewise, the UlaR-DNA complex could be released by l-ascorbate-6-phosphate, indicating that this phosphorylated compound also acts as an effector of UlaR. In this case, concentrations higher by 1 order of magnitude than those required to release YiaJ were required to dissociate the UlaR-DNA complex, indicating that there is a significant difference in the affinities of the two proteins for this effector molecule. The l-ascorbate-6-phosphate concentration determined for UlaR dissociation is similar to the concentration determined for other effector molecules recognized by members of the DeoR family (30). The inability of the products of the yiaK-S system to phosphorylate l-ascorbate, together with the poor growth on l-ascorbate in the absence of the ulaA function, suggests that intracellular formation of l-ascorbate-6-phosphate is mediated only by the UlaABC phosphotransferase transport function. Based on these results, a model for the coordinated expression of the ulaA-G and yiaK-S genetic systems involving l-ascorbate-6-phosphate is proposed here (Fig. 6).
Both systems are repressed by the presence of glucose in the medium. Although the cAMP-CRP complex appears to participate in this regulation, it is worth mentioning that other cAMP-CRP-independent mechanisms could be involved in this control mechanism. For instance, the presence of glucose inhibits the ability of the UlaABC phosphotransferase system to carry l-ascorbate and simultaneously phosphorylate it into l-ascorbate-6-phosphate. The lack of synthesis of l-ascorbate-6-phosphate by the ula system in the presence of glucose leads to a lack of induction of Φ(yiaK-lacZ), Φ(ulaG-lacZ), and Φ(ulaA-lacZ) and hence to blockage of the cross-induction of the two genetic systems, conditions required by K. pneumoniae for efficient utilization of l-ascorbate in the presence of oxygen.
Although there is no doubt concerning the requirement for yiaK-S for the metabolism of l-ascorbate under aerobic conditions, the functions of the yiaK-S gene products in this metabolism remain elusive, as we could not measure any kinase or oxidase activity with this compound in crude extracts of K. pneumoniae cells grown in the presence of l-ascorbate (data not shown). Assignment of functions to the yiaK-S products has been the objective of several groups in recent years. Plantinga et al. (21, 22) reported that YiaMNO recognizes l-xylulose as a substrate. However, using different techniques, Thomas et al. (29) demonstrated that 2,3-diketogulonate, the breakdown product of l-ascorbate, rather than l-xylulose is the substrate recognized by the extracytoplasmic solute receptor protein YiaO, which, along with the YiaMN membrane proteins, belongs to the group containing the tripartite ATP-independent periplasmic transporters (14, 29). It has also been reported that deletion of the yiaMNO genes in E. coli K-12 strain MC4100 results in remarkable changes in the transition from exponential growth to the stationary phase, high-salt survival, and biofilm formation (23). Moreover, Ly et al. (18), on the basis of sequence similarity, pointed out that YiaMNO of E. coli may be orthologous with the TRAP transporter TeaABC, an osmoregulatory ectoine transporter in Halomonas elongata (9), and a putative role for the YiaMNO transporter in the uptake of an unknown osmoprotectant has been proposed. In this regard, bacteria that cause food- and waterborne diseases face diverse and changing environments during their life cycles (1, 11, 28). The stress factors to which these bacteria are subjected may include, among other factors, exposure to reactive oxygen species that may be generated in the presence of l-ascorbate in the medium (6).
Analysis of the yiaK-S system in several Enterobacteriaceae revealed significant variations between the taxa. Although a number of strains do not have this genetic system, when the system is present, the main differences are (i) in the alternative presence of the yiaMNO or yiaX2 genes, which appear to encode transport proteins with different substrate affinities, and (ii) in the presence or absence of yiaX1, which encodes a protein displaying similarity to the CheA protein of Salmonella enterica serovar Typhimurium, which has been reported to be involved in chemotaxis. The high variability of this system is also shown by the observation that S. enterica serovar Typhimurium LT2 displays a genetic organization distinct from that described for E. coli and K. pneumoniae (Fig. 1). In this case, in addition to yiaMNO, yiaX1 is also present, indicating the diversification of this system. The diversity observed in the genetic organization of yiaK-S system could indicate that this system is beneficial only under some conditions.
Acknowledgments
This work was supported by grant BFU2007-63090 from the Ministerio de Educación y Ciencia (Spain) and by the Comissionat per Universitats i Recerca de la Generalitat de Catalunya. L.D.L.R. and F.G. were recipients of predoctoral fellowships from the Generalitat de Catalunya.
We gratefully acknowledge Miquel Regué for providing K. pneumoniae strain C3.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Abee, T., and J. A. Wouters. 1999. Microbial stress response in minimal processing. Int. J. Food Microbiol. 5065-91. [DOI] [PubMed] [Google Scholar]
- 2.Baillon, M. L. A., A. H. M. van Vliet, J. M. Ketley, C. Constantinidou, and C. W. Penn. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 1814798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boronat, A., and J. Aguilar. 1979. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos, E., J. Aguilar, L. Baldoma, and J. Badia. 2002. The gene yjfQ encodes the repressor of the yjfR-X regulon (ula), which is involved in l-ascorbate metabolism in Escherichia coli. J. Bacteriol. 1846065-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos, E., L. Baldoma, J. Aguilar, and J. Badia. 2004. Regulation of expression of the divergent ulaG and ulaABCDEF operon involved in l-ascorbate dissimilation in Escherichia coli. J. Bacteriol. 1861720-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos, E., C. Montella, F. Garces, L. Baldoma, J. Aguilar, and J. Badia. 2007. Aerobic l-ascorbate metabolism and associated oxidative stress in Escherichia coli. Microbiology 1533399-3408. [DOI] [PubMed] [Google Scholar]
- 7.Esselen, W. B., and J. E. Fuller. 1939. The oxidation of ascorbic acid as influenced by intestinal bacteria. J. Bacteriol. 37501-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua, W. C. 1992. And improved chloramphenicol resistance gene cassette for site-directed marker replacement mutagenesis. BioTechniques 12223-225. [PubMed] [Google Scholar]
- 9.Grammann, K., A. Volke, and H. J. Kunte. 2002. New type of osmoregulated solute transporter identified in halophilic members of the Bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581T. J. Bacteriol. 1843078-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunstein, M., and D. S. Hogness. 1975. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. USA 723961-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock, D., T. Besser, J. Lejeune, M. Davis, and D. Rice. 2001. The control of VTEC in the animal reservoir. Int. J. Food Microbiol. 6671-78. [DOI] [PubMed] [Google Scholar]
- 12.Ibañez, E., E. Campos, L. Baldomà, J. Aguilar, and J. Badia. 2000. Regulation of expression of the yiaKLMNOPQRS operon for carbohydrate utilization in Escherichia coli: involvement of the main transcriptional factors. J. Bacteriol. 1824617-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibañez, E., R. Gimenez, T. Pedraza, L. Baldoma, J. Aguilar, and J. Badia. 2000. Role of the yiaR and yiaS genes of Escherichia coli in metabolism of endogenously formed l-xylulose. J. Bacteriol. 1824625-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly, D. J., and G. H. Thomas. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25405-424. [DOI] [PubMed] [Google Scholar]
- 15.Liao, M. L., S. Y. Wang, C. Chung, Y. T. Liang, and P. A. Sejb. 1988. Synthesis of l-ascorbate-6-phosphate. Carbohydr. Res. 17673-77. [Google Scholar]
- 16.Liu, Q., and R. A. Bender. 2007. Complex regulation of urease formation from the two promoters of the ure operon of Klebsiella pneumoniae. J. Bacteriol. 1897593-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-273. [PubMed] [Google Scholar]
- 18.Ly, A., J. Henderson, A. Lu, D. E. Culham, and J. M. Wood. 2004. Osmoregulatory systems of Escherichia coli: identification of betaine-carnitine-choline transporter family member BetU and distributions of betU and trkG among pathogenic and nonpathogenic isolates. J. Bacteriol. 186296-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez, J., L. Martínez, M. Rosemblueth, J. Silva, and E. Martinez-Romero. 2004. How are gene sequence analyses modifying bacterial taxonomy? The case of Klebsiella. Int. Microbiol. 4261-268. [PubMed] [Google Scholar]
- 20.Nunoshiba, T., E. Hidalgo, C. F. Amabile-Cuevas, and B. Demple. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 1746054-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plantinga,T. H., C. van der Does, and A. J. M. Driessen. 2004. Transporter's evolution and carbohydrate metabolic clusters. Trends Microbiol. 124. [DOI] [PubMed] [Google Scholar]
- 22.Plantinga, T. H., C. Van Der Does, J. Badia, J. Aguilar, W. N. Konings, and A. J. Driessen. 2004. Functional characterization of the Escherichia coli K-12 yiaMNO transport protein genes. Mol. Membr. Biol. 2151-57. [DOI] [PubMed] [Google Scholar]
- 23.Plantinga, T. H., C. van der Does, D. Tomkiewicz, G. van Keulen, W. N. Konings, and A. J. Driessen. 2005. Deletion of the yiaMNO transporter genes affects the growth characteristics of Escherichia coli K-12. Microbiology 1511683-1689. [DOI] [PubMed] [Google Scholar]
- 24.Rabus, R., D. L. Jack, D. J. Kelly, and M. H. Saier, Jr. 1999. TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology 1453431-3445. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Sanchez, J. C., R. Gimenez, A. Schneider, W.-D. Fessner, L. Baldomà, J. Aguilar, and J. Badia. 1994. Activation of a cryptic gene encoding a kinase for l-xylulose opens a new pathway for the utilization of l-lyxose by Escherichia coli. J. Biol. Chem. 26929665-29669. [PubMed] [Google Scholar]
- 27.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 28.Sleator, R. D., and C. Hill. 2001. Bacterial osmoadaptation: the role of osmolytes in bacteria stress and virulence. FEMS Microbiol. Rev. 7311-23. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, G. H., T. Southworth, M. R. León-Kempis, A. Leech, and D. J. Kelly. 2006. Novel ligands for the extracellular solute receptors of two bacterial TRAP transporters. Microbiology 152187-198. [DOI] [PubMed] [Google Scholar]
- 30.van Rooijen, R. J., K. J. Dechering, C. Niek, J. Wilmink, and W. M. de Vos. 1993. Lysines 72, 80 and 213 and aspartic acid 210 of the Lactococcus lactis LacR repressor are involved in the response to the inducer tagatose-6-phosphate leading to induction of lac operon expression. Protein Eng. 6201-206. [DOI] [PubMed] [Google Scholar]
- 31.Volk, W. A., and L. Larsen. 1962. β-l-Gulonic acid as an intermediate in the bacterial metabolism of ascorbic acid. J. Biol. Chem. 2372454-2457. [PubMed] [Google Scholar]
- 32.Yew, W.-S., and J. A. Gerlt. 2002. Utilization of l-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J. Bacteriol. 184302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young, R. M., and L. H. James. 1942. Action of intestinal microorganisms on ascorbic acid. J. Bacteriol. 4475-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, Z., M. Aboulwafa, M. H. Smith, and M. H. Saier, Jr. 2003. The ascorbate transporter of Escherichia coli. J. Bacteriol. 1852243-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]