Abstract
The increasing occurrence of multidrug-resistant pathogens of clinical and agricultural importance is a global public health concern. While antimicrobial use in human and veterinary medicine is known to contribute to the dissemination of antimicrobial resistance, the impact of microbial communities and mobile resistance genes from the environment in this process is not well understood. Isolated from an industrially polluted aquatic environment, Escherichia coli SMS-3-5 is resistant to a record number of antimicrobial compounds from all major classes, including two front-line fluoroquinolones (ciprofloxacin and moxifloxacin), and in many cases at record-high concentrations. To gain insights into antimicrobial resistance in environmental bacterial populations, the genome of E. coli SMS-3-5 was sequenced and compared to the genome sequences of other E. coli strains. In addition, selected genetic loci from E. coli SMS-3-5 predicted to be involved in antimicrobial resistance were phenotypically characterized. Using recombinant vector clones from shotgun sequencing libraries, resistance to tetracycline, streptomycin, and sulfonamide/trimethoprim was assigned to a single mosaic region on a 130-kb plasmid (pSMS35_130). The remaining plasmid backbone showed similarity to virulence plasmids from avian-pathogenic E. coli (APEC) strains. Individual resistance gene cassettes from pSMS35_130 are conserved among resistant bacterial isolates from multiple phylogenetic and geographic sources. Resistance to quinolones was assigned to several chromosomal loci, mostly encoding transport systems that are also present in susceptible E. coli isolates. Antimicrobial resistance in E. coli SMS-3-5 is therefore dependent both on determinants acquired from a mobile gene pool that is likely available to clinical and agricultural pathogens, as well, and on specifically adapted multidrug efflux systems. The association of antimicrobial resistance with APEC virulence genes on pSMS35_130 highlights the risk of promoting the spread of virulence through the extensive use of antibiotics.
Antibiotic resistance in bacterial pathogens is a major public health challenge worldwide, with increased morbidity and mortality due to bacterial infections and associated costs in billions of dollars in the United States alone (48, 57). While commensal bacteria are thought to function as reservoirs of antimicrobial resistance genes for pathogens in host-associated communities (74, 77, 95), further research is needed to address questions about the origin, presence, and persistence of antimicrobial resistance genes in the environment and their potential transfer from environmental to clinical settings.
Most clinically relevant antibiotics originate from soil-dwelling actinomycetes (43). Accordingly, microbial communities in the soil environment, where resistance is necessary to counteract allelopathic compounds, have been shown to function as reservoirs of resistance to natural and artificial antimicrobials (20, 21, 70). Comparative sequence analysis of different types of antimicrobial resistance genes suggests that they originated and diversified in environmental communities, from which they were mobilized and propagated into taxonomically and ecologically distant bacterial populations (2). Plasmid exchange between environmental and host-associated bacterial communities is a recognized source for the rapid spread of antimicrobial resistance phenotypes (91). The potential significance of plasmids in disseminating antimicrobial resistance genes is further accentuated by the association of plasmids with mobile genetic elements, such as transposons, integrons, and insertion (IS) elements (49, 88). To better understand the evolution and dissemination of resistance phenotypes from clinical, agricultural, and environmental settings, it is therefore necessary to perform comparative sequence analysis of resistant isolates on three different levels, comparing whole genomes, single plasmids, and individual resistance gene cassettes.
The gammaproteobacterium Escherichia coli is a ubiquitous member of the normal gastrointestinal microflora of humans and animals (25). Since it is also widely present in the environment, E. coli could function as a mediator for gene flow between environmental and clinical settings. Highly adapted pathogenic E. coli strains have acquired specific virulence attributes, and successful combinations of these genetic factors persist in E. coli pathotypes that cause disease in humans and agriculturally important animals (42). Pathogenic E. coli strains fall into two main categories, with intestinal isolates typically causing enteric/diarrheal syndromes and extraintestinal isolates causing urinary tract infections and sepsis/meningitis. The emergence of fluoroquinolone-resistant E. coli strains from lineages associated with relatively low virulence potentials (36) has led to the hypothesis that the use of antibiotics could accelerate the evolution of formerly commensal strains toward virulence (1). Previously sequenced E. coli genomes originated mainly from pathogenic and host-associated isolates. In comparison to these strains, the genome sequences of environmental E. coli isolates with antimicrobial resistance phenotypes could provide insights into the evolutionary relationship between antimicrobial resistance, the adaptation to pathogenic lifestyles, and virulence.
E. coli SMS-3-5 was isolated from an industrial, metal-contaminated coastal environment and was found to be resistant to a record number of antimicrobials, in many cases at record high concentrations. Surprisingly, this environmental isolate is resistant to ciprofloxacin and moxifloxacin, two front-line fluoroquinolones. In all, E. coli SMS-3-5 is resistant to six quinolone compounds, including narrow-, extended-, and broad-spectrum drugs, with MICs among the highest ever recorded for E. coli (34). To our knowledge, E. coli SMS-3-5 is the first nonclinical E. coli isolate found to be resistant to extended- and broad-spectrum quinolones. In the present study, the genome of E. coli SMS-3-5 was sequenced and analyzed, with emphasis on both resistance- and virulence-associated functions, and compared to the genomes of seven previously sequenced intestinal and extraintestinal E. coli strains, as well as to the laboratory-adapted E. coli K-12. In addition, recombinant vector clones from sequencing libraries were used to phenotypically characterize selected antimicrobial resistance genes and to improve functional gene assignments.
MATERIALS AND METHODS
Bacterial strain.
E. coli strain SMS-3-5 was isolated from sediments of Shipyard Creek, a tidal system in the harbor of Charleston, SC (32°50′32"N, 79°56′59"W), on 23 March 2005. The creek drains the Macalloy Superfund Site and is heavily contaminated with toxic metals and other industrial waste (94). After aseptic collection, the sediment samples were stored in sterile plastic bags on ice and processed within 24 h. Serial dilutions of sediment slurries were spread plated in triplicate onto 15- by 100-mm petri dishes containing m-FC agar (Difco), following standard protocols for fecal coliform counts in water (29). To select E. coli isolates, individual blue colonies (fecal coliforms) were streaked to nutrient agar with 4-methylumbelliferyl-β-d-glucuronide (Difco Laboratories), incubated overnight at 37°C, and checked for fluorescence under UV irradiation. Fluorescent isolates were considered to be E. coli. PCR amplification and sequencing of the 16S rRNA gene was performed on a small subset of isolates for unambiguous species verification. Positively identified isolates were inoculated into cryovials containing 80% tryptic soy broth and 20% glycerol. The inoculated cryovials were coded to conceal their sources, vortexed, and frozen (−80°C) for later use in antimicrobial resistance screening assays.
Serotyping and ECOR grouping.
Serotyping was performed at the Gastroenteric Disease Center, Pennsylvania State University, University Park. E. coli SMS-3-5 was classified according to the E. coli reference group (ECOR) system (32) by in silico analysis of the complete genome sequence based on the rapid-phylogenetic-grouping PCR system technique (17).
Genome sequencing and annotation.
Whole-genome random shotgun plasmid insert libraries of 3 to 5 kbp and 10 to 12 kbp and fosmid insert libraries of 30 to 40 kbp were constructed as previously reported (68) and sequenced using a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). Recombinant plasmid vector clones were constructed using the pUC-derived pHOS2 vector system (50) in combination with Thunderbolt GC10 electrocompetent cells (Sigma-Aldrich, St. Louis, MO). Assembly and closure were performed as previously described (68).
Comparative sequence analysis.
Coding sequences (CDS) shared between different chromosomes or plasmids were determined using BLAST score ratio analysis (67). In brief, for each predicted peptide sequence in a reference chromosome, the BLASTP raw score was collected for alignment against itself (REF_SCORE) and the most similar protein (QUE_SCORE) in each of the query genomes analyzed. These scores were normalized by dividing the QUE_SCORE obtained for each query plasmid protein by the REF_SCORE. In general, CDS with a normalized ratio of >0.4 are identical over 30% of their full-length sequences.
Antimicrobial susceptibility testing.
E. coli isolates from Shipyard Creek and recombinant clones from the sequencing libraries were screened for susceptibility to a panel of 26 antibiotics by using microdilution onto commercial dehydrated 96-well MicroScan panels (Dade Behring, Sacramento, CA) in cation-adjusted Mueller-Hinton broth (56a) according to the manufacturer's instructions. The antimicrobials present on the MicroScan panels were chosen based on their modes of action, propensities for resistance, and clinical relevance. The tested antimicrobials from nine structural groups included aminoglycosides (amikacin, 8 to 64 mg/liter; gentamicin, 2 to 16 mg/liter; streptomycin, 16 to 128 mg/liter; and apramycin, 8 to 32 mg/liter), β-lactams (ampicillin, 4 to 32 mg/liter; amoxicillin, 4 to 32 mg/liter; penicillin, 16 to 128 mg/liter; imipenem, 2 to 16 mg/liter; meropenem, 2 to 16 mg/liter; ceftriaxone, 8 to 64 mg/liter; cefoxitin, 8 to 32 mg/liter; cephalothin, 16 to 128 mg/liter; and cephalexin, 16 to 128 mg/liter), folate synthesis inhibitors (trimethoprim, 2 to 16 mg/liter, and trimethoprim-sulfamethoxazole, 2 and 38 to 4 and 76 mg/liter), sulfa agents (sulfathiazole, 250 to 500 mg/liter), nitrofurantoins (nitrofurantoin, 16 to 128 mg/liter), tetracyclines (oxytetraycline, 4 to 32 mg/liter, and tetracycline, 4 to 32 mg/liter), quinolones (ciprofloxacin, 1 to 4 mg/liter; moxifloxacin, 0.25 to 4 mg/liter; nalidixic acid, 4 to 32 mg/liter; and ofloxacin, 1 to 8 mg/liter), macrolides (azithromycin, 2 to 8 mg/liter, and erythromycin, 16 to 128 mg/liter), and chloramphenicols (chloramphenicol, 8 to 32 mg/liter). All concentration used in the analysis were successive doublings ranging from the minimum to the maximum levels. In addition to the antibiotics used on the MicroScan panels, resistances to the antimicrobial agents kanamycin, piperacillin, levofloxacin, norfloxacin, sparfloxacin, and rifampin were also assessed using liquid medium microdilution as previously described (56a). For antibiotics with high-level resistance, successive doublings of antimicrobial concentrations were used until inhibition was reached or until saturation of antimicrobials in liquid media over the course of the susceptibility testing was evident. Thunderbolt GC10 electrocompetent cells were used for the construction of plasmid vector sequencing libraries, and a pHOS2 clone with a recombinant insert spanning a 3.7-kbp region from the ribosomal-protein gene cluster served as a negative control for MIC tests.
Swarming-motility assays.
The swarming motility of E. coli SMS-3-5 was compared to that of E. coli K-12 and assayed under different growth conditions. Liquid cultures were freshly inoculated from glycerol stocks and grown overnight in LB medium. Aliquots of 20 to 50 μl were dropped onto plates with 0.5%, 1.0%, 1.5%, or 2.0% agar supplemented with LB, brain heart infusion, or marine broth medium (Difco).
Nucleotide sequence accession numbers.
The genome sequence of E. coli SMS-3-5 has been deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank) under accession numbers CP000970 (chromosome), CP000971 (pSMS35_130), CP000972 (pSMS35_8), CP000973 (pSMS35_4), and CP000974 (pSMS35_3).
RESULTS
The environmental E. coli isolate SMS-3-5.
E. coli strain SMS-3-5 was isolated from Shipyard Creek, an industrially contaminated tidal system in the southeastern United States. The isolate was obtained as part of a large-scale coastal E. coli screening program and was selected for in-depth analysis due to its exceptional level of antibiotic resistance. Of the 433 isolates that were analyzed for this study, 12% were resistant to five or more antimicrobial compounds (C. Baker-Austin, unpublished data). E. coli SMS-3-5 is tolerant of or resistant to high numbers of antibiotics (32 out of 33 tested compounds [Table 1]) from 11 structural classes, with many of the MICs above previously documented levels or at maximum drug solubility. Extreme resistance was found to diverse aminoglycosides, penicillins, cephalosporins, fluoroquinolones, macrolides, phenicoles, sulfonamides, and tetracyclines. E. coli SMS-3-5 is resistant to exceptionally high concentrations of nalidixic acid (>1,000 μg/ml); the expanded-spectrum quinolones ciprofloxacin (200 μg/ml), norfloxacin (>500 μg/ml), and ofloxacin (>1,000 μg/ml); and the broad-spectrum quinolones levofloxacin (125 μg/ml) and moxifloxacin (45 μg/ml). To our knowledge, the MIC for moxifloxacin is the highest and that for ciprofloxacin is the second highest on record (34). According to the ECOR typing system (32), E. coli SMS-3-5 belongs to group D, and its serotype is O19:H34.
TABLE 1.
Resistance levels of E. coli SMS-3-5
| Antibiotic | MIC (μg/ml) | CLSI MIC breakpoint (μg/ml) |
|---|---|---|
| β-Lactams | ||
| Amoxicillin | >5,000 | NDa |
| Ampicillin | >2,500 | ≥32b |
| Cefoxitin | 64 | ≥32b |
| Ceftriaxone | 32 | ≥64b |
| Cephalexin | 200 | NDa |
| Cephalothin | 64 | ≥32b |
| Imipenem | 2 | ≥16b |
| Meropenem | 2 | ≥16b |
| Penicillin | 250 | NDa |
| Piperacillin | >10,000 | ≥128b |
| Aminoglycosides | ||
| Amikacin | 8 | ≥64b |
| Apramycin | 8 | NDa |
| Gentamicin | 2 | ≥16b |
| Kanamycin | >2,500 | ≥64b |
| Streptomycin | >5,000 | NDa |
| Quinolones | ||
| Ciprofloxacin | 200 | ≥4b |
| Levofloxacin | 125 | ≥8b |
| Moxifloxacin | 45 | NDa |
| Nalidixic acid | >1,000 | ≥32b |
| Norfloxacin | >500 | ≥16b |
| Ofloxacin | >1,000 | ≥8b |
| Sparfloxacin | 125 | NDa |
| Macrolides | ||
| Azithromycin | 2 | NDa |
| Erythromycin | 16 | NDa |
| Rifampin | 25 | NDa |
| Sulfonamides | ||
| Sulfathiazole | >5,000 | ≥512b |
| Trimethoprim | >500 | ≥16b |
| Trimethoprim/Sulfamethoxazole | >500 | ≥4/76b |
| Tetracyclines | ||
| Doxycycline | 60 | ≥16b |
| Oxytetracycline | 250 | NDa |
| Tetracycline | >512 | ≥16b |
| Miscellaneous | ||
| Chlorampenicol | 256 | ≥32b |
| Nitrofurantoin | <16 | ≥128b |
The E. coli SMS-3-5 genome.
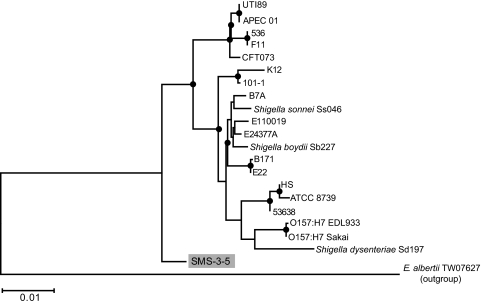
The E. coli SMS-3-5 genome consists of five replicons, a 5,068,389-bp chromosome and four plasmids (pSMS35_130, 130,440 bp; pSMS35_8, 8,909 bp; pSMS35_4, 4,074 bp; and pSMS35_3, 3,565 bp). The general features of the E. coli SMS-3-5 genome compared to those of seven previously sequenced E. coli genomes are shown in Table 2. The phylogenetic relationship of E. coli SMS-3-5 to 20 previously sequenced E. coli and Shigella strains was investigated by multilocus sequence typing (MLST) using the approach described by Wirth et al. (93). This MLST schema assigns E. coli SMS-3-5 to a phylogenetic clade that is closer to the last common ancestor and separates the isolate from any of the previously sequenced E. coli and Shigella strains (Fig. 1). According to the comparison of E. coli SMS-3-5 to the MLST database of 462 E. coli isolates (http://web.mpiib-berlin.mpg.de/mlst), the strain belongs to the ST354 complex, which contains several isolates from urinary tract infections (three out of five isolates).
TABLE 2.
General features of the genomes of E. coli SMS-3-5 and seven previously sequenced E. coli strains
| Strain and replicon(s)a | Size (bp) | GC (%) | No. of CDS | Reference |
|---|---|---|---|---|
| SMS-3-5 | ||||
| Chr | 5,068,389 | 50 | 4,743 | |
| pSMS35_130 | 130,440 | 50 | 153 | |
| pSMS35_8 | 8,909 | 46 | 10 | |
| pSMS35_4 | 4,074 | 49 | 4 | |
| pSMS35_3 | 3,565 | 43 | 3 | |
| K-12 | 10 | |||
| Chr | 4,639,675 | 50 | 4,131 | |
| EDL933 | 65 | |||
| Chr | 5,528,445 | 50 | 5,324 | |
| pO157 | 92,077 | 47 | 99 | |
| Sakai | 30 | |||
| Chr | 5,498,450 | 50 | 5,253 | |
| pO157 | 92,721 | 47 | 85 | |
| pOSAK1 | 3,306 | 43 | 3 | |
| APEC O1 | 39 | |||
| Chr | 5,082,025 | 50 | 4,457 | |
| pAPEC-O1-R | 241,342 | 47 | 225 | |
| pAPEC-O1-ColBM | 174,242 | 50 | 198 | |
| UTI89 | 13 | |||
| Chr | 5,065,741 | 50 | 5,044 | |
| pUTI89 | 114,230 | 51 | 145 | |
| CFT073 | 90 | |||
| Chr | 5,231,428 | 50 | 5,379 | |
| 536 | 33 | |||
| Chr | 4,938,920 | 50 | 4,629 |
Chr, chromosome.
FIG. 1.
Maximum likelihood phylogenetic tree of sequenced Escherichia and Shigella strains based on MLST. The alignments were produced with ClustalW (14). Modeltest 3.5 (66) was used to determine the most appropriate evolutionary model. The tree was created with PAUP* (83). Nodes marked with circles have ≥70% neighbor-joining bootstrap support. Similar results were obtained for trees calculated by neighbor-joining and parsimony methods.
(i) The E. coli SMS-3-5 chromosome.
Comparative sequence analysis was carried out to identify unique genomic islands (GIs) and to further investigate the evolutionary relationship of E. coli SMS-3-5 to seven previously sequenced E. coli strains. The genome sequence of the nonpathogenic E. coli strain K-12 served as a reference for this analysis (10). Other genome sequences included those of the avian-pathogenic (APEC) E. coli strain APEC O1 (39) and five human-pathogenic E. coli strains: three uropathogenic E. coli (UPEC) (UTI89 [13], CFT073 [90], and 536 [33]) and two enterohemorrhagic E. coli (EHEC) (O157:H7 EDL933 [65] and O157:H7 Sakai [30]) strains. Overall, the 5.06-Mbp chromosome of E. coli SMS-3-5 showed high sequence similarity and gene synteny with these previously sequenced E. coli genomes (Fig. 2). Of the 4,743 CDS that are carried on the E. coli SMS-3-5 chromosome, 341 CDS (7.2%) are unique to the strain, whereas 3,230 CDS (68.1%) are shared with all seven previously sequenced E. coli chromosomes (see Table S1 in the supplemental material). Unique features of the chromosome are associated with GIs of deviating nucleotide composition patterns, IS elements, and CDS with divergent codon usages, indicating that these regions have been acquired through horizontal gene transfer (Fig. 2).
FIG. 2.
Circular representation of the E. coli SMS-3-5 chromosome. From outside to inside: circles 1 and 2, strand-dependent depiction of all CDS; circle 3, IS elements (red) and laterally transferred CDS (dark green), predicted with SigiHMM (87); circle 4, trinucleotide sequence composition; circles 5 to 11, NUCmer (47) sequence comparisons (>95% nucleotide identity) with the chromosomes of E. coli strains K-12 (circle 5), O157:H7 EDL933 (circle 6), O157:H7 Sakai (circle 7), APEC O1 (circle 8), UTI89 (circle 9), CFT073 (circle 10), and 536 (circle 11) (sequence matches in the same orientation are shown in red and those in inverted orientation in blue); circle 12, cumulative GC skew. Selected GIs on the SMS-3-5 chromosome are highlighted, depending on functional annotations, in light green (virulence factors [Table 3]), gold (surface structures), orange (phages), blue (metabolism), and gray (unknown functions). Genetic loci involved in quinolone resistance are marked with black arrowheads with designations corresponding to those in Table 5. Abbreviations (clockwise, with gene names in italics): Col, colicin; Fim1 to Fim7, fimbriae; mhpRABCDEF, 3-hydroxyphenylpropionate degradation; Glu, glutamate fermentation (mutE and mamA); LPS1 to LPS3, lipopolysaccharide biosynthesis; lsrRABCDFGK, LuxS-regulated autoinducer uptake and modification; puuRABCD, putrescine utilization; hyfRABCDEFGHI, hydrogenase 4; cas, CRISPR-associated proteins; scrRABKY, sucrose uptake and degradation; rafRABDY, raffinose uptake and utilization; sitABCD, iron/manganese transport; iucABCD and iutA, aerobactin biosynthesis and transport; kpsCDEFMTUS and neuABCDES, polysialic acid capsule biosynthesis; gspCDEFGHIJKLM, general secretion pathway; glcABCDEFG, glycolate utilization; PTS, carbohydrate-specific PTS system; TTS, Salmonella type III secretion system (sicA, sipBD, and hilA); alsABCEK, allose uptake and utilization; HRA-1, heat-resistant agglutinin 1; GimA, carbon source-regulated cell invasion (35).
On the E. coli SMS-3-5 chromosome, the distribution patterns of the GIs that are shared by different subsets of previously sequenced E. coli chromosomes confirm the close evolutionary relationship previously reported between APEC and UPEC strains (39). They also indicate similarities in the presence or absence of GIs between the two EHEC strains and K-12 (Fig. 2). A large number of GI-located CDS from the E. coli SMS-3-5 chromosome are shared either with the chromosomes of the APEC and UPEC strains (104, 2.2%) or with the chromosomes of the EHEC strains and E. coli K-12 (133, 2.8%). For a complete list of conserved CDS, see Table S1 in the supplemental material. GIs shared between E. coli SMS-3-5 and UPEC/APEC strains tend to encode virulence-associated functions (i.e., colicin, polysialic acid capsule biosynthesis, and the general secretion pathway) (Table 3), whereas GIs shared between E. coli SMS-3-5 and EHEC/K-12 include a higher number of gene clusters involved in metabolic functions (i.e., the hydroxyphenylproprionate pathway, the first step of glutamate fermentation [absent from K-12], and the putrescine utilization pathway). The hyf operon encoding the biosynthesis of hydrogenase 4, absent from the APEC/UPEC strains, is also present in E. coli SMS-3-5, the EHEC strains, and K-12.
TABLE 3.
Putative virulence-associated functions encoded on the large resistance plasmid pSMS35_130 and in genomic islands of the SMS-3-5 chromosome
| Namea | CDS | Location | Function | Gene(s) | Distributionb | Best hitc |
|---|---|---|---|---|---|---|
| hlyF | EcSMS35_A0098 | pSMS | Hemolysin gene from avian E. coli isolate (56) | hlyF | APEC O1 (pAPEC-O1-ColBM) | P1658/97 (plasmid) |
| ompT | EcSMS35_A0099 | pSMS | Outer membrane protease gene (72) | ompT | APEC O1 (pAPEC-O1-ColBM) | pAPEC-O2-ColV (plasmid) |
| colB′ | EcSMS35_A0106-A0107 | pSMS | Colicin-B biosynthesis and immunity proteins (75, 76) | cba′, cbi′d | APEC O1 (pAPEC-O1-ColBM) | pAPEC-O1-ColBM (plasmid) |
| colM | EcSMS35_A0111-A0112 | pSMS | Colicin-M biosynthesis and immunity proteins (44, 62) | cma, cmi | APEC O1 (pAPEC-O1-ColBM) | pAPEC-O1-ColBM (plasmid) |
| Col | EcSMS35_0116-0121 | Chre | Uropathogen-specific colicin biosynthesis and immunity proteins | UPEC, APEC O1 | UTI89 | |
| Flag-2 | EcSMS35_0241-0284 | Chr | Lateral flagellar operon (69) | lafABCDEFKSTUVWZ, lfgABCDEFGHIJKLMN, lfhAB, lfiEFGHIJMNPQR | E. coli O42 | |
| sit-sitABCD | EcSMS35_3170-3173 (chromosome) | pSMS/Chr | Iron/manganese transport system | sitABCD | APEC O1 (pAPEC-O1-ColBM) | pAPEC-O2-ColV (plasmid) |
| EcSMS35_A0081-A0084 (pSMS35) | gene clusters (73) | |||||
| iut/iuc | EcSMS35_3178-3182 | Chr | Aerobactin biosynthesis and transport system gene cluster (22) | iucABCD, iutA | APEC O1 (pAPEC-O1-ColBM) | pAPEC-O1-ColBM (plasmid) |
| kps | EcSMS35_3219-3235 | Chr | Polysialic acid capsule biosynthesis operon (3) | kpsCDEFMTUS, neuABCDES | APEC O1, UPEC | UTI89 |
| gsp | EcSMS35_3238-3248 | Chr | General secretion pathway operon | gspCDEFGHIJKLM | APEC O1, UTI89, 536 | UTI89 |
| TTS | EcSMS35_4018-4024 | Chr | Putative type III secretion system, chaperone and effector components | Shigella dysenteriae Sd197 (20%)f | ||
| HRA-1 | EcSMS35_4755 | Chr | Heat-resistant agglutinin 1 (51) | UTI89, CFT073 | E. coli O9:H10:K99 | |
| GimA | EcSMS35_4856-4869 | Chr | Cell invasion and carbon metabolism (35) | ibeRAT, cglDECT, gcxCIKR, ptnCKE | APEC O1, UTI89 | E. coli A0 34/86 |
| Fim1 | EcSMS35_0146-52 | Chr | Putative fimbrial biosynthesis operons | (K12, APEC, UPEC)g | ||
| Fim2 | EcSMS35_0575--80 | K12, EHEC | Sakai | |||
| Fim3 | EcSMS35_2175-85 | K12, O157:H7 | Sakai | |||
| Fim4 | EcSMS35_2491-97 | Shigella sonnei Ss046 (50%) | ||||
| Fim5 | EcSMS35_2509-13 | CFT073 | CFT073 | |||
| Fim6 | EcSMS35_3921-29 | |||||
| Fim7 | EcSMS35_4093-96 | Shigella flexneri 5b 8401 | ||||
| LPS1 | EcSMS35_1020-28 | |||||
| LPS2 | EcSMS35_2255-68 | Chr | Putative lipopolysaccharide biosynthesis operons | |||
| LPS3 | EcSMS35_3959-67 | UPEC, APEC O1 | 536 |
Among completed E. coli strains (K-12, EDL933, Sakai, APEC O1, UTI89, CFT073, and 536) or pathotypes (UPEC, UTI89, CFT073, and 536; EHEC, EDL933 and Sakai).
According to nucleotide sequence BLAST comparison with NCBI nr database.
Truncated genes.
Chr, chromosome.
Percentage of query region covered by best hit.
Gene cluster with weak similarity present in non-EHEC strains.
Table 3 shows some of the GI-located CDS found in a subset of the other E. coli genomes and directly implicated in virulence-associated functions. These GIs encode, among others, the biosynthesis of the polysialic acid capsule (3), a cell invasion system (35), and the heat-resistant agglutinin 1 (51). Ten chromosomal GIs are predicted to code for the biosynthesis of fimbrial (Fim1-Fim7) or lipopolysaccharide (LPS) structures that could be linked to virulence or could reflect adaptations of E. coli SMS-3-5 to its environmental habitat. Except for a single fimbrial gene cluster, which is present in E. coli SMS-3-5 and the UPEC strain CFT073 (Fim5), all other surface-related gene clusters are either unique in E. coli SMS-3-5 (Fim1, Fim4, Fim6, Fim7, and LPS1 to LPS3) or shared with the EHEC strains and E. coli K-12 (Fim2-Fim3).
E. coli SMS-3-5 carries the complete Flag-2 gene cluster that encodes a novel flagellar system and that was first described in the enteroaggregative E. coli strain O42 (69). This cluster is absent from all the other E. coli strains analyzed, except for the two outermost genes, lfhA and lafU (see Fig. S1 in the supplemental material [modified after reference 55]). Flag-2 was shown to be present in only 15 of the 72 ECOR strains tested by Ren et al. (69). In E. coli SMS-3-5, codon usage or deviating nucleotide sequence analyses do not indicate integration of horizontally transferred DNA at this chromosomal site, making it likely that this cluster was deleted in other E. coli genomes. The Flag-2 gene clusters from E. coli SMS-3-5 and E. coli O42 are nearly identical (>94% nucleotide sequence identity) and display similarities to the corresponding gene clusters from Aeromonas hydrophila (11) and Vibrio parahaemolyticus (81) (see Fig. S1 in the supplemental material). Interestingly, the E. coli O42 Flag-2 gene cluster carries a single frameshift mutation in the chaperone protein lfgC, which is consistent with the failure to elicit swarming motility in E. coli O42. While the Flag-2 gene cluster appears to be intact and complete in E. coli SMS-3-5, we were not able to reproducibly induce swarming motility in the strain (see Materials and Methods).
The E. coli SMS-3-5 chromosome carries two unique metabolic islands that are likely to increase the growth substrate spectrum of this strain compared to previously sequenced pathogenic E. coli strains. These islands code for sucrose (scrRABKY) and raffinose (rafRABDY) uptake and degradation systems (Fig. 2). While rafRABDY was previously described as carried on plasmids (5), in E. coli SMS-3-5 it is located within a mosaic region of the chromosome containing the highest density of IS elements in the entire genome. In this IS-rich region, E. coli SMS-3-5 also harbors CDS for the synthesis and uptake of the siderophore aerobactin (iucABCD and iutA) (22) and for an iron/manganese transport system (sitABCD) (73). Both transport systems have been associated with virulence in APEC strains (72) in which corresponding gene clusters are located on virulence plasmids, such as pAPEC-O1-ColBM (38) and pAPEC-O2-ColV (41). A second copy of the sitABCD cluster in the E. coli SMS-3-5 genome is located on the large plasmid pSMS35_130 (Fig. 3). Both clusters are highly conserved (95% nucleotide identity over the 3.5-kbp gene cluster), but the similarity is limited to the sitABCD operon and does not include the surrounding sequences. To our knowledge, this is the first time that both the aerobactin and the iron/manganese transport systems have been found chromosomally encoded and that sitABCD is present in two different copies within the same genome.
FIG. 3.
Circular representation of pSMS35_130. pSMS35_130 is shown as composed of transfer, virulence, and resistance regions. From outside to inside: circles 1 and 2 (from different regions), strand-specific depiction of all CDS, color coded according to predicted functions in gray (replication), dark green (transfer and maintenance), brown/gold/red (transposition and recombination), light blue (APEC virulence), pink (colicin biosynthesis and immunity), and green (antibiotic resistance) (IS1 elements are depicted in gold, IS26 elements in red, and all other CDS in black); circles 3 to 6, NUCmer sequence comparisons (>90% nucleotide identity) with the E. coli plasmids p1658/97 (circle 3), pAPEC-O1-ColBM (circle 4), pAPEC-O2-ColV (circle 4), pAPEC-O2-R (circle 6), and pSC138 from S. enterica serovar Choleraesuis (circle 6) (sequence matches in the same orientation are shown in red and those in inverted orientation in blue); circle 7, laterally transferred CDS (black); circle 8, trinucleotide sequence composition. Abbreviations (clockwise, with gene names in italics): tra-trb (traABCDEFGHIJKLMNPQRSTUVWXY and trbABCDEHIJ), type IV secretion system-like conjugative transfer system; parB′, ParB-like partition protein; ssb, single-strand binding protein; sitABCD, iron/manganese transport system; ompT, outer membrane protease 7; colB′, truncated colicin-B biosynthesis and immunity proteins; colM, colicin-M biosynthesis and immunity proteins; cmlA, chloramphenicol transferase 2; dhfrV, dihydrofolate reductase type V; sul2, dihydropteroate synthase type 2; strAB, aminoglycoside resistance proteins A and B; blaT, beta-lactamase TEM; aadA, aminoglycoside adenylyltransferase; tetRA, tetracycline resistance repressor and resistance protein, class A; aph, aminoglycoside 3′-phosphotransferase.
(ii) The E. coli SMS 3-5 plasmids.
The largest plasmid from the E. coli SMS-3-5 genome, pSMS35_130, shares highly similar (>80% nucleotide identity) sequence regions with the two APEC virulence plasmids pAPEC-O1-ColBM (38) (62% coverage) and pAPEC-O2-ColV (41) (49% coverage) (Fig. 3, circles 1 and 2). In addition, pSMS35_130 shares a subset of those regions with the resistance plasmids p1658/97, harbored by an E. coli strain isolated from the human trachea (98) (61% coverage), and pAPEC-O2-R (40) (47% coverage), from an APEC isolate (Fig. 3, circles 3 and 4). Based on the comparative sequence analysis, pSMS35_130 can be divided into three regions that show distinct degrees of conservation among resistance- and virulence-associated plasmids. They encode functions related to plasmid transfer and maintenance, virulence, and antibiotic resistance (Fig. 3).
Antimicrobial resistance is encoded within a single continuous region on pSMS35_130 (∼42 kb) that is characterized by a high density of predicted horizontally transferred DNA and a deviating nucleotide composition. Furthermore, a total of 32 transposase and integrase genes, including 10 copies of IS26, are spread across the resistance region, in most cases marking the borders of syntenic regions shared by pSMS35_130 and related plasmids (Fig. 3). The resistance region of pSMS35_130 shows the highest overall similarity to the resistance plasmid pSC138 from Salmonella enterica serovar Choleraesuis (16) (>80% similarity; 64% coverage) and to a chromosomal GI from Acinetobacter baumannii AYE (26) (>80% similarity; 53% coverage). In both cases, the syntenic stretches are shorter than 5 kbp and limited to the resistance region of pSMS35_130. Individual resistance gene cassettes are composed of resistance genes and adjacent mobile genetic elements and show widespread distribution among taxonomically and biogeographically distinct drug-resistant isolates (Table 4). The pSMS35_130 resistance region codes for resistance to chloramphenicol (cmlA), trimethoprim (dhfrV), sulfonamides (sul2), aminoglycosides (strAB, aadA, and aph), beta-lactams (blaT), and tetracyclines (tetA). Sequencing clones harboring several of these genetic elements were shown to confer antibiotic resistance on susceptible E. coli strains (Table 5).
TABLE 4.
Comparison of resistance gene cassettes from pSMS35_130 to the NCBI nonredundant nucleotide database (http://www.ncbi.nlm.nih.gov)
| Resistance phenotypea | Gene compositionb | Length (kb) | No. of BLAST Hits | Bacterial isolatec | Accession no. | Source of isolate |
|---|---|---|---|---|---|---|
| Cp | IS26-cmlA-chp.-IS26 | 3.0 | 3 | Salmonella sp. strain TC67, pl. TC67 | AB262968 | Japan (2006)d |
| Klebsiella pneumoniae NK245, pl. pK245 | DQ449578 | Clinical isolate, Taiwan (2006)d | ||||
| S. enterica serovar Choleraesuis SC-B67, pl pSC138 | AY509004 | Septic patient, Taiwan (2002) | ||||
| Tm | Int.-dhfrV | 1.9 | 8 | K. pneumoniae NK245, pl. pK245 | DQ449578 | Clinical isolate, Taiwan (2006)d |
| S. enterica serovar Typhi CT18, pl. pHCM1 | AL513383 | Mekong River delta, Vietnam (1993) | ||||
| Uncultured bacterium, pl. pRSB107 | AJ851089 | Wastewater plant, Germany (2004)d | ||||
| Achromobacter xylosoxidans | DQ393569 | Cystic fibrosis patient, France (2006)d | ||||
| E. coli, pl. pHSH1 | AY259085 | Clinical isolate, China (2000-2001) | ||||
| Aeromonas salmonicida, pl. pRAS1 | AJ517790 | Wild salmon, Norway (1989) | ||||
| Su, Am | IS5075-chp.-sul2-strAB | 4.0 | 1 | Salmonella enteritidis, pl. pCAST2 | AF542061 | Korea (2002)d |
| Su, Am | Chp.-sul2-strAB | 2.9 | 13 | Vibrio cholerae MO10, cTn SXT | AY034138 | Clinical isolate, India (1992) |
| K. pneumoniae NK245, pl. pK245 | DQ449578 | Clinical isolate, Taiwan (2006)d | ||||
| S. enterica serovar Newport SL254, pl. pSN254 | CP000604 | Retail meat, United States (2002) | ||||
| Aeromonas bestiarum 5S9, pl pAb5S9 | EF495198 | River sediment, France (2007)d | ||||
| S. enterica serovar Typhi CT18, pl. pHCM1 | AL513383 | Mekong River delta, Vietnam (1993) | ||||
| β-L | blaT-tnpR′-IS26 | 2.3 | 1 | S. enterica serovar Typhi CT18, pl. pHCM1 | AL513383 | Mekong River delta, Vietnam (1993) |
| β-L | blaT-tnpR′ | 1.3 | 11 | K. pneumoniae 2412097A, pl. pRMH760 | AY123253 | Clinical isolate, Australia (1997) |
| S. enterica serovar Parathyphi A AKU12601, pl. pAKU_1 | AM412236 | Clinical isolate, Pakistan (2002) | ||||
| Uncultured bacterium, pl. pRSB107 | AJ851089 | Wastewater plant, Germany (2004)d | ||||
| S. enterica serovar Choleraesuis L2454, pl. pMAK1 | AB366440 | Japan (2007)d | ||||
| E. coli 690FNR, pl. pLEW517 | DQ390455 | Primate feces, United States (1991-93) | ||||
| Stt, Am | sat-hyd.-aadA | 3.0 | 8 | S. enterica serovar Choleraesuis SC-B67, pl pSC138 | AY509004 | Septic patient, Taiwan (2002) |
| E. coli | EF560797 | Pig cecum, United Kingdom (2007)d | ||||
| E. coli CVM1562, pl. pCVM1562 | AY816216 | Diarrheic pig, United States (1998-99) | ||||
| Enterobacter cloacae, pl. pQC | DQ019420 | Clinical isolate, The Netherlands (2002) | ||||
| S. enterica serovar Typhimurium BF272 | EF051039 | Portugal (2002-04) | ||||
| T | Tnp.(Tn1721)′-chp.-tetRA-trp.-chp.-tnp.′ | <6.2 | 6 | S. enterica serovar Dublin L-789, pl. pMAK2 | AB366441 | Japan (2007)d |
| Uncultured bacterium, pl. pB10 | AJ564903 | Wastewater plant, Germany (2003)d | ||||
| E. coli JM109, Tn1721 | X61367 | Germany (1992)d | ||||
| S. enterica serovar Dublin, pl. pIE321 | EF633507 | Spain (2007)d | ||||
| Aeromonas salmonicida, pl. pRAS1 | AJ517790 | Wild salmon, Norway (1995) | ||||
| T | (Tnp.)-chp.-tetRA-trp.-chp.-(tnp.′) | <5.7 | 8 | Aeromonas caviae HG5B, pl. pRAS1 | CR376602 | Clinical isolate, United Kingdom (1997) |
| A.r baumannii AYE | CT025832 | France (2006)d | ||||
| Bordetella bronchiseptica, pl. pKBB4037 | AJ877266 | Germany (2005)d | ||||
| S. enterica serovar Typhimurium, pl. pFPTB1 | AJ634602 | Italy (2004) | ||||
| E. coli, pl. pC15-1a | AY458016 | Canada (2000-02) | ||||
| Am | IS26′-aphA-IS26 | 2.2 | 8 | Pseudomonas aeruginosa 2293E | L36547 | 1994d |
| S. enterica serovar Typhimurium G8430, pl. pU302L | AY333434 | Clinical isolate, United States (2006)d | ||||
| Uncultured bacterium, pl. pRSB107 | AJ851089 | Wastewater plant, Germany (2004)d | ||||
| Shigella flexneri SH595, TnSF1 | AF188331 | Taiwan (1999)d | ||||
| pl. NTP16 | L05392 | United Kingdom (1992)d | ||||
| - | IS26 | 0.8 | 108 | Gammaproteobacteriae | NAf | NA |
| Corynebacterium sp. (Actinobacteria) |
Cp, chloramphenicol; Tm, trimethoprim; Su, sulfonamide; Am, aminoglycoside; β-L, beta-lactam; Stt, streptothricin; T, tetracycline.
Abbreviations: chp, conserved hypothetical protein; int, integrase/recombinase; hyd, hydrolase; tnp, transposase; trp, transporter. Gene names are in italics. Truncated elements are marked with ′. Genes in parentheses are not present in all hits.
Only continuous regions showing gene synteny and high nucleotide sequence identity (>90%) are shown as individual hits. pl., plasmid; cTn, conjugative transposon.
Estimated year of isolation based on submission of publication.
Similar IS26 elements (100% coverage, >99% nucleotide identity) have been identified in species of the following genera: Acinetobacter, Citrobacter, Enterobacter, Escherichia, Klebsiella, Pasteurella, Photobacterium, Providencia, Pseudomonas, Salmonella, Serratia, Shigella, Vibrio, and Yersinia.
NA, not applicable.
TABLE 5.
Resistance profiles of E. coli SMS-3-5, the laboratory E. coli strain GC10 without plasmid vector, and selected recombinant plasmid vector clones of GC10 from the whole-genome shotgun sequencing library of SMS-3-5
| Strain/Clone | Clone insertsa
|
Distributionb
|
MICc [μg/ml]
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (kbp) | No. of CDS | Gene(s)d | Replicone | K-12 | Other E. coli strains | N | Cf | O | Ot | T | St | Tm/Sm | Cp | |
| SMS-3-5 | 32 (1,024) | 4 | 8 | 32 | 32 (512) | 500 (>5000) | 4/76 (>500, >500f) | 32 (256) | ||||||
| GC10g | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| EC1BG86 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| EC2A028 | 9.6 | 11 | dhfrV, cmlA, IS26 | Pl | 0 | 0 (13-23) | 0 | 0 | 0 | 0 | 0 | 0 | 4/76 (>500,76e) | 32 (256) |
| EC1DP02 | 3.2 | 3 | sul2, IS5075 | Pl | 0 | 0-2 (0-6) | 0 | 0 | 0 | 0 | 0 | 500 (500) | 0 | 0 |
| EC1AK08 | 4.1 | 4 | aadA, sat-1, IS26 | Pl | 0 | 0 (0-59) | 0 | 0 | 0 | 0 | 0 | 500 (500) | 0 | 0 |
| EC1BD74 | 4.4 | 5 | tetRA, IS26, tnp-Tn1721 | Pl | 0 | 0 (0-55) | 0 | 0 | 0 | 32 | 32 (512) | 0 | 0 | 0 |
| EC2AN65 | 10.0 | 7 | fsr | Chr | 99 | 98-99 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| EC2A927 | 12.0 | 9 | (eefRABCD) | Chr | 34 | 99-100 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| EC2CI57 | 9.3 | 7 | ompF | Chr | 100 | 100 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| EC2AW38 | 11.6 | 7 | gyrA (MFS, GntR) | Chr | 66 | 66-67 | 32 (32) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| EC2CO68 | gyrBh | Chr | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| EC2AN16 | 9.3 | 7 | (MFS∼nor, LysR) | Chr | 75 | 75-99 (EHEC) | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| EC2AC67 | 11.2 | 10 | pmrD, yfbW | Chr | 100 | 100 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
A complete list of all clone inserts with corresponding CDS and coordinates can be found in Table S2 in the supplemental material.
Distribution (%) according to BLAST comparison. Other E. coli strains, O157:H7 EDL, Sakai, APEC O1, UTI89, CFT053, and 536. Fractions present on plasmids are in parentheses.
As determined on 96-well MicroScan panels. MICs for individual clones and antimicrobials, determined using liquid medium microdilution, are in parentheses. N, nalidixic acid (quinolone); Cf, ciprofloxacin (fluoroquinolone); O, ofloxacin (fluoroquinolone); Ot, oxytetracycline (tetracycline); T, tetracycline; St, sulfathiazole (sulfonamide); Tm/Sm, trimethoprim-sulfamethoxazole; Cp, chloramphenicol.
fsr, fosmidomycin resistance protein; CPA-2, monovalent cation:proton antiporter 2; Bcr/Acr, multidrug resistance gene cluster with similarity to bicyclomycin and acriflavin resistance proteins; ompF, outer membrane protein F; gyrA, DNA gyrase subunit A; GntR, transcriptional regulator, GntR family; MFS∼nor, transporter with weak similarity (25% amino acid identity) to quinolone resistance protein NorA from S. aureus; LysR, transcriptional regulator, LysR family; pmrD, polymyxin B resistance protein; yfbW, small multidrug resistance family protein; tnp, transposase. Genes not present in K-12 are shown in parentheses.
Pl, plasmid; Chr, chromosome.
MIC determinations for trimethoprim and sulfamethoxazole in liquid media were performed separately.
Thunderbolt GC10 electrocompetent cells (Sigma-Aldrich) and a sequencing clone (EC1BG86) spanning a region from the ribosomal protein gene cluster were used as negative controls.
Other clones without a resistance phenotype to any of the antibiotics listed carried inserts with parC-parE, soxRS, mdtABC-mdtD-mdtEF-mdtG-mdtH-mdtIJ-mdtK-mdtL-mdtM-mdtNOP, acrRAB-acrEF, marRAC, envRAB, mdlAB, macAB, and mdfA.
The remainder of pSMS35_130 is divided between a transfer region (∼54-kb) and a virulence region (∼33-kb). The pSMS35_130 transfer region has the most homogeneous nucleotide composition of the three regions, the lowest density of horizontally transferred DNA, and a strong bias in the orientation of the CDS (Fig. 3). It codes for type IV secretion-related conjugative transfer (traABCDEFGHIJKLMNPQRSTUVWXY and trbABCDEHIJ) and plasmid maintenance functions (hok, parB, sopAB, and pemIK). pSMS35_130 carries an incompatibility group IncFIIA origin of replication. The transfer region accounts for most of the similarities observed between pSMS35_130 and the antibiotic resistance plasmids pAPEC-O2-R and p1658/97, as well as the APEC virulence plasmids pAPEC-O1-ColBM and pAPEC-O2-ColV (Fig. 3). pAPEC-O2-ColV can be cotransferred with pAPEC-O2-R into plasmid-free laboratory E. coli strains (37). Since both plasmids and pSMS35_130 harbor similar sets of CDS for F-type conjugative transfer, it is likely that pSMS35_130 would be transferable to nonresistant strains. However, due to the broad resistance spectrum of E. coli SMS 3-5 and the resulting lack of counterselection markers available, the transferability of pSMS35_130 could not be confirmed. Interestingly, it was not possible to cure pSMS35_130 from the E. coli SMS-3-5 genome using either novobiocin treatment (up to 100 μg/ml) or growth at 40°C.
The virulence-associated region of pSMS35_130 also shows a mosaic structure with multiple inversions and loss of gene synteny compared to the APEC virulence plasmids pAPEC-O1-ColBM and pAPEC-O2-ColV or the antimicrobial resistance plasmid p1658/97. The virulence region is absent from other antimicrobial resistance plasmids and contains a subset of those virulence factors found on the APEC virulence plasmids, including the plasmid copy of the iron/manganese transport system sitABCD, an outer membrane protease gene (ompT), and a putative hemolysin F gene (hlyF). pSMS35_130 carries a colicin gene cluster that is similar to the one described on pAPEC-O1-ColBM (38) but is disrupted by insertions of IS1 and IS2. A total of five copies of IS1 are present on pSMS35_130. These elements flank the transfer region, separate some of the virulence genes in the virulence region, and are associated with breaking gene synteny between pSMS35_130 and the other plasmids analyzed. The virulence region carries a second origin of replication belonging to incompatibility group IncFIB. Interestingly, the only other resistance plasmid with similarity to the virulence region from pSMS35_130 lacks most of the putative APEC virulence factors (Fig. 3).
No evidence for the presence of additional resistance- or virulence-associated functions is found on any of the three small plasmids pSMS35_8, pSMS35_4, and pSMS35_3. pSMS35_8 shares a 6.5-kbp region (>90% nucleotide identity) with ColE1 (12), including the cea-imm gene cluster for colicin E biosynthesis and immunity. pSMS35_4 is almost identical (>74% nucleotide identity) to the two cryptic E. coli plasmids pMG828-2 (99% coverage; GenBank accession no. DQ995352) and pIGWZ12 (97) (96% coverage). No significant sequence similarity could be found between pSMS35_3 and any known plasmid sequence.
Antimicrobial resistance.
To assess the phenotypic impact of selected loci from the E. coli SMS-3-5 genome on the overall resistance phenotype of the strain, antimicrobial resistance tests were carried out (Table 5) using sequencing clones carrying genes thought to be involved in antimicrobial resistance. The MICs of tested antimicrobial compounds were determined for the recombinant vector-containing clones and compared to that of the susceptible vectorless host strain (Thunderbolt GC10 electrocompetent cells; Sigma-Aldrich, St. Louis, MO). A clone that carried a recombinant vector insert with a fragment from the ribosomal-protein gene cluster served as an additional negative control. Based on the final genome assembly, clones with short to medium insert sizes (3 to 12 kbp) were selected that contained DNA spanning regions encoding predicted antimicrobial resistance functions from the chromosome or plasmid pSMS35-5_130. The tested loci included CDS for known antimicrobial target proteins and for previously uncharacterized multidrug efflux systems (Table 5). Due to the intrinsic ampicillin resistance markers harbored by the plasmid cloning vector used to construct the sequencing library, we were unable to assay for resistance to β-lactam antibiotics. However, expression of individual and single-copy sul2, dhfrV, cmlA, and tetRA genes in this cloning system conferred resistance to sulfonamide, trimethoprim, chloramphenicol, and tetracycline, respectively, on the susceptible E. coli host strain. The resistance levels of the recombinant clones in all cases matched those established for E. coli SMS-3-5. In addition, the results revealed that a sequence region encompassing genes for aminoglycoside adenyltransferase (aadA) and a putative streptothricin acetyltransferase (sat-1) was able to confer a sulfonamide resistance phenotype on the E. coli host strain that was equivalent to the phenotypes observed for the recombinant sul2 clone and for E. coli SMS-3-5. AadA and Sat-1 have been characterized to confer resistance to aminoglycosides (15) and streptothricin (84), respectively, but not to sulfonamides. Further experiments are needed to confirm these findings and to explore the molecular mechanisms underlying the resistance.
Quinolone resistance.
The most common and best-understood mechanisms of high-level quinolone resistance in E. coli involve mutations in the genes encoding the antimicrobial targets DNA gyrase (gyrA and gyrB) and DNA topoisomerase IV (parC and parE) (34). Intrinsically involved with DNA replication, these genes are part of the basic microbial housekeeping apparatus and are carried on the chromosome. Single amino acid substitutions in GyrA have been shown to suffice for high-level nalidixic acid resistance in E. coli, but additional mutations in gyrAB or parCE are necessary for high-level fluoroquinolone resistance (31, 63, 86). Comparative sequence analysis of the gyrAB and parCE genes from E. coli SMS-3-5 with those of the quinolone-susceptible E. coli strain K-12 identified mutations that resulted in 11 amino acid substitutions, most of which were unique to E. coli SMS-3-5 (Fig. 4; see Table S2 in the supplemental material). The two amino acid substitutions most commonly identified in GyrA from quinolone-resistant E. coli strains (Ser83 and Asp87) (34) are found in E. coli SMS-3-5. Expression of a recombinant vector clone carrying a single copy of the E. coli SMS-3-5 gyrA gene in a quinolone-sensitive E. coli strain resulted in resistance to nalidixic acid (32 μg/ml, compared to 1,024 μg/ml for E. coli SMS-3-5) but not in detectable levels of resistance to ciprofloxacin and ofloxacin (Table 5). Since the same clone also carries a CDS for a transporter of the major facilitator superfamily (MFS), which is absent from K-12, the observed phenotype could also be the result of a previously unknown transport mechanism. The genes gyrB, parC, and parE carry nucleotide polymorphisms that result in a total of nine amino acid exchanges compared to their counterparts in E. coli K-12 (Fig. 4). To our knowledge, only two of these, Ser80Ile and Glu84Gly in ParC, have been related to fluoroquinolone resistance in E. coli, where they confer resistance to ciprofloxacin in combination with at least one mutation in gyrA (85). Due to the homology between the subunits of DNA gyrase and DNA topoisomerase IV, amino acid substitutions in the quinolone resistance-determining region in GyrA (Ser83 and Asp84) correspond to substitutions of Ser80 and Glu84 in ParC (46). Of the nine additional substitutions found in GyrAB and ParCE from E. coli SMS-3-5, four were also found in other protein homologs from E. coli and Shigella (data not shown). Interestingly, when expressed individually in quinolone-sensitive E. coli, parC, gyrB, or parE did not induce detectable quinolone resistance phenotypes, suggesting that a combination of these mutations might be required to induce quinolone resistance phenotypes.
FIG. 4.
Mutations in the DNA gyrase (gyrAB) and DNA topoisomerase IV (parCE) genes on the E. coli SMS-3-5 chromosome compared to those of E. coli K-12. The arrows indicate the resulting amino acid substitutions from E. coli K-12 to E. coli SMS-3-5. The chromosomal coordinates of the genes are shown. Amino acid substitutions known to be involved in quinolone resistance (34) are shaded in gray. Novel and unique amino acid substitutions are boxed.
Alterations in two major regulons, the multiple-antibiotic resistance operon marRAB and the redox-sensitive regulatory operon soxRS, can decrease membrane permeability and induce broad-range efflux pumps, thereby reducing microbial susceptibility to various toxic compounds, including quinolones (18). In E. coli, overexpression of marA has been shown to increase resistance to numerous antimicrobials, but the MICs determined for nalidixic acid (6.3 μg/ml), ofloxacin (0.1 μg/ml), and norfloxacin (0.1 μg/ml) (4) were low compared to those of E. coli SMS-3-5 (Table 1). MarA acts as a translation repressor of the outer membrane porin OmpF, which is believed to be critical in the diffusion of quinolones into the cell (18, 58). MIC testing results showed no increased levels of resistance to quinolones for recombinant clones carrying marRAB or soxRS, whereas ompF expression in a different clone could be responsible for the observed ofloxacin resistance phenotype (2 μg/ml) (Table 5). The lack of resistance phenotypes for recombinant clones expressing marRAB or soxRS could result from dominant effects that copies of the same genes from the chromosome have on the overall phenotypes of the host strain. ompF in E. coli SMS-3-5 carries several insertions/deletions (indels) compared to the corresponding gene in E. coli K-12. Resistance could therefore result from altered OmpF properties or changes in the expression of ompF. In addition to OmpF, MIC testing results associated three efflux systems with ofloxacin resistance: EefABC (4 μg/ml), an MFS transporter (2 μg/ml) with low similarity to NorA from Staphylococcus aureus (BAA14147; 25% protein sequence identity), and Fsr (2 μg/ml). The outer membrane multidrug efflux pump EefABC was identified in Enterobacter aerogenes and is functionally related to the acriflavin resistance system AcrAB-TolC (53). While AcrAB-TolC, together with MarA, was characterized in E. coli as responsible for a broad-range resistance phenotype to tetracycline, chloramphenicol, ampicillin, nalidixic acid, and rifampin (61), eefABC expression in E. aerogenes increases the levels of resistance to chloramphenicol, erythromycin, and ticarcillin, but not to quinolone compounds (52). In E. coli SMS-3-5, eefABC is part of a large gene cluster that is completely absent in E. coli K-12. Additional components in this gene cluster are a regulatory protein (EefR) and an additional membrane transport protein (EefD). The norA-related gene from E. coli SMS-3-5 confers resistance to ofloxacin alone, while norA from S. aureus confers resistance to both fluoroquinolones, ofloxacin and ciprofloxacin (96). Fsr is a distant member of the MFS, suggesting that it may function as a proton-dependent fosmidomycin efflux system (27, 60). According to our MIC testing results, fsr in E. coli SMS-3-5, in addition to mediating resistance to ofloxacin, appears to also be involved in ciprofloxacin resistance (2 μg/ml). A recombinant clone expressing the signal transduction protein gene pmrD and the MFS transporter gene yfbW also contributes to ciprofloxacin resistance (2 μg/ml). While pmrD is known to induce polymyxin resistance in some E. coli isolates (92), the function of yfbW is poorly understood. To our knowledge, this is the first direct evidence for the involvement of fsr and either pmrD or yfbW in resistance to quinolones and the first characterization of a new norA-related quinolone resistance gene. Of all the resistance factors that our MIC testing associated with quinolone resistance, codon usage analysis predicted only the norA-related gene from the E. coli SMS-3-5 chromosome to have originated from horizontal gene transfer.
DISCUSSION
The present study provides a new model for the functional characterization of selected genes and gene clusters in whole-genome sequencing projects. Using recombinant plasmid vector clones from shotgun sequencing libraries, the impact of specific genetic loci on the overall phenotypic characteristics of E. coli SMS-3-5 could be demonstrated. The expression of genes carried on sequencing plasmid vector inserts was sufficient to detect changes in the susceptibility levels of a standard laboratory E. coli strain. A methodological limitation to this approach is linked to the presence of the antibiotic resistance marker gene on the plasmid sequencing vectors, which creates background resistance.
For over a dozen agents, encompassing several different structural classes of drugs, E. coli SMS-3-5 demonstrated extreme antimicrobial resistance capabilities (Table 1), in many cases up to saturation levels of a given test agent using broth microdilution assays. Few other attempts have been made to ascertain the absolute limits of antibiotic resistance in E. coli using concentrations of individual agents that greatly exceed established antimicrobial MIC breakpoints (i.e., CLSI guidelines). Stapleton et al. (80) found high-level amoxicillin resistance with MIC levels at over 4,000 μg/ml in clinically derived E. coli strains (E. coli SMS-3-5, >5,000 μg/ml). Extreme resistance to ampicillin (>400 μg/ml), chloramphenicol (>316 μg/ml), and tetracycline (>400 μg/ml) was detected in fluoroquinolone-resistant clinically derived E. coli isolates (89), although in most cases, the resistance levels were significantly lower than in E. coli SMS-3-5 (ampicillin, >2500 μg/ml; chloramphenicol, 256 μg/ml; tetracycline, >512 μg/ml). Resistance levels to oxytetracycline (≥256 μg/ml) in agricultural E. coli isolates from pigs and pig farmers were found to be comparable to that of E. coli SMS-3-5 (250 μg/ml) (59). Mazzariol et al. (54) detected a level of resistance to ciprofloxacin in clinical E. coli isolates (>512 μg/ml) significantly higher than that determined for E. coli SMS-3-5 (200 μg/ml). To our knowledge, however, SMS-3-5 is the most fluoroquinolone-resistant environmental E. coli isolate studied to date.
The combination of whole-genome annotation and phenotypic characterization of selected gene clusters revealed that most of the resistance potential of E. coli SMS-3-5, including resistance to cephalosporin, tetracycline, chloramphenicol, sulfonamide, and trimethoprim, can be assigned to an ∼42-kb resistance region on pSMS35_130. Among the antimicrobial resistance phenotypes of E. coli SMS-3-5, high-level resistance to quinolone compounds plays a particularly noteworthy role, since it is carried solely on the chromosome and in regions without indication of recent gene transfer, with the exception of a norA-related gene for ofloxacin resistance. It was also demonstrated that quinolone resistance most likely involves both known resistance mechanisms (i.e., mutations in the quinolone target protein genes) and previously unknown or uncharacterized resistance mechanisms (i.e., porins and efflux systems). Quinolone resistance is mediated by combinations of E. coli housekeeping genes (gyrA and potentially gyrB and parCE, which contained a total of 11 amino acid substitutions) and multidrug efflux systems that have for the most part also been identified in E. coli K-12 and other sequenced E. coli isolates (fsr, eefRABCD, ompF, and pmrD-yfbW). Together with these transport systems, MarA could contribute to the high level of quinolone resistance. Resistance to different quinolone compounds is mediated by genes or gene clusters with specific, nonoverlapping resistance spectra. On one hand, gyrA and/or the adjacent MFS transporter gene induces resistance to the hydrophobic nalidixic acid, but not to the newer hydrophilic quinolones. On the other hand, the efflux systems OmpF, EefABC, Fsr, and the NorA-like protein confer resistance to extended-spectrum quinolones, but at relatively low levels and, except for Fsr, only to either ofloxacin or ciprofloxacin. Moreover, the gyrA-containing sequence fragment is responsible for only a fraction (32 μg/ml) of the total resistance phenotype to nalidixic acid (1,024 μg/ml) observed for E. coli SMS-3-5. These results demonstrate that resistance to different quinolone compounds is decoupled and that the cooperative actions of different mechanisms contribute to the overall resistance phenotype. Furthermore, we demonstrated the importance of efflux and membrane permeability factors in the high-level resistance to extended-spectrum quinolones.
At least one study suggests a relatively high biological cost of fluoroquinolone resistance in E. coli (8), implying that constant selective pressures are necessary to maintain resistance in mixed populations. However, 6 out of 433 E. coli strains isolated from Shipyard Creek expressed quinolone resistance (Baker-Austin, unpublished), showing that resistance in environmental E. coli populations at this site is rare but not unknown. Quinolone residues have been shown to persist in surface waters (45). Shipyard Creek is heavily contaminated with heavy metals, such as chromium, copper, zinc, and strontium (94), and trace metals and other industrial waste at the isolation site may have acted as indirect selection agents, e.g., by promoting microorganisms with effective broad-range efflux pumps (7, 82).
The physical origin of E. coli SMS-3-5 is unknown, making it difficult to predict the selective conditions that triggered the evolution of its exceptional antimicrobial resistance phenotype. Shipyard Creek is accessible to anthropogenic microbes from multiple sources through coastal currents and tides, as it discharges straight into the Charleston, SC, harbor. There are no known sewage sources in the creek drainage area. However, quinolones are widely used in cattle and poultry farming (64), in particular, to treat E. coli infections (78), and drug-resistant bacteria from agriculture could leach into the water through farming runoff. So far, APEC virulence plasmids have been identified only in host-associated E. coli isolates. Even though nothing is known about the prevalence of APEC-related resistance plasmids in the environment, the similarity of pSMS35_130 to APEC virulence plasmids suggests a close association of E. coli SMS-3-5 with avian E. coli strains, one possibility being transport of E. coli SMS-3-5 to Shipyard Creek by migrating birds. In any case, our study demonstrates that high-level resistance to multiple front-line quinolones, which is mediated by multiple mechanisms, is not limited to E. coli strains from clinical settings, but is also present in environmental populations.
The same site from which E. coli SMS-3-5 was isolated has also been sampled for two other studies that addressed the prevalence of multidrug-resistant strains of Vibrio vulnificus, a marine human pathogen without clinical reservoirs (6), and of mobile genetic elements (94). In these studies, unexpectedly high numbers of multidrug-resistant V. vulnificus strains were detected, with 17.3% of all isolates being resistant to eight or more antimicrobial compounds (6). However, while an increased abundance of class 1 integrons was found in samples from Shipyard Creek compared to uncontaminated sites (94), no consistent differences could be detected in the frequency of resistance between V. vulnificus isolates from this site and those from pristine environmental estuaries (6).
MLST comparison separates the environmental strain E. coli SMS-3-5 from other sequenced host-associated E. coli and Shigella isolates and suggests that it is more closely related to their last common ancestor (Fig. 1). A possible explanation for these findings would be that the genomes of host-associated E. coli and Shigella strains show signs of adaptive evolution. Pathogenesis in E. coli is believed to result from the acquisition and integration of horizontally transferred virulence factors into strain-specific chromosomal pathogenic islands or from the acquisition of plasmids harboring these genetic factors (9, 23, 42). The comparative genome analysis of E. coli SMS-3-5 with intra- and extraintestinal E. coli isolates provides clues that at least part of the adaptation of previously sequenced E. coli strains to their host-associated lifestyles was associated with genome reduction. The lateral flagellar gene cluster Flag-2 in E. coli SMS-3-5, remnants of which are still present at conserved chromosomal sites in other E. coli strains (69), provides an example of such a reductive evolutionary process. In this context, a principal analogy has been suggested for pathogenic E. coli isolates and laboratory strains, such as E. coli K-12, which has been propagated in nutrient-rich cultures for many generations (28). The single polar flagellar operon on the E. coli K-12 chromosome is a preferential target for genome decay, providing substantial savings in energy and amino acid requirements (19, 24). The same might hold true for the Flag-2 gene cluster in E. coli SMS-3-5. This cluster, which could be responsible for swarming motility under specific, not-yet-characterized conditions, might represent a phenotypic trait more advantageous to environmental E. coli populations. Further genome analysis of environmental and host-associated E. coli isolates is necessary to determine whether the lateral flagellar cluster is in fact predominantly found in the environmental fraction of the E. coli pangenome.
The plasmid pSMS35_130 is a composite plasmid. The IS elements IS26 and IS1 appear to have contributed to this mosaic structure, combining two separate regions encoding antimicrobial resistance and potential APEC virulence-related functions. Limited to the resistance region of pSMS35_130, IS26 (10 copies) is part of or separates individual conserved antimicrobial resistance gene cassettes. IS26 elements are widely distributed among gammaproteobacteria, where they are mostly associated with different resistance genes (IS finder database [http://www-is.biotoul.fr/]). Similar sequences have also been observed in gram-positive species (i.e., Corynebacterium; GenBank accession no. AM942444). A close association of IS26 with resistance genes in E. coli SMS-3-5 and other bacterial isolates is likely to facilitate the horizontal spread of antibiotic resistance. Repeated acquisition and integration events of individually acquired resistance gene cassettes mediated essentially by recombination between IS26 or other repetitive elements could therefore be the evolutionary drivers of the pSMS35_130 resistance region.
Analogously, the virulence region of pSMS35_130 can be regarded as composed of individual virulence gene cassettes that are separated from each other and from the plasmid transfer backbone by five copies of IS1 (Fig. 3). The boundaries of such gene clusters are apparent from interruptions in synteny between pSMS35_130 and the APEC virulence plasmids pAPEC-O1-ColBM and pAPEC-O2-ColV. The two origins of replication present in the transfer and virulence regions, respectively, suggest that pSMS35_130 evolved from an F-type ancestor through acquisition and integration of an additional plasmid encoding APEC-related virulence phenotypes. From an evolutionary standpoint, the combination of transfer and virulence determinants on a single plasmid would provide the selective benefit of a self-transferable virulence phenotype. Additional APEC virulence factors and IS1 elements are also located on the E. coli SMS-3-5 chromosome, suggesting that recombinatorial exchange of conserved virulence gene cassettes also involves the chromosome. However, the combined set of virulence-associated genes from the chromosome and pSMS35_130 is reduced compared to related APEC plasmids (38, 41, 72). The lack of the salmochelin biosynthesis operon (iroBCDEN) and the gene for increased serum survival (iss) could therefore be indicative of a general tendency toward the decay of the APEC phenotype.
Altogether, the combination of the resistance region and the APEC-like virulence region on plasmid pSMS35_130 has major implications for public health. Recently, such an APEC/resistance plasmid carrying a larger set of APEC virulence factors has been identified in a poultry isolate of S. enterica serovar Kentucky (W. F. Fricke, unpublished data). Should more of these plasmids be promiscuous and transfer to other strains or species, antibiotic use could play a critical role in coselecting for multidrug resistance and virulence phenotypes. Moreover, APEC-related virulence genes have been found to be upregulated during UPEC infections in humans (79), including sitAB and iucAD, both of which are present in E. coli SMS-3-5. Since it was demonstrated that a substantial overlap between plasmid-associated virulence genes from APEC and UPEC strains exists (71), it is possible to envision that APEC plasmids might function as a reservoir for human UPEC virulence factors.
The analysis of the E. coli SMS-3-5 genome sequence clearly shows that large fractions of antimicrobial resistance and potential virulence gene pools have been acquired horizontally. These include not only the mosaic plasmid pSMS35_130, but also chromosomal GIs, encoding, among others, polysialic acid capsule biosynthesis, the general secretion pathway, agglutinins, cell invasion, and a putative type III secretion system. These genes have high similarity to genes found in other bacterial isolates from diverse environmental and clinical settings.
Since the specific origin of the E. coli SMS-3-5 isolate remains obscure, it is impossible to define the impact of the environment on its evolution or to determine to what extent virulence and resistance determinants evolved in the environment. In either case, the comparative analysis of the E. coli SMS-3-5 genome presented in this study provides evidence for the recent and efficient transfer of mobile virulence and resistance elements between environmental and host-associated settings. This availability of a common virulence and resistance gene pool to environmental bacteria, as well as clinical pathogens, is of some concern and should be studied further.
Supplementary Material
Acknowledgments
This work was supported with Federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services, under NIAID contract N01-AI-30071.
Footnotes
Published ahead of print on 15 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 2006. Commensals upon us. Biochem. Pharmacol. 71893-900. [DOI] [PubMed] [Google Scholar]
- 2.Aminov, R. I., and R. I. Mackie. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271147-161. [DOI] [PubMed] [Google Scholar]
- 3.Andreishcheva, E. N., and W. F. Vann. 2006. Gene products required for de novo synthesis of polysialic acid in Escherichia coli K1. J. Bacteriol. 1881786-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asako, H., H. Nakajima, K. Kobayashi, M. Kobayashi, and R. Aono. 1997. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl. Environ. Microbiol. 631428-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslanidis, C., K. Schmid, and R. Schmitt. 1989. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J. Bacteriol. 1716753-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker-Austin, C., J. V. McArthur, A. H. Lindell, M. S. Wright, R. C. Tuckfield, J. Gooch, L. Warner, J. Oliver, and R. Stepanauskas. Multi-site analysis reveals widespread antibiotic resistance in the marine pathogen Vibrio vulnificus. Microb. Ecol., in press. [DOI] [PubMed]
- 7.Baker-Austin, C., M. S. Wright, R. Stepanauskas, and J. V. McArthur. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14176-182. [DOI] [PubMed] [Google Scholar]
- 8.Bemer, P., S. Corvec, C. Guitton, C. Giraudeau, G. Le Gargasson, E. Espaze, and H. Drugeon. 2007. Biological cost of fluoroquinolone resistance in Escherichia coli implicated in polyclonal infection. Pathol Biol. (Paris) 55288-291. [DOI] [PubMed] [Google Scholar]
- 9.Bielaszewska, M., U. Dobrindt, J. Gartner, I. Gallitz, J. Hacker, H. Karch, D. Muller, S. Schubert, M. Alexander Schmidt, L. J. Sorsa, and J. Zdziarski. 2007. Aspects of genome plasticity in pathogenic Escherichia coli. Int. J. Med. Microbiol. 297625-639. [DOI] [PubMed] [Google Scholar]
- 10.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 11.Canals, R., M. Altarriba, S. Vilches, G. Horsburgh, J. G. Shaw, J. M. Tomas, and S. Merino. 2006. Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 188852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, P. T., H. Ohmori, J. Tomizawa, and J. Lebowitz. 1985. Nucleotide sequence and gene organization of ColE1 DNA. J. Biol. Chem. 2608925-8935. [PubMed] [Google Scholar]
- 13.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 1035977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinault, A. C., V. A. Blakesley, E. Roessler, D. G. Willis, C. A. Smith, R. G. Cook, and R. G. Fenwick, Jr. 1986. Characterization of transferable plasmids from Shigella flexneri 2a that confer resistance to trimethoprim, streptomycin, and sulfonamides. Plasmid 15119-131. [DOI] [PubMed] [Google Scholar]
- 16.Chiu, C. H., P. Tang, C. Chu, S. Hu, Q. Bao, J. Yu, Y. Y. Chou, H. S. Wang, and Y. S. Lee. 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 331690-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 664555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, S. P., L. M. McMurry, D. C. Hooper, J. S. Wolfson, and S. B. Levy. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 331318-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper, T. F., D. E. Rozen, and R. E. Lenski. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 1001072-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dantas, G., M. O. A. Sommer, R. D. Oluwasegun, and G. M. Church. 2008. Bacteria subsisting on antibiotics. Science 320100-103. [DOI] [PubMed] [Google Scholar]
- 21.D'Costa, V. M., K. M. McGrann, D. W. Hughes, and G. D. Wright. 2006. Sampling the antibiotic resistome. Science 311374-377. [DOI] [PubMed] [Google Scholar]
- 22.de Lorenzo, V., A. Bindereif, B. H. Paw, and J. B. Neilands. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J. Bacteriol. 165570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobrindt, U. 2005. (Patho-)genomics of Escherichia coli. Int. J. Med. Microbiol. 295357-371. [DOI] [PubMed] [Google Scholar]
- 24.Edwards, R. J., R. E. Sockett, and J. F. Brookfield. 2002. A simple method for genome-wide screening for advantageous insertions of mobile DNAs in Escherichia coli. Curr. Biol. 12863-867. [DOI] [PubMed] [Google Scholar]
- 25.Falkow, S. 1996. The evolution of pathogenicity in Escherichia coli, Shigella and Salmonella, p. 2723-2729. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 26.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujisaki, S., S. Ohnuma, T. Horiuchi, I. Takahashi, S. Tsukui, Y. Nishimura, T. Nishino, M. Kitabatake, and H. Inokuchi. 1996. Cloning of a gene from Escherichia coli that confers resistance to fosmidomycin as a consequence of amplification. Gene 17583-87. [DOI] [PubMed] [Google Scholar]
- 28.Fux, C. A., M. Shirtliff, P. Stoodley, and J. W. Costerton. 2005. Can laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol. 1358-63. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg, A. E., L. S. Clesceri, and A. D. E. Eaton. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 30.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 811-22. [DOI] [PubMed] [Google Scholar]
- 31.Heisig, P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40879-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 1726175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochhut, B., C. Wilde, G. Balling, B. Middendorf, U. Dobrindt, E. Brzuszkiewicz, G. Gottschalk, E. Carniel, and J. Hacker. 2006. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 61584-595. [DOI] [PubMed] [Google Scholar]
- 34.Hopkins, K. L., R. H. Davies, and E. J. Threlfall. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25358-373. [DOI] [PubMed] [Google Scholar]
- 35.Huang, S. H., Y. H. Chen, G. Kong, S. H. Chen, J. Besemer, M. Borodovsky, and A. Jong. 2001. A novel genetic island of meningitic Escherichia coli K1 containing the ibeA invasion gene (GimA): functional annotation and carbon-source-regulated invasion of human brain microvascular endothelial cells. Funct. Integr. Genomics 1312-322. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, J. R., C. van der Schee, M. A. Kuskowski, W. Goessens, and A. van Belkum. 2002. Phylogenetic background and virulence profiles of fluoroquinolone-resistant clinical Escherichia coli isolates from the Netherlands. J. Infect. Dis. 1861852-1856. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, T. J., C. W. Giddings, S. M. Horne, P. S. Gibbs, R. E. Wooley, J. Skyberg, P. Olah, R. Kercher, J. S. Sherwood, S. L. Foley, and L. K. Nolan. 2002. Location of increased serum survival gene and selected virulence traits on a conjugative R plasmid in an avian Escherichia coli isolate. Avian Dis. 46342-352. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, T. J., S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 1885975-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 1893228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2005. DNA sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid. Antimicrob. Agents Chemother. 494681-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188745-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 43.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics, 1st ed. John Innes Foundation, Norwich, United Kingdom.
- 44.Kock, J., T. Olschlager, R. M. Kamp, and V. Braun. 1987. Primary structure of colicin M, an inhibitor of murein biosynthesis. J. Bacteriol. 1693358-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolpin, D. W., E. T. Furlong, M. T. Meyer, E. M. Thurman, S. D. Zaugg, L. B. Barber, and H. T. Buxton. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999-2000: a national reconnaissance. Environ. Sci. Technol. 361202-1211. [DOI] [PubMed] [Google Scholar]
- 46.Kumagai, Y., J. I. Kato, K. Hoshino, T. Akasaka, K. Sato, and H. Ikeda. 1996. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob. Agents Chemother. 40710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy, S. B. 2001. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 33(Suppl. 3)S124-S129. [DOI] [PubMed] [Google Scholar]
- 49.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433865-868. [DOI] [PubMed] [Google Scholar]
- 51.Lutwyche, P., R. Rupps, J. Cavanagh, R. A. Warren, and D. E. Brooks. 1994. Cloning, sequencing, and viscometric adhesion analysis of heat-resistant agglutinin 1, an integral membrane hemagglutinin from Escherichia coli O9:H10:K99. Infect. Immun. 625020-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masi, M., J. M. Pages, and E. Pradel. 2006. Production of the cryptic EefABC efflux pump in Enterobacter aerogenes chloramphenicol-resistant mutants. J. Antimicrob. Chemother. 571223-1226. [DOI] [PubMed] [Google Scholar]
- 53.Masi, M., J. M. Pages, C. Villard, and E. Pradel. 2005. The eefABC multidrug efflux pump operon is repressed by H-NS in Enterobacter aerogenes. J. Bacteriol. 1873894-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazzariol, A., Y. Tokue, T. M. Kanegawa, G. Cornaglia, and H. Nikaido. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 443441-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merino, S., J. G. Shaw, and J. M. Tomas. 2006. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol. Lett. 263127-135. [DOI] [PubMed] [Google Scholar]
- 56.Morales, C., M. D. Lee, C. Hofacre, and J. J. Maurer. 2004. Detection of a novel virulence gene and a Salmonella virulence homologue among Escherichia coli isolated from broiler chickens. Foodborne Pathog Dis. 1160-165. [DOI] [PubMed] [Google Scholar]
- 56a.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing; 14th informational supplement. NCCLS, Wayne, PA.
- 57.Neu, H. C. 1992. The crisis in antibiotic resistance. Science 2571064-1073. [DOI] [PubMed] [Google Scholar]
- 58.Neves, P., E. Berkane, P. Gameiro, M. Winterhalter, and B. de Castro. 2005. Interaction between quinolones antibiotics and bacterial outer membrane porin OmpF. Biophys. Chem. 113123-128. [DOI] [PubMed] [Google Scholar]
- 59.Nijsten, R., N. London, A. van den Bogaard, and E. Stobberingh. 1996. Antibiotic resistance among Escherichia coli isolated from fecal samples of pig farmers and pigs. J. Antimicrob. Chemother. 371131-1140. [DOI] [PubMed] [Google Scholar]
- 60.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 1835803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olschlager, T., and V. Braun. 1987. Sequence, expression, and localization of the immunity protein for colicin M. J. Bacteriol. 1694765-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozeki, S., T. Deguchi, M. Yasuda, M. Nakano, T. Kawamura, Y. Nishino, and Y. Kawada. 1997. Development of a rapid assay for detecting gyrA mutations in Escherichia coli and determination of incidence of gyrA mutations in clinical strains isolated from patients with complicated urinary tract infections. J. Clin. Microbiol. 352315-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedersen, K., and A. Wedderkopp. 2003. Resistance to quinolones in Campylobacter jejuni and Campylobacter coli from Danish broilers at farm level. J. Appl. Microbiol. 94111-119. [DOI] [PubMed] [Google Scholar]
- 65.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 66.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 67.Rasko, D. A., G. S. Myers, and J. Ravel. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinform. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasko, D. A., M. J. Rosovitz, O. A. Okstad, D. E. Fouts, L. Jiang, R. Z. Cer, A. B. Kolsto, S. R. Gill, and J. Ravel. 2007. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 18952-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ren, C. P., S. A. Beatson, J. Parkhill, and M. J. Pallen. 2005. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 1871430-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Riesenfeld, C. S., R. M. Goodman, and J. Handelsman. 2004. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6981-989. [DOI] [PubMed] [Google Scholar]
- 71.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 1512097-2110. [DOI] [PubMed] [Google Scholar]
- 72.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36241-256. [DOI] [PubMed] [Google Scholar]
- 73.Sabri, M., S. Leveille, and C. M. Dozois. 2006. A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152745-758. [DOI] [PubMed] [Google Scholar]
- 74.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12412-416. [DOI] [PubMed] [Google Scholar]
- 75.Schramm, E., J. Mende, V. Braun, and R. M. Kamp. 1987. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J. Bacteriol. 1693350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schramm, E., T. Olschlager, W. Troger, and V. Braun. 1988. Sequence, expression and localization of the immunity protein for colicin B. Mol. Gen. Genet. 211176-182. [DOI] [PubMed] [Google Scholar]
- 77.Schwarz, S., C. Kehrenberg, and T. R. Walsh. 2001. Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17431-437. [DOI] [PubMed] [Google Scholar]
- 78.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, and M. T. Osterholm. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 3401525-1532. [DOI] [PubMed] [Google Scholar]
- 79.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 726373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stapleton, P., P. J. Wu, A. King, K. Shannon, G. French, and I. Phillips. 1995. Incidence and mechanisms of resistance to the combination of amoxicillin and clavulanic acid in Escherichia coli. Antimicrob. Agents Chemother. 392478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stewart, B. J., and L. L. McCarter. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 1854508-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Summers, A. O. 2002. Generally overlooked fundamentals of bacterial genetics and ecology. Clin. Infect. Dis. 34(Suppl. 3)S85-S92. [DOI] [PubMed] [Google Scholar]
- 83.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 84.Tschape, H., E. Tietze, R. Prager, W. Voigt, E. Wolter, and G. Seltmann. 1984. Plasmid-borne streptothricin resistance in gram-negative bacteria. Plasmid 12189-196. [DOI] [PubMed] [Google Scholar]
- 85.Vila, J., J. Ruiz, P. Goni, and M. T. De Anta. 1996. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40491-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vila, J., J. Ruiz, F. Marco, A. Barcelo, P. Goni, E. Giralt, and T. Jimenez de Anta. 1994. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob. Agents Chemother. 382477-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waack, S., O. Keller, R. Asper, T. Brodag, C. Damm, W. F. Fricke, K. Surovcik, P. Meinicke, and R. Merkl. 2006. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinform. 7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walsh, T. R. 2006. Combinatorial genetic evolution of multiresistance. Curr. Opin. Microbiol. 9476-482. [DOI] [PubMed] [Google Scholar]
- 89.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 451515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilson, B. A., and A. A. Salyers. 2003. Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 11347-350. [DOI] [PubMed] [Google Scholar]
- 92.Winfield, M. D., and E. A. Groisman. 2004. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc. Natl. Acad. Sci. USA 10117162-17167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 601136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wright, M. S., C. Baker-Austin, A. H. Lindell, R. Stepanauskas, H. W. Stokes, and J. V. McArthur. 2008. Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J. 2417-428. [DOI] [PubMed] [Google Scholar]
- 95.Yates, C. M., D. J. Shaw, A. J. Roe, M. E. Woolhouse, and S. G. Amyes. 2006. Enhancement of bacterial competitive fitness by apramycin resistance plasmids from non-pathogenic Escherichia coli. Biol. Lett. 2463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 1726942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zaleski, P., R. Wolinowska, K. Strzezek, A. Lakomy, and A. Plucienniczak. 2006. The complete sequence and segregational stability analysis of a new cryptic plasmid pIGWZ12 from a clinical strain of Escherichia coli. Plasmid 56228-232. [DOI] [PubMed] [Google Scholar]
- 98.Zienkiewicz, M., I. Kern-Zdanowicz, M. Golebiewski, J. Zylinska, P. Mieczkowski, M. Gniadkowski, J. Bardowski, and P. Ceglowski. 2007. Mosaic structure of p1658/97, a 125-kilobase plasmid harboring an active amplicon with the extended-spectrum beta-lactamase gene blaSHV-5. Antimicrob. Agents Chemother. 511164-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.