Abstract
Salmonella pathogenicity island 1 (SPI1) enables infecting salmonellae to invade the intestinal epithelium and induce a proinflammatory response and macrophage cell death. SPI1 expression is controlled by a complex cascade with several transcriptional regulators within the island and global regulators outside it. Previously, we reported that DnaK-depleted salmonellae could neither invade epithelial cells nor secrete SPI1-encoded proteins, suggesting that DnaK is involved in the expression of SPI1. Here, we found that DnaK is involved in SPI1 expression through inhibition of σ32 protein, which directs the transcription of a group of genes in response to various global stresses. Overproduction of σ32 resulted in decreased levels of the SPI1-specific transcriptional regulators HilD and HilA. Further analysis demonstrated that the σ32-mediated system negatively regulates HilD and HilA at the posttranslational and transcriptional levels, respectively. The executioner of this negative regulation was shown to be a σ32-induced protein ATP-dependent Lon protease, which specifically degrades HilD. Since HilD can activate hilA transcription, is at the top of the hierarchical SPI1 regulatory loop, and has a dominant role, the posttranslational control of HilD by Lon is critically important for precise expression of SPI1. Consequently, we suggest that SPI1 expression is controlled by the feedback regulatory loop in which σ32 induces Lon to control turnover of HilD, and DnaK, which inhibits σ32 function, leading to the modulation of lon expression. This regulation in response to a specific combination of environmental signals would ensure that SPI1 expression is restricted to a few specific locations in the host.
Salmonella serovars are facultative intracellular pathogens that cause a range of diseases in a wide variety of hosts. Salmonella enterica serovar Typhimurium causes gastroenteritis in humans and systemic diseases similar to typhoid fever in mice. Salmonella is typically ingested in contaminated food or water. The bacteria colonize the small intestine and invade normally nonphagocytic epithelial cells to gain access to the underlying lymph tissue. Invasion is mediated by a type III secretion system (TTSS) encoded on a 40-kb island at centisome 63 of the Salmonella genome, termed Salmonella pathogenicity island 1 (SPI1) (16, 21). The SPI1 TTSS forms a multiprotein secretion apparatus, called a needle complex, through which effector proteins are injected into the host cell cytosol, where they activate signal transduction pathways, rearrange the actin cytoskeleton, and cause membrane ruffling, ultimately inducing uptake of the bacteria. In addition to invasiveness, various functions have been attributed to the SPI1 TTSS, including Salmonella-induced macrophage death (8, 25, 26), enteropathogenesis (59), and secretion of a pathogen-elicited epithelial chemoattractant that directs polymorphonuclear neutrophil movement across epithelial monolayers (30, 38).
SPI1 expression is controlled by a complex cascade with several transcriptional regulators present within the island. These regulators, HilD, HilC, HilA, and InvF, act in an ordered fashion to activate coordinated expression of the SPI1 genes (2, 10, 12, 13). The regulatory circuit converges on the expression of hilA, which activates the expression of all of the SPI1 operons encoding the TTSS apparatus, chaperones, and some effectors either directly or indirectly by activating the expression of another regulator, invF (3, 29, 33). HilD and HilC, which are members of the AraC/XylS family of transcriptional regulators, can each individually bind to the DNA immediately upstream of hilA, and it is believed that this binding leads to hilA expression (44, 45). In addition, RtsA, which also belongs to the AraC/XylS family, has been shown to activate the expression of SPI1 genes by binding upstream of hilA (14). HilD leads to transcriptional activation of hilC and rtsA, which activate themselves (13, 34, 41). Therefore, it is suggested that HilD is at the top of the hierarchy of regulation of SPI1 expression.
Control of SPI1 also extends to global regulators encoded outside the island. Several global regulators respond to a specific combination of environmental signals that presumably act as cues that the bacteria are in the appropriate anatomical location (4). The present report suggests that all of the global regulators control SPI1 expression in a HilD-dependent manner, that is, through posttranscriptional or posttranslational control of HilD, which in turn activates hilC and rtsA (15). One such regulator is SirA in the BarA/SirA two-component regulatory system. It has been demonstrated that overproduction of SirA can activate transcription from the hilA promoter only when HilD is present (13). SirA acts by inducing the expression of two small RNA molecules, CsrB and CsrC. These small RNAs are antagonistic to the posttranscriptional regulatory protein CsrA, which binds to the message of its targets and alters mRNA stability (43). Thus, SirA induction of csrBC prevents CsrA action, indirectly activating hilD expression posttranscriptionally. Recently, we have shown that the flagellum-related gene product FliZ positively regulates hilD expression at the posttranscriptional level (27). The systems that negatively regulate SPI1 expression are the two-component PhoP/PhoQ and PhoR/PhoB regulatory systems and FimZY for type 1 fimbrial expression (6, 35, 42). These seem to function primarily through HilE, which binds directly to HilD, presumably preventing its action (5, 15). We have previously demonstrated that ATP-dependent Lon protease is a powerful negative regulator of SPI1 expression; depletion of Lon increased hilA expression 40-fold and caused a 10-fold increase in the invasion of cultured epithelial cells (53). Lon regulates HilD posttranslationally by specifically degrading it (55). We have further demonstrated that the DnaK chaperone machinery is essential for invasion of epithelial cells by Salmonella depending on the SPI1 TTSS; DnaK-depleted Salmonella cells could neither invade cultured epithelial cells nor secrete any of the invasion proteins encoded in SPI1 (54).
In the present study, we demonstrated that SPI1 expression is tightly controlled in the network of the global response mediated by the σ32 factor for RNA polymerase. σ32 is the first alternative σ factor discovered in Escherichia coli and can direct the transcription of a group of genes upon heat shock stress and other general stresses (40, 62). These gene products, collectively termed heat shock proteins, include molecular chaperones such as DnaK and ATP-dependent proteases such as Lon, which constitute the cellular network for the de novo folding and quality control of proteins under normal and stress conditions (22, 57). On the basis of the present results, we gain insight into the complex network for regulating SPI1 expression modulated by the σ32-initiated regulatory loop.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All of the strains and plasmids used in this study are shown in Table 1. Bacterial cells were grown in L broth (1% Bacto tryptone, 0.5% Bacto yeast extract, 0.5% sodium chloride, pH 7.4) and L agar, which were supplemented when necessary with either chloramphenicol (20 μg ml−1), ampicillin (25 μg ml−1), or kanamycin (25 μg ml−1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesb | Reference or source |
|---|---|---|
| S. enterica serovar Typhimurium strainsa | ||
| χ3306 | Virulent strain; gyrA | 20 |
| CS2021 | dnaK::Cm in χ3306 | 54 |
| CS2022 | lon::Cm in χ3306 | 53 |
| CS2501 | CS2021 harboring pTKY608 | 54 |
| CS2937 | χ3306 harboring pKD46 | This study |
| CS3072 | χ3306 harboring pDMI1 and pUHE21-2Δfd12 | This study |
| CS3249 | hilA::Km in CS2021 | This study |
| CS3271 | CS3249 harboring pZA4lacIq and pUHE21-2Δfd12 | This study |
| CS3273 | CS3249 harboring pZA4lacIq and pTKY748 | This study |
| CS3586 | χ3306 harboring pDMI1 and pUHE212-1(rpoH) | This study |
| CS3593 | CS2022 harboring pDMI1 and pUHE21-2Δfd12 | This study |
| CS3596 | CS2022 harboring pDMI1 and pUHE212-1(rpoH) | This study |
| CS3613 | χ3306 harboring pTKY821 | This study |
| CS3614 | CS2021 harboring pTKY821 | This study |
| CS3641 | CS2021 harboring pBB535 | This study |
| CS3647 | CS3641 harboring pTKY797 | This study |
| CS3658 | ΔhilD in CS2021 | This study |
| CS3659 | CS3658 harboring pTKY797 and pZA4lacIq | This study |
| CS3660 | CS3658 harboring pTKY797 and pBB535 | This study |
| CS3756 | χ3306 harboring pDM1 and pTKY720 | This study |
| CS3800 | lon::Sp in CS2021 | This study |
| Plasmids | ||
| pTKY608 | pMW119 carrying the dnaK-dnaJ operon of strain χ3306; Ap | 54 |
| pTKY651 | pUHE21-2Δfd12 with 1,050-bp hilD fragment; Ap | 55 |
| pTKY720 | pUHE21-2Δfd12 with 2,352-bp lon gene | This study |
| pTKY748 | pUHE21-2Δfd12 with 1,836-bp hilA gene | This study |
| pTKY797 | pMPM-A4 carrying hilD gene; Ap | This study |
| pTKY821 | pCB182 with 347-bp fragment containing lon promoter; Ap | This study |
| pBB535 | p15A derivative with PA1/lacO-1-dnaKJ operon; Sp | 57 |
| pUHE212-1-rpoH | pUHE212-1 carrying rpoH gene; Ap | 17 |
| pCB182 | Promoter cloning vector; Ap | 47 |
| pKD4 | Km | 11 |
| pKD46 | λ Red (γ, β, exo) | 11 |
| pMPM-A4 | Cloning vector with arabinose-inducible promoter; Ap | 37 |
| pUHE212-1 | N-terminal His tag vector; Ap | 17 |
| pUHE21-2Δfd12 | PA1/lacO-1 system vector; Ap | 17 |
| pZA4lacIq | lacIq; Sp | Our collection |
All Salmonella derivatives are originally from strain χ3306.
Ap, ampicillin resistance; Km, kanamycin resistance, Sp, spectinomycin resistance.
Construction of plasmids.
Plasmid pTKY720, in which the expression of lon is controlled by the PA1/lacO-1 promoter, was constructed by amplifying a BglII-HindIII fragment carrying lon by PCR with the lon-F1 (5′-GAAAAGCGGATCCGTAATCGTGT-3′) and lon-R1 (5′-CCGAAAAAGCTTGCCAGCCC-3′) primers and subsequent cloning of the fragment into pUHE21-2Δfd12 cleaved with BamHI and HindIII. Plasmid pTKY748, in which the expression of hilA is controlled by the PA1/lacO-1 promoter, was constructed by amplifying a BglII-PvuII fragment carrying hilA by PCR with the hilA-F (5′-GAGAGTACACTAGATCTATGCCACAT-3′) and hilA-R (5′-TCATCGCCGATTCCAGCTGGGCGATA-3′) primers and subsequent cloning of the fragment into pUHE21-2Δfd12 cleaved with BamHI and PvuII. Plasmid pTKY797, in which the expression of hilD is controlled by the ParaBAD promoter, was constructed by cloning the EcoRI-PstI fragment carrying hilD from pTKY651 into plasmid pMPM-A4 cleaved with EcoRI and PstI. To construct plasmid pTKY821 containing the lon promoter, the lon promoter locus was amplified from the chromosome of strain χ3306 with primers lon-F2 (5′-AAACAGGATCCGCAGGCTTCT-3′) and lon-R2 (5′-CGTAGAAGCTTCCAGACAACG-3′). The 347-bp fragment generated was cleaved with BamHI at the 5′ end and HindIII at the 3′ end and then cloned into the vector pCB182 cleaved with BamHI and HindIII.
Construction of a hilA::Km mutant and a dnaK::Cm ΔhilD double mutant.
Insertion of a kanamycin resistance (Km) cassette flanked by a FLP recombination sequence into hilA on the chromosome of χ3306 was accomplished by λ Red-mediated recombination essentially as described by Datsenko and Wanner (11). PCR products used to construct gene replacements were generated with template plasmid pKD4 and the hilAP1-F (5′-TATTATAACTTTTCACCCTGTAAGAGAATACACTATTATCGTGTAGGCTGGAGCTGCTTC-3′) and hilAP2-R (5′-ACGATGATAAAAAAATAATGCATATCTCCTCTCTCAGATTCATATGAATATCCTCCTTA-3′) primers. The 1,476-bp fragment generated was purified and then introduced into strain CS2937 carrying plasmid pKD46, encoding the λ Red recombinase, by transformation. The Km cassette insertion in hilA was verified by PCR amplification of the chromosomal DNA with the pKD4-P1 (5′-GTGGTAGGCTGGAGCTGCTTC-3′) and pKD4-P2 (5′-CATATGAATATCCTCCTTAG-3′) primers and by Southern blotting.
To construct the dnaK::Cm ΔhilD double mutant CS3658, bacteriophage P22 was propagated on a Salmonella mutant in which hilD was replaced with the construct in which hilD is disrupted by a single-crossover event (55), and the resultant lysates were used to infect CS2021 (dnaK::Cm). The transductants were selected for ampicillin resistance. Subsequently, a double-crossover event in the ΔhilD mutant was assessed by its resistance to sucrose and sensitivity to ampicillin. Disruption of the hilD gene was checked by PCR with hilD-F (5′-CCGGGGATCCATGGAAAATGTAACCTTTG-3′) and hilD-R (5′-CAATCTGCAGGAATAGCCTCCCATCCTG-3′).
Immunoblotting analysis.
To prepare whole-cell proteins, bacterial cells were harvested by centrifuging the culture and then suspended in sample buffer (28). Gel electrophoresis was carried out according to the method of Laemmli (28), with a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and Coomassie brilliant blue staining. The separated proteins were transferred onto Immun-Blot polyvinylidene difluoride membranes (Bio-Rad) and reacted with rabbit anti-HilD serum (1:12,500), anti-HilA serum(1:25,000), and anti-SipC serum (1:25,000) and mouse anti-DnaK monoclonal antibody (1:25,000; Stressgen), followed by alkaline phosphatase-conjugated anti-rabbit or anti-mouse immunoglobulin G. The enzymatic reactions were performed in the presence of 0.3 mg ml−1 nitroblue tetrazolium (Wako) and 0.15 mg ml−1 5-bromo-4-chloro-3-indolylphosphate (Sigma). Anti-HilD, -HilA, and -SipC sera were previously established in our laboratory (50, 55).
Assay for invasion of epithelial cells.
Intestine-407 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Cells (2 × 105) were seeded into 24-well tissue culture plates to obtain about 90% confluent monolayers on the following day. Bacterial cultures were grown to an optical density at 600 nm (OD600) of 0.5, and hilA expression was induced by adding 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubating the mixture for 2 h. The bacterial cells were washed with Hanks' balanced salt solution (HBSS) and used to inoculate monolayers previously washed with HBSS at a multiplicity of infection of 10. The monolayers were centrifuged at 500 × g for 5 min, incubated for 2 h at 30°C, washed thoroughly with HBSS, and further incubated for 3 h in Dulbecco modified Eagle medium containing 100 μg gentamicin ml−1 to eliminate extracellular bacteria before lysis with Triton X-100. Bacterial numbers were determined by plating the lysates on L agar plates after appropriate dilution.
RNA extraction and quantitative real-time RT-PCR.
Total RNAs were extracted from 1 ml of bacterial culture with the RNeasy Mini kit and the RNAprotect bacterial reagent according to the manufacturer's instructions. Reverse transcription (RT) was performed on 1 μg isolated total RNA with a QuantiTect RT kit by following the manufacturer's instructions. PCRs were performed with an Mx3000P QPCR system. For each PCR, the reaction mixture was prepared with Brilliant Sybr green QPCR Master Mix with 1 μl cDNA, forward and reverse primers (for hilA and hilD, 0.4 μM; for 16S rRNA, 1 μM), and the ROX reference dye supplied in a total of 25 μl. Thermal cycling conditions were an initial denaturation step for 10 min at 95°C, following 45 cycles of denaturing for 30 s at 95°C, annealing for 1 min at 55°C, and elongation for 30 s at 72°C. The following primer sequences were used: for hilA, sense primer CCGAGAGTCTGCATTACTCTATCGT and antisense primer TATCCTTAACACTGCGGCAGTTC; for hilD, sense primer ACTCGAGATACCGACGCAAC and antisense primer CTTCTGGCAGGAAAGTCAGG; for the 16S rRNA gene, sense primer GAATGCCACGGTGAATACGTT and antisense primer ACCCACTCCCATGGTGTGA.
Stability of HilD protein in vivo.
Cultures were grown to an OD600 of 0.5 in L broth containing 500 μM IPTG to induce dnaK expression, followed by the induction of hilD expression by adding 0.005% arabinose and incubation for 30 min. Tetracycline (100 μg ml−1) and glucose (2%) were then added to the culture to block the translation and expression of hilD. Aliquots of the cells were taken at appropriate intervals and mixed with trichloroacetic acid (final concentration, 10%), chilled on ice for 15 min, and centrifuged at 10,000 × g for 15 min. The pellets were washed with acetone and resuspended in sample buffer (28). A portion of each sample was separated on a 10% polyacrylamide gel and immunostained with anti-HilD serum (1:12,500). The levels of protein were quantified with Quantity One.
RESULTS
The inability of the dnaKJ mutant to invade is due to the marked decrease in the hilA transcript level.
In our previous study, we revealed that a dnaK::Cm mutant derived from S. enterica serovar Typhimurium lost the ability to invade cultured Intestine-407 cells (54). Several SPI1 proteins involved in invasion, such as SipA, SipC, and SipD, can be detected in the culture medium as proteins secreted by Salmonella (53). SipC and SipD are integral components of the translocation apparatus, which has been shown to be inserted into the host cell plasma membrane upon infection (46). Furthermore, SipC has been shown to initiate actin polymerization and anchor bundled filaments beneath invading bacteria (23). SipA is an effector that can bind actin directly to reduce the critical actin concentration required for polymerization (65). We have found that neither SipA nor SipC nor SipD was secreted in the culture medium of dnaK::Cm mutant cells (51, 54), suggesting that DnaK may be involved in the expression, stabilization, and/or secretion of SPI1-encoded proteins in Salmonella. The mutation was created by inserting a chloramphenicol resistance cassette including a stop codon and therefore resulted in a polar effect of dnaJ in the dnaK-dnaJ operon (54). Since DnaK functions as a molecular chaperone with the cochaperone DnaJ, we decided to use this mutant strain, CS2021, for further analysis.
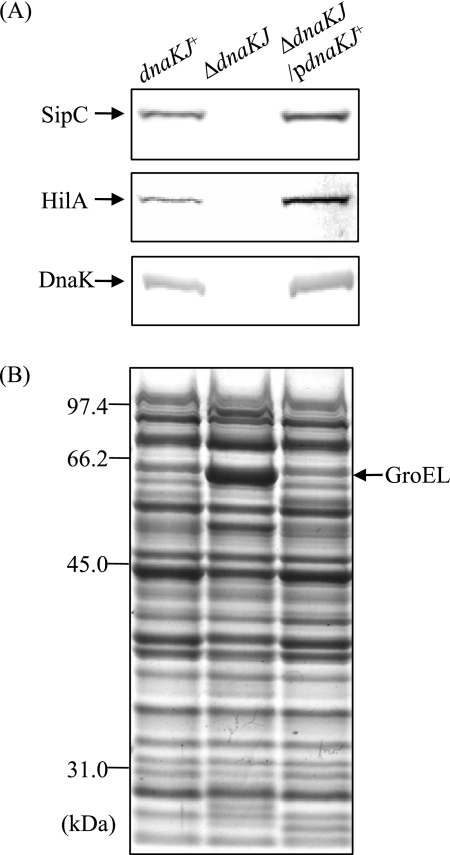
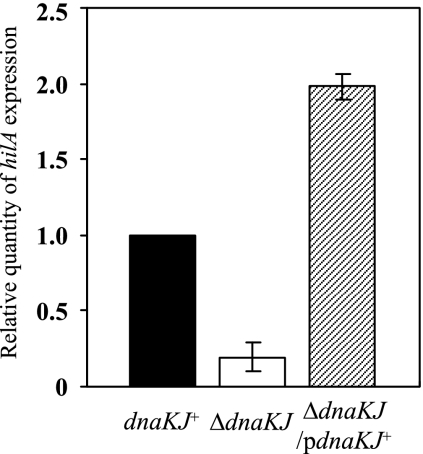
To examine whether the disappearance of SPI1 proteins secreted from the cells by dnaKJ disruption is due to loss of expression of the proteins, we initially compared the cellular levels of SipC in ΔdnaKJ cells and isogenic dnaKJ+ cells by immunoblotting. No SipC was detected in the ΔdnaKJ cells, while it was clearly detected in the dnaKJ+ cells (Fig. 1). To confirm that the absence of SipC from the whole-cell lysate of this strain is due to the disruption of dnaKJ, the mutation was complemented by a functional dnaKJ operon from χ3306 in trans and tested for SipC. As shown in Fig. 1, SipC was restored in the complemented strain, suggesting that the DnaKJ chaperone machinery is involved in producing SipC. We then examined the cellular levels of HilA, which is the central regulator in the overall scheme of SPI1 gene expression. No HilA was detected in the ΔdnaKJ cells, but this defect was fully compensated for by a functional copy of dnaKJ in trans. The effect of dnaKJ disruption on the expression of hilA was also examined by measuring the hilA transcript by quantitative real-time RT-PCR (Fig. 2). The dnaKJ disruption markedly decreased the amount of hilA transcript, and this decrease was fully restored by a functional copy of dnaKJ, suggesting that the DnaKJ chaperone machinery is probably involved in hilA expression. The hilA transcript level in the ΔdnaKJ cells complemented in trans exceeds that in the wild-type control. This is probably due to DnaKJ in excess over the normal level, as shown by immunoblotting (Fig. 1A).
FIG. 1.
Cellular levels of SPI1 proteins in wild-type cells and cells in which dnaKJ is disrupted. Bacterial cells of strains χ3306 (dnaKJ+), CS2021 (ΔdnaKJ), and CS2501 (ΔdnaKJ/pdnaKJ+) were used. (A) Immunoblotting of cellular lysates with anti-SipC, anti-HilA, and anti-DnaK sera. (B) Coomassie brilliant blue-stained SDS-10% polyacrylamide gel electrophoresis patterns of the samples used for immunoblotting. pdnaKJ+, pTKY608.
FIG. 2.
Expression of hilA in wild-type cells and cells in which dnaKJ is disrupted. Total RNA was prepared from strains χ3306 (dnaKJ+), CS2021 (ΔdnaKJ), and CS2501 (ΔdnaKJ/pdnaKJ+) grown in L broth to an OD600 of 0.5 at 30°C. The levels of hilA transcripts were measured by quantitative real-time RT-PCR and then normalized to 16S rRNA gene expression. The values represent the means and standard deviations of n-fold changes in comparison with the transcription level in χ3306. pdnaKJ+, pTKY608.
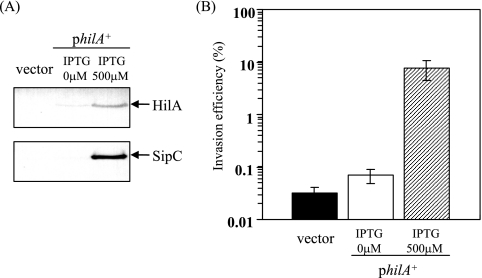
To determine whether the loss of the ability to invade epithelial cells by dnaKJ disruption is due to the great decrease in hilA transcript levels, we examined the efficiency of invasion of ΔdnaKJ cells expressing hilA under the control of the PA1/lacO-1 promoter system on plasmid pTKY748, in a ΔhilA background on the chromosome. Bacterial cells exposed to 500 μM IPTG for 2 h produced sufficient HilA to be detected even in the dnaKJ-deficient background (Fig. 3A). Furthermore, the increased HilA levels found subsequent to the activation of hilA by IPTG resulted in a sufficient amount of the SPI1 product, SipC. The invasion assay with the cultured Intestine-407 cells demonstrated that the induced HilA level enhanced the efficiency of invasion 240-fold even in the dnaKJ-deficient background (Fig. 3B). Taking these findings together, it is suggested that the DnaKJ chaperone machinery is essential for hilA expression to ensure the production of SPI1 proteins required for the capacity of Salmonella to invade epithelial cells.
FIG. 3.
Effect of overproduction of HilA on Salmonella invasiveness in a dnaKJ-deficient background. Bacterial cells of strains CS3271 (ΔdnaKJΔhilA/vector) and CS3273 (ΔdnaKJ ΔhilA/philA) were grown to an OD600 of 0.5 at 30°C in L broth, followed by the induction of hilA expression by adding 0 or 500 μM IPTG for 2 h. (A) Immunoblotting of cellular lysates with anti-HilA and anti-SipC sera. (B) Efficiency of invasion of cultured Intestine-407 cells. Bacterial cells in which hilA expression was induced by adding 500 μM IPTG for 2 h were used to inoculate monolayers. Invasion efficiency was examined as described in Materials and Methods. The data are the means and standard deviations for each strain tested in triplicate. Vector, pUHE21-2Δfd12; philA+, pTKY748.
HilD, a transcriptional regulator of SPI1 expression, is destabilized in cells in which dnaKJ is disrupted.
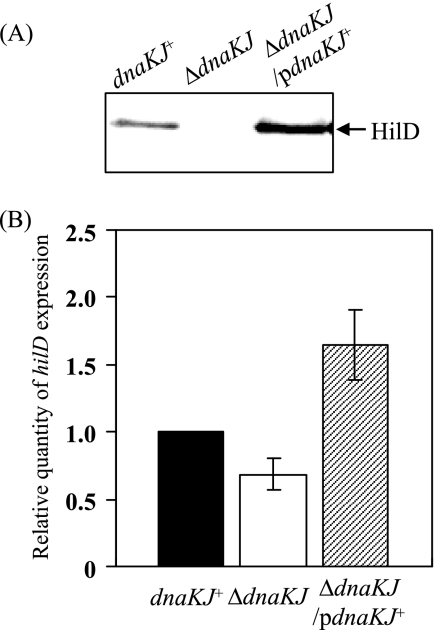
To reveal the involvement of the DnaKJ chaperone machinery in regulating hilA expression, we examined the cellular level of HilD, which has been shown to bind to the upstream sequence of hilA to activate its transcription directly (7, 13, 34). Immunoblotting of cell lysates prepared from the ΔdnaKJ mutant and isogenic dnaKJ+ cells detected no significant amount of HilD in the ΔdnaKJ cells (Fig. 4A). The dramatic decrease in HilD caused by dnaKJ disruption was fully compensated for by providing a functional dnaKJ operon in trans. Therefore, it is possible that the DnaKJ chaperone machinery is involved in regulating hilA transcription by modulating the cellular level of HilD. The effect of dnaKJ disruption on hilD expression was then examined in cells with the genetic backgrounds used for immunoblotting (Fig. 4A) by quantitative real-time RT-PCR. The results (Fig. 4B) demonstrate that dnaKJ disruption moderately decreased the expression of hilD, notwithstanding the marked decrease in the amount of HilD protein caused by this mutation. It has been demonstrated that HilD is at the top of the hierarchy of the SPI1 regulatory loop and also activates its own promoter, leading to amplification of the regulatory loop (13). Therefore, the slightly lower level of hilD transcript in the ΔdnaKJ mutant cells compared to that in the isogenic dnaKJ+ cells could be due to the absence of amplification of the regulatory loop by HilD in the ΔdnaKJ cells. The levels of both HilD protein and hilD transcript in the ΔdnaKJ cells complemented in trans exceed those in wild-type control cells. These are probably due to excess DnaKJ over the normal level, as shown in Fig. 1A. Taken together, these findings suggest that the DnaKJ chaperone machinery is possibly involved in the posttranscriptional and/or posttranslational regulation of hilD.
FIG. 4.
Cellular levels of HilD and relative levels of hilD expression. (A) Whole-cell lysates were prepared from strains χ3306 (dnaKJ+), CS2021 (ΔdnaKJ), and CS2501 (ΔdnaKJ/pdnaKJ+) grown in L broth to an OD600 of 1.0 at 30°C and then separated on an SDS-10% polyacrylamide gel. The separated proteins were immunostained with anti-HilD serum. (B) Total RNAs were prepared from the strains used in panel A grown in L broth to an OD600 of 0.5 at 30°C. The levels of hilD transcripts were measured by quantitative real-time RT-PCR and then normalized to 16S rRNA gene expression. The values represent the means and standard deviations of n-fold changes in comparison with the transcription level in χ3306. pdnaKJ+, pTKY608.
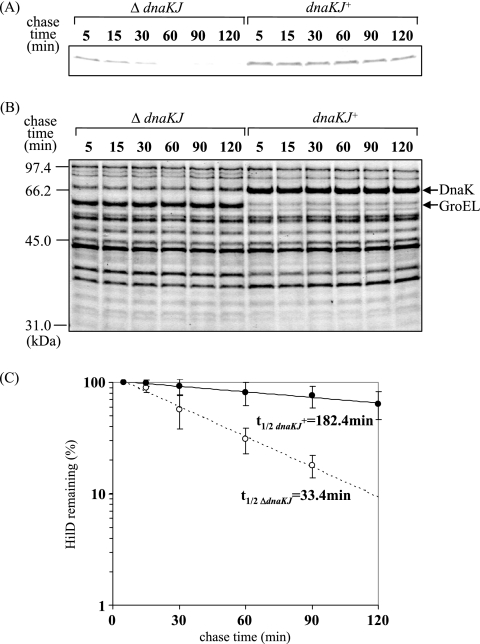
To test whether the DnaKJ chaperone machinery is involved in the posttranslational control of HilD, we decided to determine the in vivo half-life of HilD in the presence or absence of DnaKJ. In DnaKJ-producing cells, the dnaKJ genes were expressed under the control of the PA1/lacO-1 promoter system. The expression of hilD was initiated from the ParaBAD promoter on plasmid pTKY797 in a ΔhilD background on the chromosome by adding arabinose. The results (Fig. 5A) show the cellular levels of HilD at the indicated times after addition of tetracycline to prevent de novo synthesis of proteins. Whereas HilD disappeared at 60 min after the arrest of de novo synthesis in cells in which dnaKJ is disrupted, it was clearly detectable up to 120 min in cells producing DnaKJ. The half-lives of HilD in the cells in which dnaKJ is disrupted and the dnaKJ-expressing cells were 33.4 and 182.4 min, respectively (Fig. 5C). These results indicate that the DnaK chaperone machinery is involved in the control of HilD turnover.
FIG. 5.
In vivo stabilities of HilD protein in wild-type cells and cells in which dnaKJ is disrupted. (A) The bacterial strains used were CS3659 (ΔdnaKJ) and CS3660 (dnaKJ+). Cells were grown to an OD600 of 0.5 at 30°C in L broth containing 500 μM IPTG to induce dnaKJ expression, followed by the induction of hilD expression by adding 0.005% arabinose for 30 min. Tetracycline (100 μg ml−1) and glucose (2%) were added, and samples were added to prechilled trichloroacetic acid (final concentration, 10%) at the indicated times. The proteins were separated on an SDS-10% polyacrylamide gel and then immunostained with anti-HilD antibody. (B) Coomassie brilliant blue-stained gel patterns of the same samples used for immunoblotting. (C) Quantification of the precipitated proteins relative to the value at 5 min. Mean values of at least three independent experiments are given. t1/2, half-life.
σ32 mediates the control of SPI1 gene expression.
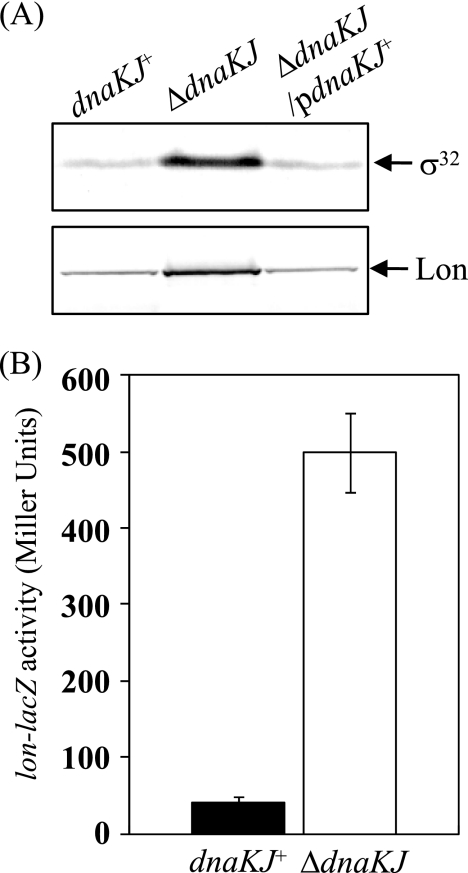
How is the DnaKJ chaperone machinery involved in turnover of HilD? It is unlikely that it directly degrades the HilD protein. We have previously reported that HilD is degraded by ATP-dependent Lon protease, leading to downregulation of SPI1 gene expression (55). The genes for the Lon protease and the DnaKJ chaperone are led by two consensus promoter sequences, each recognized by the σ70 factor involved in the transcription of most genes in cells and by σ32, an alternative σ factor the level of which is increased in response to a temperature upshift and other stresses. σ32 preferentially binds to core RNA polymerase to form an Eσ32 holoenzyme complex, so the increased σ32 results in the accelerated transcription of σ32-dependent genes. In E. coli, it has been demonstrated that the DnaKJ chaperone machinery negatively controls both the amount and the activity of σ32, creating a feedback loop of the σ32 regulon (31, 49, 56, 64). In S. enterica serovar Typhimurium, we observed that the amount of σ32 was greatly increased by dnaKJ disruption (Fig. 6A), suggesting negative modulation of σ32 by the DnaKJ machinery. We also observed that the dnaKJ disruption stimulated the transcription from the lon promoter 10-fold (Fig. 6B) and significantly increased the cellular level of Lon in S. enterica serovar Typhimurium (Fig. 6A). The finding that the dnaKJ disruption resulted in an increase in the amount of σ32 and therefore stimulated the transcription of lon in S. enterica serovar Typhimurium raises the possibility that the cellular level of HilD may be controlled by the σ32-initiated regulatory loop and this regulates SPI1 expression.
FIG. 6.
Effect of dnaKJ disruption on expression of σ32 and lon in Salmonella cells. (A) Whole-cell extracts were prepared from strains χ3306 (dnaKJ+), CS2021 (ΔdnaKJ), and CS2501 (ΔdnaKJ/pdnaKJ+) grown in L broth to an OD600 of 1.0 at 30°C and separated on an SDS-10% polyacrylamide gel. The separated proteins were immunostained with anti-σ32 serum and anti-Lon serum. pdnaKJ+, pTKY608. (B) The expression levels of lacZ fusion to the lon promoter in cells harboring pTKY821 were assayed for β-galactosidase activity. The values represent the means and standard deviations of samples tested at least in triplicate. The strains used were CS3613 (dnaKJ+) and CS3614 (ΔdnaKJ).
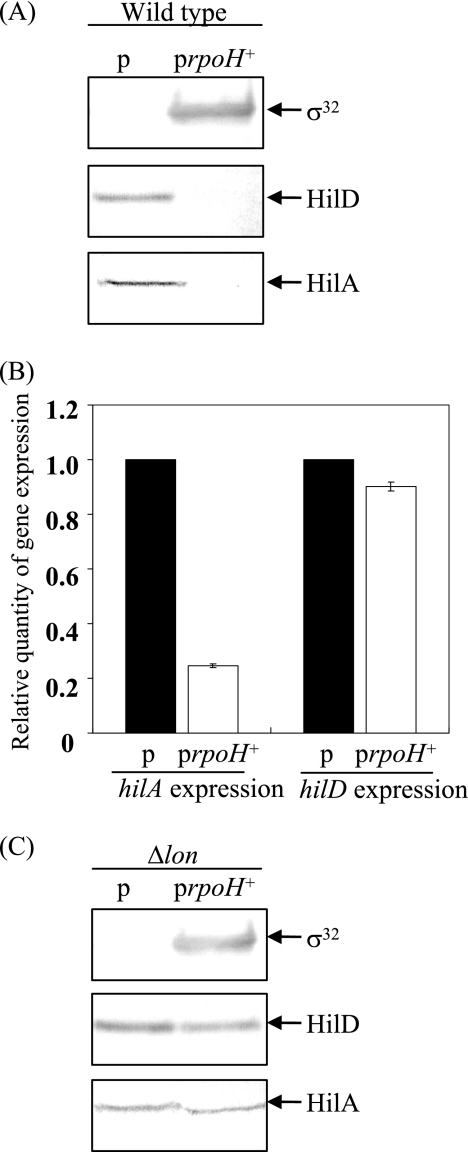
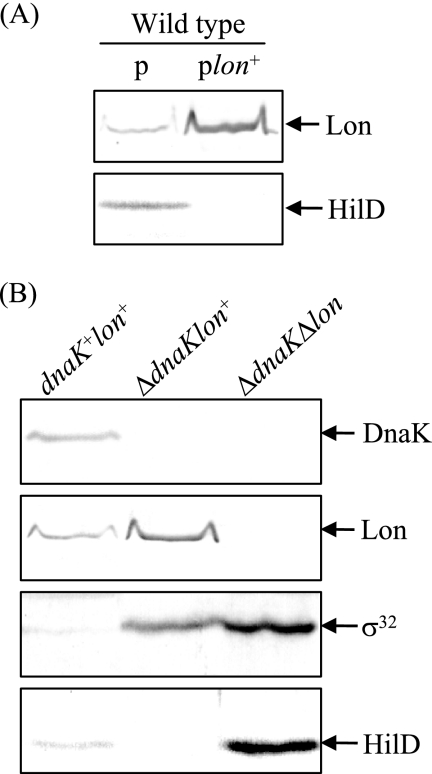
To examine this possibility, the cellular level of HilD was determined in σ32-overproducing cells in which rpoH, encoding σ32, is induced under the control of the PA1/lacO-1 promoter system. Immunoblotting with lysates prepared from wild-type cells with or without PA1/lacO-1-rpoH on the plasmid showed that overproduction of σ32 resulted in a marked decrease in the cellular level of HilD (Fig. 7A). Owing to the dramatic decrease in HilD, HilA disappeared from the σ32-overproducing cells. On the other hand, the effect of overproducing σ32 on the transcription of hilD and hilA was comparatively determined by quantitative real-time RT-PCR. The results (Fig. 7B) demonstrate that overproduction of σ32 did not significantly affect hilD transcription but dramatically reduced hilA transcription. The decrease in hilA transcription could be due to the marked decrease in the cellular level of HilD. To check whether σ32 modulates the HilD level by controlling the induction of Lon, we examined the effect of σ32 overproduction on the levels of HilD in a lon-deficient background. The results (Fig. 7C) show that the increased level of σ32 does not result in decreased HilD if Lon protease was absent from the cells, suggesting that σ32 controls the cellular level of HilD through the induction of Lon protease, which specifically recognizes and degrades it. Similarly, overproduction of σ32 did not affect the cellular level of HilA in the lon-deficient cells.
FIG. 7.
Effects of σ32 overexpression on cellular levels of HilA and HilD proteins and hilA and hilD transcripts. (A) Cultures of strains CS3072 (p) and CS3586 (prpoH+) were grown in L broth to an OD600 of 0.5 at 37°C, followed by the induction of σ32 with 200 μM IPTG for 1 h. Whole-cell lysates were separated on an SDS-10% polyacrylamide gel and then subjected to immunoblotting with anti-σ32, anti-HilD, and anti-HilA sera. (B) Cultures of strains used in panel A were grown in L broth to an OD600 of 0.5 at 37°C, followed by the induction of σ32 with 200 μM IPTG for 30 min. The levels of hilD and hilA transcripts were measured by quantitative real-time RT-PCR and then normalized to 16S rRNA gene expression. The values represent the means and standard deviations of n-fold changes in comparison with the transcription levels of the corresponding genes in CS3072. (C) Cultures of strains CS3593 (Δlon/p) and CS3596 (Δlon/prpoH+) were grown in L broth to an OD600 of 0.5 at 37°C, followed by incubation with 200 μM IPTG for 1 h to induce σ32. Whole-cell lysates were separated on an SDS-10% polyacrylamide gel and then subjected to immunoblotting analysis. p, pUE212-1; prpoH+, pUE212-1-rpoH.
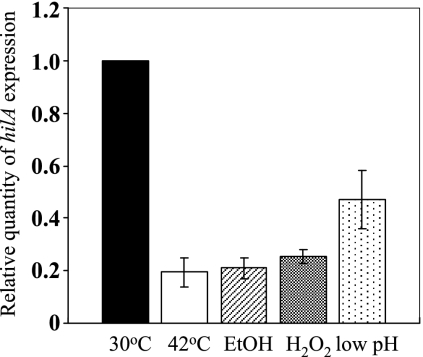
Taking these findings together, it is suggested that SPI1 gene expression is regulated by the feedback regulatory loop in which σ32 induces Lon protease to control the turnover of HilD and the DnaKJ chaperone machinery, which can inhibit σ32 function, leading to the modulation of lon expression. Therefore, the extreme accumulation of Lon due to collapse of the negative control of σ32 regulon by dnaKJ disruption could explain the disappearance of SPI1 expression from the mutant cells. We confirmed that SPI1 gene expression is controlled in response to global stresses, leading to the induction of σ32, by measuring the hilA transcription in cells exposed to a variety of stresses, including heat shock (42°C), acidic shock (pH 4.0), 5% ethanol, and 1 mM H2O2. The results (Fig. 8) demonstrate that hilA transcription significantly decreased after the exposure of cells to these stresses, suggesting that SPI1 gene expression is under the control of a σ32-mediated stress response.
FIG. 8.
Expression of hilA in Salmonella cells exposed to heat stress and other stresses. Cells of strain χ3306 grown in L broth to an OD600 of 0.5 at 30°C were exposed to heat shock (42°C), 5% ethanol (EtOH), 1 mM H2O2, and acidic shock (pH 4.0) for 10 min. Total RNAs were extracted, and hilA transcripts were measured by quantitative real-time RT-PCR and then normalized to 16S rRNA gene expression. The values represent the means and standard deviations of n-fold changes in comparison with the transcription levels of hilA in cells incubated at 30°C.
An alternative explanation for the role of the DnaKJ chaperone in the control of HilD turnover is that it is directly involved in the folding of HilD and its absence accelerates the degradation of HilD by Lon or other proteases, since unfolded proteins similar to irreversibly damaged proteins are nonspecifically degraded by proteases (24). To address this possibility, we examined whether overproduction of Lon in wild-type cells also decreases the amount of HilD similar to dnaKJ deletion. The results (Fig. 9A) demonstrate that the overproduction of Lon in wild-type cells markedly decreased the amount of HilD, as observed in ΔdnaKJ cells (Fig. 4A), suggesting that Lon can specifically degrade folded HilD. Furthermore, we determined the cellular level of HilD in a ΔdnaKJ Δlon double mutant. As shown in Fig. 9B, the decreased effect of dnaKJ disruption on the cellular level of HilD was abolished by introducing a lon mutation. The results simultaneously indicate that the lon disruption resulted in the marked accumulation of HilD even in the absence of the DnaKJ chaperone. Therefore, it is unlikely that DnaKJ is directly involved in the folding of HilD and its absence accelerates the degradation of HilD by proteases.
FIG. 9.
Effects of lon overexpression in a dnakJ+ background (A) and a ΔdnakJ Δlon double mutation (B) on cellular levels of HilD protein. (A) Cultures of strains CS3072 (p) and CS3756 (plon+) were grown in L broth to an OD600 of 0.5 at 37°C, followed by the induction of lon with 200 μM IPTG for 1 h. p, pUE21-2Δfd12; plon+, pTKY720. (B) Cultures of strains χ3306 (dnakJ+ lon+), CS2021 (ΔdnakJ lon+), and CS3800 (ΔdnakJ Δ lon) were grown in L broth to an OD600 of 0.5 at 30°C. Whole-cell lysates were separated on an SDS-10% polyacrylamide gel and then subjected to immunoblotting analysis.
DISCUSSION
The expression of SPI1 genes is regulated by a variety of positive and negative regulator genes in response to environmental conditions. In the present study, we have demonstrated that SPI1 expression is tightly regulated in the σ32-mediated regulatory loop. This is executed by the specific degradation of HilD by ATP-dependent Lon protease, a heat shock protein induced by σ32. Since HilD is at the top of the hierarchy of the SPI1 regulatory loop and has a predominant role in regulation, its degradation is essential for precise regulation of SPI1 expression. The levels of heat shock proteins are controlled primarily by σ32, which senses the cellular protein folding environment through negative feedback control mediated by molecular chaperones that are also σ32 regulon members. It has been demonstrated that each of the DnaKJ and GroELS chaperones in E. coli constitutes a negative feedback loop that couples σ32 activity to the cellular protein folding state: overproduction of either chaperone machinery decreases both the amount and the activity of σ32; conversely, chaperone depletion or overexpression of the chaperone substrates, misfolded and unfolded proteins, increases both the amount and the activity of σ32 (19, 48, 58). The homeostatic regulation model proposes that induction of the heat shock response relies on sequestering DnaKJ and GroEL by binding to the damaged proteins that accumulate during stress (19, 58). The present study has shown that the loss of the DnaKJ chaperone machinery results in the continuous expression of Lon because the negative feedback control is impaired (Fig. 6), therefore leading to severely diminished expression of SPI1 genes through excess degradation of HilD. Consequently, it is suggested that the expression of SPI1 genes is negatively regulated by a σ32-mediated stress response. On the other hand, the question of whether any of the SPI1 genes are under the positive control of σ32 remains unanswered.
The heat shock proteins, which are alternatively called stress proteins, are believed to be induced during various stages of bacterial infection because pathogens are exposed to a variety of environmental stress conditions such as sudden elevated temperature and stomach acidity before reaching the low-oxygen, hyperosmotic environment of the small intestine and the bactericidal mechanisms associated with the host immune system. Actually, heat shock proteins such as DnaK, GroEL, and GroES have been identified among the proteins induced during the growth of bacteria, including Salmonella (9), Yersinia (61), Legionella (1), and Brucella (32), within macrophages. To cause systemic infection, Salmonella must grow inside macrophages and must overcome exposure to oxidative stress, acid pH, cationic peptides, and nutrient deprivation, suggesting that the σ32-controlled regulon of Salmonella indeed responds to the hostile environment in the macrophage phagosome. We have previously reported direct evidence that the σ32 heat shock regulon is involved in the pathogenesis of S. enterica serovar Typhimurium; that is, the heat shock proteins DnaK, Lon, ClpX, and ClpP are essential for intracellular growth within macrophages and the systemic infection of mice (52, 54, 60).
In addition to its ability to grow within infected macrophages, Salmonella has been shown to induce macrophage cell death by mechanisms depending on caspase 1 but not by the classical mechanism depending on caspase 3, which is a key executioner caspase in the proteolytic cascade leading to cell death (36, 39). Rapid cell death is independent of intracellular bacterial multiplication but dependent on the SPI1-encoded SipB effector protein (25). Caspase 1-dependent programmed cell death is distinct from other forms of classical apoptosis that depend on caspase 3. One characteristic is that caspase 1 is a proinflammatory enzyme that cleaves the inactive precursors of interleukin-1β and -18 into their active cytokines (63). In contrast, we have found that the lon disruption-containing mutant of S. enterica serovar Typhimurium induces rapid, large-scale cell death by a mechanism involving both caspases 1 and 3 (50). Furthermore, we have demonstrated that lon disruption leads to the continuous expression of SPI1 genes within macrophages, where they are normally repressed, and that derepression of the SPI1 genes causes massive macrophage apoptosis. A recent report demonstrates that liver phagocytes can undergo apoptotic caspase 3-mediated cell death in vivo, with apoptosis being a rare event, more prevalent in heavily Salmonella-infected cells (18). Once Salmonella has established a systemic infection, excess macrophage apoptosis would be detrimental to the pathogen because it utilizes macrophages as vectors for systemic dissemination throughout the host. Thus, SPI1 gene expression must be suppressed to allow sufficient time for the bacteria to replicate, escape, and invade new macrophages. Induction of the σ32-mediated heat shock regulon by S. enterica serovar Typhimurium that has met the hostile environment within macrophages would be necessary to restrict SPI1 expression. That is quite important for suppressing apoptosis sufficiently to allow time for Salmonella to replicate within macrophages. Since the increased DnaKJ and GroES chaperone machineries directly regulate σ32 by using a chaperone network (19), the heat shock response would ensure an appropriate level of Lon as a negative regulator of SPI1 expression. In addition, the induced levels of chaperone machineries would be required to cope with the accumulation of partially unfolded or denatured proteins in cells exposed to the intracellular stresses associated with phagocytosis. Bacterial pathogenesis generally depends on the environmental conditions inside host cells. Interaction of specific virulence factors, e.g., the SPI1 TTSS in Salmonella, with global regulators such as σ32 responding to environmental signals would contribute to the spatiotemporal regulation of multistage pathogenesis.
Acknowledgments
We thank A. Tokumitsu for technical assistance.
This work was supported by grants-in-aid for scientific research (17390125) and research on priority areas (19041015) from the Ministry for Education, Culture, Sports, Sciences and Technology of the Japanese Government.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 611320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47715-728. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18715-727. [DOI] [PubMed] [Google Scholar]
- 4.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22703-714. [DOI] [PubMed] [Google Scholar]
- 5.Baxter, M. A., and B. D. Jones. 2005. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 731377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 1754475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 3831-40. [DOI] [PubMed] [Google Scholar]
- 9.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248730-732. [DOI] [PubMed] [Google Scholar]
- 10.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 201850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichelberg, K., and J. E. Galán. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 674099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57691-705. [DOI] [PubMed] [Google Scholar]
- 14.Ellermeier, J. R., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1855096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 1024-29. [DOI] [PubMed] [Google Scholar]
- 16.Galán, J. E. 1999. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr. Opin. Microbiol. 246-50. [DOI] [PubMed] [Google Scholar]
- 17.Gamer, J., H. Bujard, and B. Bukau. 1992. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor σ32. Cell 69833-842. [DOI] [PubMed] [Google Scholar]
- 18.Grant, A. J., M. Sheppard, R. Deardon, S. P. Brown, G. Foster, C. E. Bryant, D. J. Maskell, and P. Mastroeni. 20 February 2008. Caspase-3-dependent phagocyte death during systemic Salmonella enterica serovar Typhimurium infection of mice. Immunology 12528-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guisbert, E., C. Herman, C. Z. Lu, and C. A. Gross. 2004. A chaperone network controls the heat shock response in E. coli. Genes Dev. 182812-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulig, P., and R. Curtiss III. 1987. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 552891-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3549-559. [DOI] [PubMed] [Google Scholar]
- 22.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381571-580. [DOI] [PubMed] [Google Scholar]
- 23.Hayward, R. D., and V. Koronakis. 1999. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 184926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman, C., and R. D'Arit. 1998. Proteolysis and chaperones: the destruction/reconstruction dilemma. Curr. Opin. Microbiol. 1204-209. [DOI] [PubMed] [Google Scholar]
- 25.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 962396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jesenberger, V., K. J. Procyk, J. Yuan, S. Reipert, and M. Baccarini. 2000. Salmonella-induced caspase-2 activation in macrophages: a novel mechanism in pathogen-mediated apoptosis. J. Exp. Med. 1921035-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kage, H., A. Takaya, M. Ohya, and T. Yamamoto. 2008. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J. Bacteriol. 1902470-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 891847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, C. A., M. Silva, A. M. Siber, A. J. Kelly, E. Galyov, and B. A. McCormick. 2000. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. USA 9712283-12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberek, K., and C. Georgopoulos. 1993. Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc. Natl. Acad. Sci. USA 9011019-11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, J., and T. A. Ficht. 1995. Protein synthesis in Brucella abortus induced during macrophage infection. Infect. Immun. 631409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 31281-1291. [DOI] [PubMed] [Google Scholar]
- 34.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1832733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucas, R. L., C. P. Lostroh, C. C. Dirusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1821872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundberg, U., U. Vinatzer, D. Berdnic, A. von Gabain, and M. Baccarini. 1999. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J. Bacteriol. 1813433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer, M. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 16341-46. [DOI] [PubMed] [Google Scholar]
- 38.McCormick, B. A., C. A. Parkos, S. P. Colgan, D. K. Carnes, and J. L. Madara. 1998. Apical secretion of a pathogen-elicited epithelial chemoattractant activity in response to surface colonization of intestinal epithelia by Salmonella typhimurium. J. Immunol. 160455-466. [PubMed] [Google Scholar]
- 39.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 939833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morimoto, R. I. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 123788-3796. [DOI] [PubMed] [Google Scholar]
- 41.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1844148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pegues, D. A., M. J. Hantman, I. Behlan, and S. I. Miller. 1995. PhoP/Q transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17169-181. [DOI] [PubMed] [Google Scholar]
- 43.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1754744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schechter, L., and C. A. Lee. 2001. AlaC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 401289-1299. [DOI] [PubMed] [Google Scholar]
- 45.Schechter, L., S. Damrauer, and C. A. Lee. 1999. Two AlaC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32629-642. [DOI] [PubMed] [Google Scholar]
- 46.Scherer, C. A., E. Cooper, and S. I. Miller. 2000. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 371133-1145. [DOI] [PubMed] [Google Scholar]
- 47.Schneider, K., and C. Beck. 1986. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene 4237-48. [DOI] [PubMed] [Google Scholar]
- 48.Straus, D., W. Walter, and C. A. Gross. 1989. The activity of σ32 is reduced under conditions of excess heat shock protein production in Escherichia coli. Genes Dev. 32003-2010. [DOI] [PubMed] [Google Scholar]
- 49.Straus, D., W. Walter, and C. A. Gross. 1990. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of σ32. Genes Dev. 42202-2209. [DOI] [PubMed] [Google Scholar]
- 50.Takaya, A., A. Suzuki., Y. Kikuchi, M. Eguchi, E. Isogai, T. Tomoyasu, and T. Yamamoto. 2005. Derepression of Salmonella pathogenicity island 1 genes within macrophages leads to rapid apoptosis via caspase-1- and caspase-3-dependent pathways. Cell. Microbiol. 779-90. [DOI] [PubMed] [Google Scholar]
- 51.Takaya, A., M. Matsui, T. Tomoyasu, M. Kaya, and T. Yamamoto. 2006. The DnaK chaperone machinery converts the native FlhD2C2 hetero-tetramer into a functional transcriptional regulator of flagellar regulon expression in Salmonella. Mol. Microbiol. 591327-1340. [DOI] [PubMed] [Google Scholar]
- 52.Takaya, A., M. Suzuki, H. Matsui, T. Tomoyasu, H. Sashinami, A. Nakane, and T. Yamamoto. 2003. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar Typhimurium infection of mice. Infect. Immun. 71690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takaya, A., T. Tomoyasu, H. Matsui, and T. Yamamoto. 2004. The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infect. Immun. 721364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takaya, A., Y. Kubota, E. Isogai, and T. Yamamoto. 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol. Microbiol. 55839-852. [DOI] [PubMed] [Google Scholar]
- 56.Tilly, K., J. Speuce, and C. Georgopoulos. 1989. Modulation of stability of the Escherichia coli heat shock regulatory factor σ32. J. Bacteriol. 1711585-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomoyasu, T., A. Mogk, H. Langen, P. Goloubinoff, and B. Bukau. 2001. Genetic dissection of the roles of changes and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40397-413. [DOI] [PubMed] [Google Scholar]
- 58.Tomoyasu, T., T. Ogura, T. Tatsuya, and B. Bukau. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30567-581. [DOI] [PubMed] [Google Scholar]
- 59.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36997-1005. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto, T., H. Sashinami, A. Takaya, T. Tomoyasu, H. Matsui, Y. Kikuchi, T. Hanawa, S. Kamiya, and A. Nakane. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 693164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto, T., T. Hanawa, and S. Ogata. 1994. Induction of Yersinia enterocolitica stress proteins by phagocytosis with macrophage. Microbiol. Immunol. 38295-300. [DOI] [PubMed] [Google Scholar]
- 62.Yura, T., H. Nagai, and H. Mori. 1993. Regulation of the heat shock response in bacteria. Annu. Rev. Microbiol. 47321-350. [DOI] [PubMed] [Google Scholar]
- 63.Zeuner, A., A. Eramo, C. Peschle, and R. De Maria. 1999. Caspase activation without death. Cell Death Differ. 61075-1080. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, K., M. Liu, and R. R. Burgess. 2005. The global transcriptional response of Escherichia coli to induced σ32 protein involves σ32 regulon activation followed by inactivation and degradation of σ32 in vivo. J. Biol. Chem. 28017758-17768. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, D., M. S. Mooseker, and J. E. Galán. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 2832092-2095. [DOI] [PubMed] [Google Scholar]