Abstract
The expression of genes involved in photosystem development in Rhodobacter sphaeroides is dependent upon three major regulatory networks: FnrL, the PrrBA (RegBA) two-component system, and the transcriptional repressor/antirepressor PpsR/AppA. Of the three regulators, PpsR appears to have the narrowest range of physiological effects, which are limited to effects on the structural and pigment biosynthetic activities involved in photosynthetic membrane function. Although a PrrA− mutant is unable to grow under photosynthetic conditions, when a ppsR mutation was present, photosynthetic growth occurred. An examination of the double mutant under anaerobic-dark-dimethyl sulfoxide conditions using microarray analysis revealed the existence of an “extended” PpsR regulon and new physiological roles. To characterize the PpsR regulon and to better ascertain the significance of degeneracy within the PpsR binding sequence in vivo, we adapted the chromatin immunoprecipitation technique to R. sphaeroides. We demonstrated that in vivo there was direct and significant binding by PpsR to newly identified genes involved in microaerobic respiration and periplasmic stress resistance, as well as to photosynthesis genes. The new members of the PpsR regulon are located outside the photosynthesis gene cluster and have degenerate PpsR binding sequences. The possible interaction under physiologic conditions with degenerate binding sequences in the presence of other biologically relevant molecules is discussed with respect to its importance in physiological processes and to the existence of complex phenotypes associated with regulatory mutants. This study further defines the DNA structure necessary for PpsR binding in situ.
Rhodobacter sphaeroides 2.4.1 and other representatives of the purple nonsulfur photosynthetic bacteria are able to adapt and grow under a variety of environmental conditions (42, 44). The transition from high to low oxygen tension alone is an environmental alteration sufficient to induce the reversible physiological changes which accompany the formation of the intracytoplasmic membrane (ICM) (22). The ICM houses the pigment protein complexes used to gather light quanta, as well as the photosynthetic electron carriers that together constitute the photosynthetic apparatus (28). Formation of the ICM is precisely controlled via transcriptional and posttranslational regulatory processes. Such changes are accompanied by increased expression of genes involved in photosystem (PS) development (22). All of these genes, which encode the various structural components of the PS, except puc2BA (27, 51) are located in the same region of chromosome I of R. sphaeroides, the photosynthesis gene cluster (PGC) (5, 6).
In R. sphaeroides, transcriptional regulation of the expression of the PS genes is dependent upon the oxygen tension, and the following three major regulatory pathways are responsible for gene induction and/or repression: the FnrL regulatory protein, the PrrBA two-component system, and the PpsR/AppA repressor/antirepressor. The PrrBA two-component redox sensing pathway plays a critical role in the formation of the photosynthetic apparatus and also serves as a global regulator of gene expression when the oxygen tension decreases (11, 21). The cbb3 cytochrome c oxidase regulates PS gene expression via the PrrBA two-component system by monitoring O2 levels through sensing the rate of transfer and volume of electrons that travel through the oxidase on their way to O2 (24, 25, 37-39, 41). The same redox flow has been proposed to play a role in the control of carotenoid accumulation (35, 36, 40). The Prr system can act as both a transcriptional inducer and a repressor, and although a PrrA− mutant strain of R. sphaeroides is unable to grow under photosynthetic conditions, it is able to grow under anaerobic-dark-dimethyl sulfoxide (DMSO) conditions, as well as under aerobic conditions (10).
The PpsR transcription factor has been considered a “master” regulator, repressing only genes involved in PS development under aerobic growth conditions in R. sphaeroides (33). It has been proposed that two palindromes (TGTcN10gACA) corresponding to the refined PpsR consensus binding sequence must be present for functional repression (33). Two mechanisms have been described as mechanisms that are responsible for the control of binding by PpsR: (i) oxidation-reduction of two conserved cysteine residues (Cys251 and Cys424) (13, 31), which requires further investigation (4), and (ii) the interaction between PpsR and the AppA antirepressor protein, which has the unique property of being able to integrate oxygen and light signals (3). Since AppA is apparently present under all growth conditions at various levels, its effect on PpsR and the ultimate role of PpsR in PS gene expression is very complex. AppA has been shown to act as an antirepressor of PpsR in vivo (16), and direct interaction of AppA with PpsR has been demonstrated in vitro (30). In the presence of blue light illumination and/or a high oxygen tension, the AppA-PpsR2 complex is dissociated, making PpsR available for DNA binding and repression of target genes (3). AppA is able to sense blue light via the bound flavin adenine dinucleotide chromophore at its amino-terminal BLUF (sensor of blue light using flavin adenine dinucleotide) domain (14, 17). Recently, a novel heme binding SCHIC (sensor containing heme instead of cobalamin) domain located in the central region of AppA was discovered and shown to be involved in the oxygen-sensing capacity of AppA (34). Further, the likely presence of an iron-sulfur center bound to the carboxy-terminal region of AppA (19) complicates our understanding of the role of AppA as an antirepressor of PpsR. Inactivation of the PpsR protein either by mutation or through intervention of the antirepressor AppA leads to derepression of PS genes under aerobic conditions, and such a mutant strain is very unstable under these conditions, as well as under anaerobic dark-DMSO growth conditions (16).
Here, we used a new approach to characterize both the PrrA and PpsR regulons: examination of suppressor mutations of a PrrA null strain, leading to recovery of a wild-type-like phenotype in a double-mutant R. sphaeroides 2.4.1 strain (33; J. M. Eraso, unpublished data). We performed microarray analyses of the PrrA− PpsR− double-mutant strain, which is considerably more stable than the strain harboring the ppsR mutation alone, and compared our results to the transcriptome profiles of the wild-type and PRRA2 (PrrA− mutant) strains using common growth conditions (anaerobic-dark-DMSO conditions). We developed a new way to study the role of PpsR and established that there are new target genes located outside the PGC, at least doubling the previously reported number of genes regulated by PpsR (33). We also showed that there is tripartite regulation, not described previously, of the cco operon and the rdxB gene cluster by PpsR and PrrA in addition to FnrL. Because the AppA protein is structurally complex, as revealed through its numerous roles as a redox regulator of PpsR function, and because PpsR shows even greater degeneracy in its DNA binding sequence than previously described, we adapted and used for the first time the chromatin immunoprecipitation (ChIP) technique with R. sphaeroides, using anti-PpsR antibody (13). This technique has the advantage of showing in vivo the potential for direct regulation by PpsR of newly identified members of the “extended” PpsR regulon, as well as the significance of PpsR binding under conditions that are not possible to accurately reproduce in vitro. The observation that the PpsR regulatory protein could interact with DNA regions comprised of degenerate binding sequences emphasizes the possibility that hierarchal binding of DNA binding proteins could be used to modulate cellular physiology.
MATERIALS AND METHODS
Strains and growth conditions.
The R. sphaeroides strains and plasmid used in this study are shown in Table 1. R. sphaeroides strains were grown aerobically in Sistrom's minimal medium A containing succinate with a gas mixture containing 30% O2, 69% N2, and 1% CO2 and anaerobically in the dark in Sistrom's minimal medium A supplemented with yeast extract (0.1%, wt/vol) and DMSO (0.5%, vol/vol) with a gas mixture containing 95% N2 and 5% CO2 (45, 52). Aerobic and anaerobic cells were harvested at optical densities at 600 nm of 0.2 ± 0.05 and 0.45 ± 0.05, respectively. When necessary, antibiotics were used at the following concentrations: kanamycin, 25 μg/ml; streptomycin, 50 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 1 μg/ml.
TABLE 1.
Bacterial strains and plasmid
Quantitative analysis of spectral complexes.
Harvested cells of R. sphaeroides 2.4.1 and the PrrA− PpsR− mutant grown under anaerobic-dark-DMSO conditions were resuspended in 10 mM KH2PO4/K2HPO4, 1 mM EDTA (pH 7.2) and broken by three passages through a French pressure cell. Samples were centrifuged at 20,000 × g and 4°C for 15 min in order to remove unbroken cells and debris, as described elsewhere (33). Extracts containing equal amounts of protein (as determined with a bicinchoninic acid assay kit from Pierce used as recommended by the manufacturer) were used to determine the spectral data with a UV2450 spectrophotometer (Shimadzu Corp., Columbia, MD). The B800-850 and B875 complex levels were determined using the data collected, as reported previously (52).
ChIP.
The ChIP experiments were performed using an Active Motif ChIP shearing kit (ChIP-IT) without controls as described by the manufacturer (www.activemotif.com), with the following modifications. Cells of R. sphaeroides strains were grown as described above. Cross-linking was performed by adding formaldehyde (final concentration, 1%) directly to the medium for 10 min and was terminated by adding glycine Stop-Fix solution and incubating the preparation for 10 min at room temperature with gentle agitation. Cells were harvested, washed twice with ice-cold phosphate-buffered saline, resuspended in 1 ml of lysis solution supplemented with 5 μl of phenylmethylsulfonyl fluoride and 5 μl of protease inhibitor cocktail, and incubated on ice for 30 min. The resuspended cells were broken by three passages through a French pressure cell. One milliliter of digestion buffer supplemented with 5 μl of phenylmethylsulfonyl fluoride and 5 μl of protease inhibitor cocktail was added to the lysate and preheated for 5 min at 37°C. Fifty microliters of an enzymatic shearing mixture (200 U/ml) was added, and this was followed by 28 to 30 min of incubation at 37°C with periodic agitation. The reaction was stopped by addition of 20 μl of 0.5 M EDTA and incubation for 10 min on ice. After centrifugation at 20,000 × g and 4°C for 10 min, the supernatant was recovered, and the shearing efficiency was checked as described by the manufacturer. DNA fragments that were between 0.2 and 1 kb long were obtained. Preclearing of chromatin samples, input recovery, immunoprecipitation with or without anti-PpsR antibody (13), addition of protein G beads, washing, elution of DNA-protein complexes, reverse cross-linking, RNA removal, and proteinase K treatment were performed by following the manufacturer's instructions. The DNA fragments were eluted with water using a QIAquick PCR purification kit from Qiagen.
RNA manipulation.
An optimized procedure for isolation of intact mRNA for DNA microarrays has been described previously (29, 44).
Microarray experiments and data analysis.
The R. sphaeroides 2.4.1 Affymetrix GeneChip has been described by Pappas et al. (42). Total RNA from three independent cultures of the R. sphaeroides PrrA− PpsR− strain grown under anaerobic-dark-DMSO conditions was used. The methods used for cDNA synthesis, fragmentation, labeling, and hybridization were adapted from the methods optimized for the GeneChip designed for the Pseudomonas aeruginosa genome array by Affymetrix Inc. (http://www.affymetrix.com/support/technical/manuals.affx) and have been described elsewhere (29, 44). The mean of triplicate measurements (three Affymetrix GeneChips) was used to describe the expression level of a gene for each R. sphaeroides strain. For group comparisons, microarray data for the wild-type (21, 28) and PrrA− (PRRA2) (11, 21) strains of R. sphaeroides grown under anaerobic-dark-DMSO conditions were used. The filtering criterion for the group means was a 1.2-fold change using the 90% confidence boundary for no change, which was calculated using the standard error of the group means (44). The threshold for the absolute difference between the two group means when two transcriptome profiles were compared was 100. The Pearson correlation coefficient (r value) was calculated using the Microsoft Excel program, and for three experiments using the PrrA− PpsR− strain of R. sphaeroides grown under anaerobic-dark-DMSO conditions, it ranged from 0.992 to 0.996. The expression data have been deposited in the Gene Expression Omnibus database (www.ncbi.nih.gov/projects/geo) under platform GPL162.
qRT-PCR.
For each strain of R. sphaeroides tested, aliquots of RNA samples were extracted from two cultures grown independently under anaerobic-dark-DMSO conditions. RNA samples from the R. sphaeroides 2.4.1, PRRA2, and PrrA− PpsR− strains were used previously for microarray experiments. Reverse transcription was performed as described previously (29, 44), and equivalent amounts of cDNAs were used for quantitative real-time PCR (qRT-PCR) experiments (Applied Biosystems 7500). The RSP0154 gene encoding a 3-hydroxyisobutyrate dehydrogenase and exhibiting equal levels of expression in the different R. sphaeroides strains after microarray analysis was used for normalization (data not shown). The SYBR green PCR master mixture (Applied Biosystems) was used with the appropriate amounts of cDNA samples. This method was also used to monitor ChIP results for two independent cultures of the R. sphaeroides 2.4.1, 2.4.1(pPNs), and PrrA− PpsR− strains grown under aerobic or anaerobic-dark-DMSO conditions. The amount of PCR product was estimated for different genomic regions using input DNA (i.e., the total sheared DNA prior to immunoprecipitation) and immunoprecipitated DNA with and without anti-PpsR antibody as the matrix. Immunoprecipitation efficiencies (IE) were determined by dividing the values obtained for immunoprecipitated DNA samples by the values obtained for the input DNA. Enrichment (IE with antibody/IE without antibody) was then calculated. The SYBR green PCR master mixture (Applied Biosystems) was used with the appropriate amounts of ChIP samples. For each reaction a standard curve and a dissociation curve were drawn. The slope of the standard curve allowed the efficiency of each qRT-PCR experiment to be calculated. We selected only results obtained for reactions having efficiencies between 90 and 110%, which corresponded to slopes ranging from −3.6 to −3.1. A fusion curve was drawn for each reaction to make sure that only one specific PCR product was obtained. Each reaction was performed in duplicate, and results were obtained from at least two independent experiments.
RESULTS
Spectral complex analysis of R. sphaeroides strains grown under anaerobic-dark-DMSO conditions.
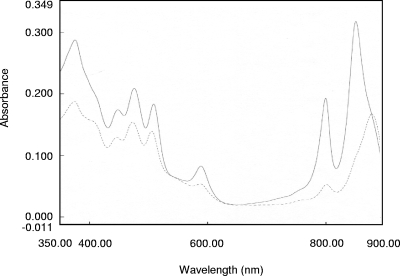
In R. sphaeroides, mutation of the ppsR gene in a PrrA− genetic background leads to restoration of photosynthetic growth (33; Eraso, unpublished), which suggests that the PrrBA and AppA-PpsR pathways interact as previously demonstrated (33). The double-mutant strain is useful because it is stable enough to examine the full range of PpsR regulation. DNA analyses of R. sphaeroides 2.4.1 and the PpsR− PrrA− double-mutant strain were performed to confirm (i) disruption of the ppsR gene by insertion of a ΩKmr cartridge and (ii) replacement of the prrA gene after insertion of an ΩSmr Spr cartridge (9, 10) (data not shown). The 2.4.1 wild-type strain of R. sphaeroides grown under anaerobic-dark-DMSO conditions produced a significant amount of photosynthetic complexes (Fig. 1), although the photosynthetic apparatus is not required for growth of the cells under these gratuitous conditions. In contrast to the wild type, the PRRA2 mutant strain of R. sphaeroides does not produce photosynthetic complexes under anaerobic-dark-DMSO conditions (10, 33), while a PpsR null mutant contains larger amounts of the B800-850 complex under anaerobic growth conditions (16). As shown in Fig. 1, the PrrA− PpsR− double-mutant strain of R. sphaeroides is able to produce photosynthetic complexes. An examination of the amounts of light-harvesting (LH) complexes in the wild-type and PrrA− PpsR− strains showed that, compared to the wild-type strain, in the PrrA− PpsR− strain (i) there was an approximately 30% increase in the amount of the B875 complex and (ii) there was an approximately 94% decrease in the amount of the B800-850 complex. Similar results were obtained when a PrrA− PpsR− double-mutant strain of R. sphaeroides was grown under photosynthetic conditions (33), confirming that the double-mutant strain has a wild-type-like phenotype and therefore is an ideal subject for assessing the full extent of the PpsR regulon.
FIG. 1.
Spectral analysis of photosynthetic complexes. Extracts from R. sphaeroides strains 2.4.1 (solid line) and PrrA− PpsR− (dashed line) containing equal amounts of protein and grown under anaerobic-dark-DMSO conditions were used.
Microarray analysis of wild-type and mutant strains of R. sphaeroides grown under anaerobic-dark-DMSO conditions.
We compared the transcriptome profiles for (i) the PrrA− PpsR− (this study) and PRRA2 (11, 21) strains, showing the effect of the ppsR interruption; (ii) the PrrA− PpsR− (this study) and wild-type (21) strains, showing the combined effects of the ppsR interruption and a prrA deletion; (iii) and the PRRA2 (11, 21) and wild-type (21) strains, showing the effect of the prrA deletion, as described in Materials and Methods. Our approach permitted characterization of genes likely to be regulated by PpsR alone or by both the PrrA and PpsR regulatory proteins. Using cutoff values for transcriptomic studies of 1.2-, 2.0-, and 5.0-fold changes, we observed that the expression levels of approximately 700, 443, and 152 genes, respectively, were under direct or indirect control of PpsR and PrrA, while the expression levels of 669, 414, and 60 genes, respectively, were directly or indirectly controlled by PpsR alone. These results imply that a larger number of genes than previously reported are targets for direct or indirect regulation by PpsR. Moreover, the values subsequently determined by transcriptome analysis were strongly related to one another. The results obtained for any pairwise comparison could be retrieved with a high degree of confidence (at least 99%) using the changes determined for the remaining pairwise comparisons. Although these pairwise comparisons had a common denominator, the level of agreement is remarkable and validates our methodology, since the microarray experiments were performed separately by different experimenters (11, 21) using independent cultures involving the various strains of R. sphaeroides and were performed at different times.
PpsR represses genes involved in PS development under anaerobic-dark-DMSO conditions.
Table 2 shows that under anaerobic-dark-DMSO conditions, the genes located in the PGC (bch, crt, puf, puc1, and puhA) are positively regulated by PrrA and significantly repressed by PpsR, as observed under various growth conditions. The expression of genes coding for proteins thought to be regulators of PS development, such as PpaA and TspO, was significantly affected by deletion of prrA (−8.07- and −3.74-fold changes, respectively) and by interruption of ppsR (22.83- and 13.43-fold changes, respectively). The genes in this category also include the RSP0292 to RSP0295 genes. In Rhodobacter capsulatus and R. sphaeroides, the open reading frames located immediately downstream of puhA have been extensively studied and are likely to be involved in the posttranslational assembly of the photosynthetic apparatus (1, 46, 49). Finally, the expression of the RSP0296 gene coding for cytochrome c2, a periplasmic redox protein (8), was affected when prrA was deleted (−3.49-fold change), as observed previously (10), and was derepressed when PpsR was absent (19.44-fold change). The latter finding was unexpected and indicates that PpsR has a role in cycA regulation. Although selected aspects of some of these results have been reported previously (33), the results obtained in this study established the validity of this approach.
TABLE 2.
Changes in expression of genes under anaerobic-dark-DMSO conditions
| Gene | Gene product | Change (n-fold) in gene expression in R. sphaeroides PrrA− PpsR− compared to PRRA2a | Change (n-fold) in gene expression in R. sphaeroides PrrA− PpsR− compared to wild typeb | Change (n-fold) in gene expression in R. sphaeroides PRRA2 compared to wild typec |
|---|---|---|---|---|
| Genes playing a role in PS development affected by prrA and/or ppsR mutation and located in the PGC | ||||
| RSP0254 | DxsA | 26.29 | 2.69 | −9.76 |
| RSP0255 | PufX | 66.07 | 2.42 | −27.26 |
| RSP0256 | PufM | 112.64 | 2.52 | −44.62 |
| RSP0257 | PufL | 119.79 | 2.58 | −46.4 |
| RSP0258 | PufA | 47.5 | 3.72 | −12.76 |
| RSP0259 | PufQ | 134.47 | 3.93 | −34.22 |
| RSP0260d | BchZ | 99.07 | 6.97 | −14.22 |
| RSP0261d | BchY | 89.25 | 10.74 | −8.31 |
| RSP0262d | BchX | 198.65 | 13.19 | −15.06 |
| RSP0263d | BchC | 88.9 | 9.41 | −9.45 |
| RSP0264d | CrtF | 13.23 | 2.96 | NDe |
| RSP0265d | CrtE | 14.34 | ND | −12.28 |
| RSP0266d | CrtD | 7.86 | 2.62 | −3 |
| RSP0267d | CrtC | 7.34 | 1.8 | −4.07 |
| RSP0269d | TspO | 13.43 | 3.59 | −3.74 |
| RSP0270d | CrtB | 18.3 | 4.3 | −4.26 |
| RSP0271d | CrtI | 21.79 | 3.56 | −6.12 |
| RSP0272d | CrtA | 38.85 | 2.02 | −19.19 |
| RSP0273 | BchI | 11.51 | 1.29 | −8.89 |
| RSP0274 | BchD | 2.8 | ND | −3.18 |
| RSP0276 | IdI | 12.52 | 3.1 | −4.04 |
| RSP0277d | BchP | 17.35 | 2.86 | −8.43 |
| RSP0278d | RSP0278 | 21.33 | 2.83 | −7.52 |
| RSP0279d | BchG | 19.33 | 2.3 | −8.42 |
| RSP0280d | BchJ | 7.99 | ND | −6.83 |
| RSP0281d | BchE | 19.34 | 1.57 | −12.34 |
| RSP0282 | PpsR | −18.84 | −16.94 | ND |
| RSP0283d | PpaA | 22.83 | 2.83 | −8.07 |
| RSP0284d | BchF | 95.93 | 9.58 | −10.01 |
| RSP0285d | BchN | 30.07 | 3.88 | −8.91 |
| RSP0286d | BchB | 44.49 | 4.3 | −10.36 |
| RSP0287 | BchH | 63.84 | 3.88 | −16.47 |
| RSP0288 | BchL | 74.58 | 4.22 | −17.67 |
| RSP0289 | BchM | 53.46 | 6.63 | −8.06 |
| RSP0290 | RSP0290 | 39.17 | 1.97 | −19.9 |
| RSP0291 | PuhA | 40.86 | 2.06 | −19.8 |
| RSP0292 | Hypothetical protein | 44.99 | 2.78 | −16.18 |
| RSP0293 | Hypothetical protein | 43.39 | 4.87 | −8.91 |
| RSP0294 | Hypothetical protein | 33.55 | 8.32 | −4.03 |
| RSP0295 | Hypothetical protein | 24.87 | 5.7 | −4.37 |
| RSP0296 | CycA | 19.44 | 5.57 | −3.49 |
| RSP0314d | Puc1B | 404.47 | 2.77 | −145.08 |
| RSP0315d | Puc1C | +17.82 | 1.77 | −10.05 |
| Genes playing a role in PS development affected by prrA and/or ppsR mutation and located outside the PGC | ||||
| RSP0692 | RdxB | 1.98 | ND | −2.31 |
| RSP0693 | CcoP | 4.36 | 1.82 | −2.4 |
| RSP0694 | CcoQ | 5 | 1.8 | −2.78 |
| RSP0695 | CcoO | 5.32 | 1.83 | −2.91 |
| RSP0696 | CcoN | 5.28 | 1.83 | −2.89 |
| RSP0679d | HemC | 1.87 | 1.81 | ND |
| RSP0680d | HemE | 4.4 | 2.4 | −1.84 |
| RSP1556d | Puc2B | 24.83 | −1.79 | −44.33 |
| RSP1557d | Puc2A | 14.06 | −3.49 | −49.02 |
| RSP1518 | PrrA | ND | −92.07 | −282.22 |
| RSP1518 copy 1 | PrrA | ND | −28.42 | −258.16 |
| RSP1518 copy 2 | PrrA | ND | −45.06 | −236.18 |
| RSP1518 copy 3 | PrrA | ND | −47.67 | −164.02 |
| RSP1518 copy 4 | PrrA | ND | −89.49 | −275.86 |
| Genes affected by prrA and/or ppsR mutation and located outside the PGC | ||||
| RSP0381 | PhaP | 11.68 | 3.1 | −3.77 |
| RSP0382 | PhaC | 2.32 | ND | −2.39 |
| RSP0383 | PhaZ | 2.47 | ND | −2.15 |
| RSP2122 | Putative dimethylamine corrinoid protein | 27.21 | 19.05 | ND |
| RSP2768 | MetH homologue | 4.47 | 3.15 | ND |
| RSP2769 | MetF homologue | 17.36 | 10.49 | ND |
| RSP2770 | Conserved hypothetical protein | 21.7 | 8.56 | ND |
| RSP3241 | Partial transcriptional regulatory protein with C-terminal homology to OmpR | 2.5 | 1.64 | ND |
| RSP3242 | Putative trypsin-like serine protease | 23.31 | 25.92 | ND |
Gene known or predicted to be directly regulated by PpsR. The pufBA and puc1BA genes are represented by one probe set on the GeneChip (42).
ND, not detectable.
When R. sphaeroides was grown under anaerobic-dark-DMSO conditions, we also observed that PpsR regulated other genes involved in PS development but located outside the PGC (Table 2), such as the RSP0679 (hemC) and RSP0680 (hemE) genes involved in the early stages of protoporphyrin IX biosynthesis (33) and the puc2BA operon comprised of the RSP1556 and RSP1557 genes, which has been implicated in LH II photosynthetic complex biosynthesis (51), as observed previously when cells were grown under aerobic conditions (33). We also observed that deletion of prrA led to alteration of the expression of all of these genes except the RSP0679 (hemC) gene.
New members of the PpsR regulon and unraveling the relationship between PpsR and PrrBA.
Having established the validity of our experimental approach, we used the microarray data obtained with the double-mutant strain to search for other genes likely to be targets for PpsR regulation in order to better define the role of PpsR in R. sphaeroides. Expression of genes located outside the PGC, such as the RSP0692 gene (encoding RdxB) and the RSP0696 to RSP693 genes (encoding the cbb3 cytochrome c oxidase), appeared to be affected in the various mutant strains (Table 2). Another group of genes potentially subject to dual regulation (Table 2) is comprised of the RSP0381 to RSP383 genes. The RSP0381 gene codes for a hypothetical protein showing 95% identity with the phasin protein sequence encoded by the phaP gene of R. sphaeroides ATCC 17025. We could also identify phaC (RSP0382 gene) and phaZ (RSP0383 gene).
We detected the following genes whose expression is repressed by PpsR alone. The RSP2768, RSP2769, and RSP2770 genes code for a MetH homologue, a MetF homologue, and a hypothetical conserved protein with an undetermined function, respectively. The RSP2122 gene encodes a product that is thought to be a homologue of MtbC and contains a cobalamin (vitamin B12) binding domain like the RSP2768 protein or the PpaA regulatory protein. The RSP3241 gene is located on chromosome II of R. sphaeroides 2.4.1 and encodes a response regulator protein belonging to the OmpR family. The RSP3242 gene, which is located upstream of the RSP3241 gene and is transcribed in the opposite direction, encodes a putative trypsin-like serine protease. The levels of expression of the RSP3241 and RSP3242 genes were higher in the PrrA− PpsR− double-mutant strain than in the wild-type strain (1.64- and 25.92-fold, respectively) or in the PrrA− strain (2.5- and 23.31-fold, respectively) of R. sphaeroides.
This very brief description of the extended PpsR regulon more than doubles the number of genes considered to be regulated by PpsR in R. sphaeroides 2.4.1, which encode a greater diversity of physiologic effects.
qRT-PCR experiments validate the microarray analysis.
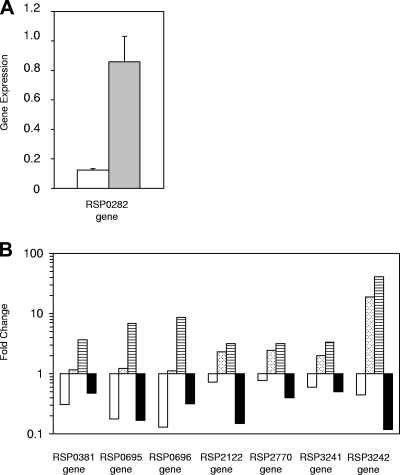
Figure 2A shows the levels of expression of ppsR in the R. sphaeroides 2.4.1 and 2.4.1(pPNs) strains grown under anaerobic-dark-DMSO conditions, and Fig. 2B shows the changes in the expression levels for several genes in the R. sphaeroides 2.4.1, PRRA2, PrrA− PpsR−, and 2.4.1(pPNs) strains grown under anaerobic-dark-DMSO conditions. The qRT-PCR results (Fig. 2B) coincide with the results obtained with the DNA microarrays. As previously observed, deletion of prrA resulted in a significant decrease in expression of the RSP0381, RSP0695, and RSP0696 genes, which was precisely counterbalanced by interruption of ppsR, so that the change was not significant when the expression of these genes in the PrrA− PpsR− strain was compared to the expression in the wild type. The increased expression of the RSP2122, RSP2770, RSP3241, and RSP3242 genes observed in the PrrA− PpsR− double-mutant strain compared to the wild-type and PRRA2 strains suggests that there is regulation by PpsR. The sevenfold-higher level of expression of ppsR in the 2.4.1(pPNs) strain than in the wild type (Fig. 2A) resulted in a concomitant decrease in the expression of all of the genes tested in the ppsR-overexpressing strain, suggesting that the level of PpsR is not saturating under normal physiologic conditions, as demonstrated below.
FIG. 2.
Validation of microarray data by qRT-PCR. (A) Expression of the ppsR (RSP0282) gene measured by qRT-PCR. Experiments were performed with R. sphaeroides 2.4.1 (open bar) and 2.4.1(pPNs) (shaded bar) grown under anaerobic-dark-DMSO conditions. (B) Changes in expression of selected genes measured by qRT-PCR. Changes (expressed as relative changes on a logarithmic scale) were calculated using average values normalized with the RSP0154 gene from two independent experiments with standard deviations that did not exceed 15%. All strains were grown under anaerobic-dark-DMSO conditions. The R. sphaeroides PrrA− strain was compared to the 2.4.1 strain (open bars), the R. sphaeroides PrrA− PpsR− strain was compared to strain 2.4.1 (bars with dots), the R. sphaeroides PrrA− strain was compared to the PrrA− PpsR− strain (bars with horizontal stripes), and R. sphaeroides 2.4.1(pPNs) was compared to strain 2.4.1 (filled bars).
ChIP technique reveals direct in vivo repression by PpsR.
In order to determine in vivo whether the regulation of genes by PpsR is direct or indirect, we adapted the ChIP technique to R. sphaeroides. This in situ approach not only demonstrates repressor interactions but also is defined by taking place within the full context of the DNA binding sequence with the antirepressor AppA and other DNA binding elements present. The sensitivity of the rabbit antibody directed against PpsR (13) was tested by performing a Western blot analysis using soluble fractions from the 2.4.1 strain, the 2.4.1(pPNs) strain overexpressing PpsR, and the PrrA− PpsR− double-mutant strain of R. sphaeroides grown under anaerobic-dark-DMSO conditions (data not shown). No PpsR was detected in the PrrA− PpsR− double-mutant strain of R. sphaeroides. The intensity of the band corresponding to the PpsR monomer was determined using the ImageJ software (1.38×; National Institutes of Health [http://rsb.info.nih.gov/ij/]), and a minimal 2.7- ± 0.1-fold increase in the level of the PpsR monomer in the 2.4.1(pPNs) strain compared to the wild type was observed.
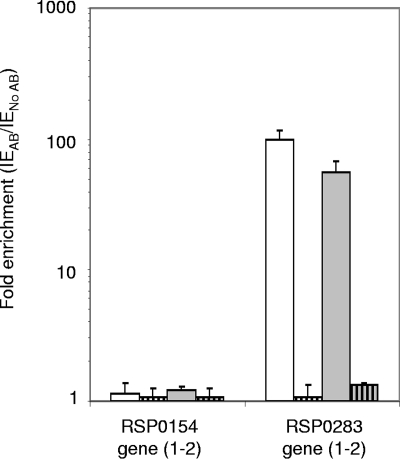
The ChIP experiments and qRT-PCRs were performed as described in Materials and Methods, and the results obtained are shown in Fig. 3. A fragment of the coding region of the RSP0154 gene, without any apparent PpsR binding sites (Table 3; see Fig. S1A in the supplemental material), was examined by performing qRT-PCR using ChIP samples as the matrix. The enrichment for this gene corresponded to the background for the ChIP experiment. The RSP0283 (ppaA) gene, located inside the PCG, is regulated by PpsR (12), and a fragment of the regulatory region of this gene, bounded by two perfect PpsR binding sequences (Table 3; see Fig. 1B in the supplemental material), was quantified using ChIP samples as the matrix. We observed strong binding of PpsR to the regulatory region of the RSP0283 (ppaA) gene under anaerobic-dark-DMSO growth conditions and an increase in the PpsR binding (∼1.7-fold) under aerobic conditions, under which PpsR was suggested to be more active, perhaps due to the lower level of AppA, as suggested by the microarray data. For the PrrA− PpsR− strain of R. sphaeroides grown under aerobic or anaerobic-dark-DMSO conditions, no significant enrichment values higher than the background values were observed for the genes tested. These results were expected since the ppsR gene is disrupted in this strain and PpsR is not present.
FIG. 3.
ChIP analysis of in vivo binding of PpsR inside the PGC. qRT-PCR was performed with immunoprecipitated samples of the wild-type (open bars) and PrrA− PpsR− double-mutant (bars with vertical stripes) strains of R. sphaeroides grown under aerobic (open bars) and anaerobic-dark-DMSO conditions (shaded bars). Enrichment (expressed as relative changes on a logarithmic scale) was calculated as described in Materials and Methods. The error bars indicate standard deviations. AB, antibody.
TABLE 3.
Predicted PpsR binding sites for ppaA and degenerate PpsR binding sites detected for the new members of the ppsR regulon
| Gene | Gene product | PpsR TGTCN10GACA sequencea | Distance from ATG (bp) |
|---|---|---|---|
| RSP0154 | 3-Hydroxyisobutyrate dehydrogenase | None | |
| RSP0283 | PpaA | TGTCAATTCTGACTTACA | −296 |
| TTTTGCGGCGAGAGCACA | −276 | ||
| TGTCAATTTTCTTTGACA | −152 | ||
| RSP0696 | CcoN | TGCTCCACATCTTCAACA | 691 |
| AGTGGTGGTACGGCCACA | 784 | ||
| RSP0695 | CcoO | TGTGGGTCTCGGGCATCA | −249 |
| TGAACGCCTTCGCCGACA | −186 | ||
| TGTGGAAAACCGTGACCA | −81 | ||
| AGTGAAGATAAGGGGACA | −21 | ||
| TGTTCTACCTCGAGAACA | 106 | ||
| RSP2122 | Putative dimethylamine corrinoid protein | TGTCGCAAACCGATGATG | −75 |
| TGTCCGAACTCGATGACG | 25 | ||
| RSP3241 | Partial transcriptional regulatory protein with | CGCGGGCCTTGGGGGACA | −181 |
| C-terminal homology to OmpR | GGGCCTTGGGGGACAACA | −178 | |
| CGTCGATGGAGAATGACA | −17 |
Underlining indicates residues that are less conserved.
Direct in vivo binding of PpsR to newly identified members of the PpsR regulon.
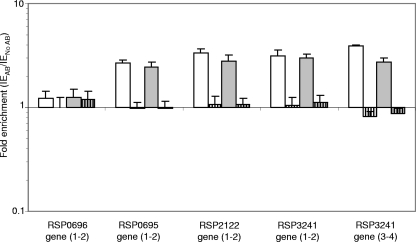
The mechanisms by which PpsR represses the expression of genes remain elusive; this is particularly true for genes showing a significant change in expression after inactivation of ppsR but for which no PpsR binding site or one or several possible PpsR binding sites have been detected in their regulatory regions. The absence of a perfect refined consensus PpsR binding sequence (TGTcN10gACA) (33) suggests that either there is indirect regulation of these genes by PpsR or there is further degeneracy in the PpsR binding sequence. We investigated whether there are at least two PpsR binding sequences with up to three mismatches in the regulatory and coding regions of the newly suggested members of the “extended” PpsR regulon. We selected the regulatory regions of the RSP0695, RSP2122, and RSP3241 genes and an internal fragment of the coding region of the RSP0696 gene (Table 3) for ChIP studies.
We selected different primers for the qRT-PCR—ChIP analysis, which are located in regions containing at least two possible PpsR binding sequences (see Fig. S1C to F in the supplemental material). Figure 4 shows that when we used ChIP samples from the wild-type strain of R. sphaeroides (grown under aerobic or anaerobic-dark-DMSO conditions) there was significant binding of PpsR (the levels were substantially above the background level) in the selected regions of the RSP0695, RSP2122, and RSP3241 genes but not in the coding region of the RSP0696 gene. Under oxic conditions, the binding affinity of PpsR was equivalent to or higher than, but not lower than, the binding affinity observed under anaerobic-dark-DMSO growth conditions for regions of the RSP0695, RSP2122, and RSP3241 genes. In the case of the RSP3241 gene, two pairs of primers were used for the qRT-PCR-ChIP analysis, and the enrichment values were very similar. For the PrrA− PpsR− strain of R. sphaeroides under both growth conditions, no significant binding of PpsR was observed for any gene tested, as expected.
FIG. 4.
ChIP analysis of in vivo binding of PpsR outside the PGC. Selected DNA regions of newly identified members of the PpsR regulon were examined by performing qRT-PCR with immunoprecipitated samples of the wild-type (open bars) and PrrA− PpsR− double-mutant (bars with vertical stripes) strains of R. sphaeroides as the matrix. The strains were grown under aerobic (open bars) and anaerobic-dark-DMSO (shaded bars) conditions. Enrichment (expressed as relative changes on a logarithmic scale) was calculated as described in Materials and Methods. The error bars indicate standard deviations. AB, antibody.
In order to determine the physiological significance of the binding of the PpsR protein for the RSP0695, RSP2122, and RSP3241 genes, the ChIP experiment was performed using the 2.4.1(pPNs) strain of R. sphaeroides, which overexpresses PpsR, under anaerobic-dark-DMSO growth conditions. Table 4 shows the results of enrichment comparisons for the 2.4.1(pPNs) and wild-type strains for each selected DNA region. The data for the negative control (RSP0154 gene) and the coding region of the RSP0696 gene, which does not exhibit significant binding of PpsR, show that there was a large increase in enrichment [enrichment for 2.4.1(pPNs)/enrichment for 2.4.1, 4.93 and 4.89, respectively] of the background, which corresponded to an increase in the nonspecific interaction due to overexpression of the plasmid DNA. The increase in the enrichment for the selected DNA region of the positive control (RSP0283 gene) reflected saturation of the PpsR binding sites in the 2.4.1(pPNs) strain and therefore the likely absence of PpsR saturation in the wild-type strain. For the RSP0695, RSP2122, and RSP3241 genes, comparison of the enrichment for the 2.4.1(pPNs) strain and the enrichment for the wild-type strain showed that the increases were close to the increase observed for the positive control (RSP0283 gene) (2.72, 2.56, and 2.28, respectively). These results reflect saturation of the PpsR binding sites in these DNA regions and support the hypothesis that PpsR plays a physiological regulatory role in the expression of these genes. They also speak directly to the role of PpsR in vivo and suggest that under standard growth conditions PpsR binding is not saturating and that one possible reason for this is the presence of the antirepressor AppA or the presence of degenerate binding sequences or both. These results clearly extend the PpsR regulon and, importantly, show that ChIP can be used to study a regulatory protein in R. sphaeroides.
TABLE 4.
Comparison of in vivo binding of PpsR in the 2.4.1 and 2.4.1(pPNs) strains of R. sphaeroides under anaerobic-dark-DMSO conditions
| Gene (primers) | Enrichment (with antibody/without antibody) (n-fold)
|
Enrichment for 2.4.1(pPNs)/enrichment for 2.4.1 (n-fold) | |
|---|---|---|---|
| 2.4.1 | 2.4.1(pPNs) | ||
| RSP0154 (1-2) | 1.22 ± 0.05 | 6.02 ± 0.87 | 4.93 |
| RSP0283 (1-2) | 56.72 ± 10.7 | 124.1 ± 15.6 | 2.19 |
| RSP0696 (1-2) | 1.24 ± 0.27 | 6.06 ± 1.16 | 4.89 |
| RSP0695 (1-2) | 2.43 ± 0.32 | 6.61 ± 0.37 | 2.72 |
| RSP2122 (1-2) | 2.8 ± 0.38 | 7.17 ± 1.27 | 2.56 |
| RSP3241 (3-4) | 2.76 ± 0.23 | 6.29 ± 0.61 | 2.28 |
DISCUSSION
The AppA-PpsR regulatory pathway plays a major role in regulating the formation of the photosynthetic apparatus under a variety of growth conditions, especially in the presence of light and oxygen (50). A PpsR mutant strain of R. sphaeroides containing a point mutation, which expresses a less-active repressor, was used for characterization of this protein as the “master repressor” of PS development under aerobic conditions (33). On the other hand, the PrrBA two-component system is essential for formation of the ICM housing the photosynthetic apparatus under photosynthetic conditions and regulates both positively and negatively a broad set of target genes (11). A PrrA− mutant strain of R. sphaeroides can grow only under aerobic conditions or under anaerobic-dark-DMSO conditions. The latter growth conditions are sufficient to induce gratuitous formation of the ICM in the presence of the alternate electron acceptor DMSO. In a PrrA− genetic background, disruption of ppsR generates a double-mutant strain that is able to grow stably under photosynthetic conditions, allowing further analysis of the PpsR regulon. Pairwise comparisons of these different strains of R. sphaeroides require that cells be grown under anaerobic-dark-DMSO conditions in order to assess the relative interactions between the PrrA and PpsR regulatory networks, as well as to assess the “extent” of the PpsR regulon, which has previously been limited to the PGC and closely related genes involved in porphyrin synthesis. It is worth noting that the PrrBA two-component system is essential for expression of appA encoding the antirepressor, and thus, in the absence of PrrA, appA is not transcribed and the repressor PpsR is fully functional. In addition to being an activator of AppA expression, PrrA has recently been shown to be a repressor of PpsR gene expression (11). Therefore, PrrA can be considered a protein that has a dual role; it directs activation of PGC gene expression, and it has an indirect effect on the regulation of PpsR expression and activity.
Photosynthetic complex production in the PrrA− PpsR− double-mutant strain grown under anaerobic-dark-DMSO conditions.
Similar to the results obtained under anaerobic phototrophic conditions (33), the level of production of LH I in the double-mutant strain of R. sphaeroides grown under anaerobic-dark-DMSO conditions was higher than in the wild type (∼30% increase), while the level of the LH II photosynthetic complex was significantly lower than in the wild type (∼94% decrease) (Fig. 1). Interruption of ppsR in the PRRA2 mutant strain led to significant increases in the expression of most of the genes involved in PS development, including the RSP0679 (hemC) and RSP0680 (hemE) genes, and the levels were higher than or equivalent to those in the wild type (Table 2). However, the requirement for heme under aerobic conditions suggests that the latter genes are subjected to additional regulatory processes. These observations strengthen the hypothesis that PpsR plays a critical role in the synthesis of the photosynthetic apparatus and also reveal that there is antagonism between the PrrA, PpsR (16), and AppA (37) regulatory proteins as an inducer and a negative regulator of PS development, respectively. From the work of Eraso et al. (11) and the results presented here, it is evident that the AppA-PpsR and PrrBA systems coregulate PS gene expression due to the role of PrrA in PpsR expression and function.
Although the mRNA levels of the puc1 operon in the PrrA− PpsR− strain were higher than those in the wild type, the actual amount of the LH II photosynthetic complex was very small, suggesting that additional regulatory processes are critical for the formation of wild-type amounts of LH photosynthetic complexes. Removal of the PpsR repressive effect was not sufficient to fully restore puc2A and puc2B gene expression (Table 2). It was reported previously that the ultimate cellular levels of LH II were dependent upon puc2BA expression in some subtle manner (51). Therefore, the lower level of expression of puc2BA in the PrrA− PpsR− double-mutant strain could explain in part the low level of LH II photosynthetic complexes. In addition, interruption of the ppsR gene in the PRRA2 mutant strain of R. sphaeroides resulted in increased expression of the cco operon (RSP0693 to RSP0696 genes) and rdxB (RSP0692 gene) at levels equal to or greater than the levels of expression in the wild-type strain (Table 2). These genes are transcribed as ccoNOQP, ccoNOQP-rdxBH, rdxBH, and rdxIS specific transcripts, and all the genes of the ccoNOQP operon are regulated coordinately (43). A higher level of the cbb3 cytochrome c oxidase could result in an increase in reductant flow under anaerobic conditions, disturbing the balance between spheroidene and spheroidenone, leading to posttranslational repression of LH II formation (40).
The “extended” PpsR regulon and unraveling the relationship between the AppA-PpsR and PrrBA regulatory pathways.
Until now, the only known role of the PpsR protein has been repression of genes involved in PS development. Pairwise comparisons of the transcriptome profiles of the wild-type, PRRA2, and PrrA− PpsR− mutant strains of R. sphaeroides revealed a broader set of target genes that are regulated directly or indirectly by PpsR and/or PrrA. The expression levels of genes encoding global regulators such as H-NS proteins involved in condensation of the bacterial chromosome are significantly affected in the absence of PpsR (RSP1388 and RSP4056) or in the absence of both PrrA and PpsR (RSP002 and RSP1517), which could have a broad effect on gene expression outside the PGC.
Microarray analysis and qRT-PCR showed that control of transcription of the cco operon involves the AppA-PpsR regulatory pathway and the PrrBA system. Thus, the cco DNA region is subject to precise transcriptional control involving the PpsR, PrrA (21), and FnrL (43) proteins, and it is the product of this operon which monitors reductant flow to O2 and hence PS gene expression.
The RSP0380 (phaR), RSP0381 (phaP), RSP0382 (phaC), and RSP0383 (phaZ) genes are located outside the PGC and code for a regulatory protein, a phasin protein, a poly-3-hydroxybutyrate polymerase, and a poly-3-hydroxybutyrate depolymerase, respectively (49a). The expression of these proteins has been found to affect redox control (1a) and hemA expression (11a).
Our results also suggest that there are additional target genes that are regulated by the AppA-PpsR system alone. The RSP2768, RSP2769, and RSP2770 genes are adjacent to one another, are oriented in the same direction, and code for MetH and MetF homologues and a hypothetical conserved protein with an undetermined function, respectively. These results led to formulation of a new role for the AppA-PpsR pathway in cobalamin (vitamin B12)-dependent biosynthesis of methionine and in tetrahydrofolate metabolism. Tetrahydrofolate metabolism is known to play an important role in C-1 metabolism and an indirect role in the formation of 5-aminolevulinic acid (42a), the starting point for tetrapyrrole synthesis (1b).
Thus, detailed analyses of the extended PpsR regulon have brought into sharper focus the importance of the AppA-PpsR regulatory system in ancillary metabolic activities important in tetrapyrrole biosynthesis that until now have not been described.
Extension of the PpsR regulon to the RSP3241 and RSP3242 genes, which are located on chromosome II of R. sphaeroides, further enhances the potential roles of the AppA-PpsR system. The RSP3242 gene codes for a putative trypsin-like serine protease with an N-terminal catalytic (trypsin) domain and two PDZ C-terminal domains. This organization is the same as that of DegP and DegQ belonging to the HtrA family of Escherichia coli proteins. This family of proteins plays a critical role in the control of protein quality in the periplasm of gram-negative bacteria (23). We detected immediately upstream of the RSP3242 gene the RSP3240 and RSP3241 genes, which are oriented in the direction opposite that of the RSP3242 gene and code for a periplasmic sensor signal transduction histidine kinase and a response regulator containing a DNA binding domain showing homology to CpxR, respectively. We therefore propose that the RSP3240 and RSP3241 genes encode a two-component system comparable to CpxRA of E. coli, which activates the transcription of the RSP3242 gene. These results suggest a new role for the AppA-PpsR pathway in response to periplasmic stress, perhaps when the cells of R. sphaeroides switch from aerobic to anaerobic growth, inducing formation of the ICM housing the photosynthetic apparatus. It is interesting that two genes coding for two PpsR proteins have been identified in the closely related bacteria Bradyrhizobium and Rhodopseudomonas palustris (2, 20). In the Bradyrhizobium strain, the PpsR1 protein plays an unexpected activator role in gene expression, and PpsR2 corresponds to the classical repressor protein. It has been shown that in this microorganism the DNA recognition process is more flexible for PpsR2 than for PpsR1, suggesting that there is a broader set of target genes for PpsR2, as observed in this study.
ChIP as a tool to study, in vivo, direct regulation of gene expression by PpsR.
Both in vitro (4, 15, 32) and bioinformatic approaches (29) enabled detailed studies of PpsR repressor activity under different growth conditions to be performed. Also, gel mobility shift analyses allowed studies of in vitro binding of PpsR to the puc promoter of R. sphaeroides to be performed under oxidizing and reducing conditions in the absence of the antirepressor AppA and other cellular molecules (4, 30). In addition, the use of transcriptional fusions and a heterologous expression system demonstrated that PpsR directly represses the transcription of puc and bchF in R. sphaeroides (15). Although extremely useful, such approaches can tell only part of the story and provide no information about cellular growth conditions and their role in gene expression. In order to determine in vivo under different growth conditions regulation mediated by PpsR for a larger number of target genes in the full context of the target region, as well as in the presence of important biologically confounding factors, the ChIP technique was adapted for use with R. sphaeroides.
ChIP experiments were first performed for the DNA regulatory region of ppaA (RSP0283), a gene known to be under direct PpsR control and containing two perfectly conserved PpsR binding sequences (12) (Table 3). The results obtained confirmed that there was strong binding of the PpsR protein to the DNA region tested when the wild-type strain of R. sphaeroides was grown not only under aerobic conditions but also under anaerobic-dark-DMSO conditions, emphasizing that PpsR has a regulatory role in the expression of genes involved in PS development. An approximately 1.7-fold increase in binding to the canonical PpsR sites was observed when R. sphaeroides was grown under highly oxidizing aerobic conditions compared to cells grown under anaerobic-dark-DMSO conditions, and this increase is comparable to the 2.2-fold-higher affinity of oxidized PpsR than of reduced PpsR previously observed in vitro for the puc promoter (30). The in situ results provide an alternate interpretation for the in vitro findings, that the AppA protein does not affect the state of PpsR under highly aerobic conditions, and the conclusions drawn from the in vitro binding of PpsR to the puc region may be unrelated to in situ binding.
We demonstrated that in vivo there was significant binding of PpsR to DNA regions encompassing the regulatory regions and the ATG codon of the RSP0695, RSP2122, and RSP3241 genes when R. sphaeroides was grown under aerobic and anaerobic-dark-DMSO conditions. From the enrichment data obtained in the ChIP experiments, we could estimate the association of the PpsR protein with the DNA regions having degenerate binding sequences relative to the RSP0283(1-2) DNA fragment. Under aerobic growth conditions, the association of PpsR with the RSP0283(1-2) fragment was 36.8-, 29.0-, 31.4-, and 25.0-fold greater than the association with the RSP0695(1-2), RSP2122(1-2), RSP3241(1-2), and RSP3241(3-4) DNA fragments, respectively. This result was likely due to the presence of degenerate binding sequences for PpsR and suggests that PpsR recognizes the degenerate sequences in the RSP0695(1-2) fragment less efficiently than it recognizes any of the other DNA regions tested.
The affinity of PpsR for the DNA regions tested was not significantly enhanced when R. sphaeroides 2.4.1 was grown under aerobic conditions compared with cells grown under anaerobic-dark-DMSO conditions, as was the case with the canonical sequence. By analogy, using the “degenerate” binding sequences was equivalent to studying binding to mutant forms of the canonical sequence and had the added advantage that physiologic “pressures,” not “guess work” in the laboratory, fixed these sequences in the genome.
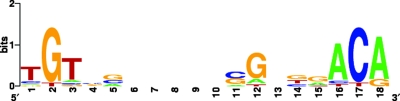
Using the 241(pPNs) strain of R. sphaeroides grown under anaerobic-dark-DMSO conditions, we observed increases in the binding of PpsR ranging from 2.28- to 2.72-fold for the RSP0283, RSP0695, RSP2122, and RSP3241 genes, indicating that the binding of PpsR to these regions has a physiological role and that it is not just the result of a fortuitous interaction between the protein and DNA, emphasizing that there is direct regulation of these genes by PpsR. Use of the ChIP technique led to another important in situ observation: while the binding of PpsR to the regulatory region of the RSP0283 gene containing the canonical PpsR binding sequence was elevated under aerobic growth conditions (97.9-fold enrichment), saturation did not occur. In fact, when PpsR was similarly overexpressed and R. sphaeroides was grown under anaerobic-dark-DMSO conditions, under which the AppA antirepressor was more likely to be present, the binding of PpsR to this region was greater (124.1-fold enrichment). These results suggest that using oxidized PpsR instead of reduced PpsR does not provide a reliable estimate of the interaction between PpsR and the DNA, which can be assessed only in the full context of gene expression. Under aerobic and anaerobic-dark-DMSO growth conditions, PpsR directly interacts with and thereby regulates the expression of genes located outside the PGC containing at least two TGTcN10gACA PpsR binding sequences, which have up to three mismatches in their regulatory regions and which likely encompass the ATG start site. Figure 5 shows the results of an alignment of the degenerate PpsR binding sites detected in the DNA regions where the ChIP experiments revealed a significant PpsR interaction in vivo (7). When we compared each degenerate sequence with the canonical PpsR binding sites, we observed that natural mutations occurred frequently at nucleotide positions 1, 4, 5, 14, and 15, resulting in a decrease in the affinity of PpsR binding without total elimination of the interaction with DNA. Interestingly, in the TGTN12ACA sequence the spacing of the 12 nucleotides is always preserved and the most conserved nucleotides are those surrounding positions 2 and 17, suggesting that these nucleotides constitute the minimal requirement from which a palindromic DNA structure sufficient for PpsR binding is generated. As previously observed for the RSP0679 (hemC) and RSP0680 (hemE) genes (33), PpsR binding sites were detected for the RSP0695 (ccoO) and RSP2122 genes within the coding regions of the genes (106 and 25 bp downstream of the start codon, respectively) and also for the RSP3241 gene, overlapping its likely ATG codon (Table 3).
FIG. 5.
PpsR binding site degeneration. The diagram was created after ChIP analysis using PpsR binding sequences shown in Table 3 and the WebLogo program (7; http://weblogo.berkeley.edu/).
This work markedly increased our understanding of the complexity that lies beneath the regulation of gene expression by the DNA binding protein PpsR. A recurring feature of ChIP analysis for sequence-specific transcription factors is that regulatory proteins can bind in vivo to sites that do not have a good match with the consensus sequence (48). In fact, this feature was previously observed for proteins such as CtrA (26), LexA (47), and FNR (18), strongly suggesting that site-specific transcription factors can bind to targets having degenerate binding sites. Cooperative interaction between multiple transcription factors, reducing the requirement for highly conserved consensus sequences, or local DNA topology can explain this phenomenon. These are conditions which are not easily applied to in vitro DNA binding studies but which are inherent to in situ studies. Today, there are postgenomic tools for determining in vivo direct or indirect interactions of DNA binding proteins with DNA sequences (i.e., DNA binding flexibility) and therefore refining our understanding of regulatory networks.
Supplementary Material
Acknowledgments
This work was supported by grant NIH GM15590 to Samuel Kaplan.
We thank M. Gomelsky (Department of Molecular Biology, University of Wyoming) for providing R. sphaeroides strain 2.4.1(pPNs) and rabbit anti-PpsR antibody.
Footnotes
Published ahead of print on 8 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aklujkar, M., R. C. Prince, and J. Thomas Beatty. 2006. The photosynthetic deficiency due to puhC gene deletion in Rhodobacter capsulatus suggests a PuhC protein-dependent process of RC/LH1/PufX complex reorganization. Arch. Biochem. Biophys. 45459-71. [DOI] [PubMed] [Google Scholar]
- 1a.Anderson, A. J., and E. A. Dawes. 1990. Occurence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b.Bolt, E. L., L. Kryszak, J. Zeilstra-Ryalls, P. M. Shoolingin-Jordan, and M. J. Warren. 1999. Characterization of the Rhodobacter sphaeroides 5 aminolaevulinic acid synthase isoenzymes, HemA and HemT, isolated from recombinant Escherichia coli. Eur. J. Biochem. 265290-299. [DOI] [PubMed] [Google Scholar]
- 2.Braatsch, S., J. R. Bernstein, F. Lessner, J. Morgan, J. C. Liao, C. S. Harwood, and J. T. Beatty. 2006. Rhodopseudomonas palustris CGA009 has two functional ppsR genes, each of which encodes a repressor of photosynthesis gene expression. Biochemistry 4514441-14451. [DOI] [PubMed] [Google Scholar]
- 3.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45827-836. [DOI] [PubMed] [Google Scholar]
- 4.Cho, S. H., S. H. Youn, S. R. Lee, H. S. Yim, and S. O. Kang. 2004. Redox property and regulation of PpsR, a transcriptional repressor of photosystem gene expression in Rhodobacter sphaeroides. Microbiology 150697-706. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary, M., and S. Kaplan. 2000. DNA sequence analysis of the photosynthesis region of Rhodobacter sphaeroides 2.4.1. Nucleic Acids Res. 28862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coomber, S. A., M. Chaudhri, A. Connor, G. Britton, and C. N. Hunter. 1990. Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol. Microbiol. 4977-989. [DOI] [PubMed] [Google Scholar]
- 7.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohue, T. J., A. G. McEwan, and S. Kaplan. 1986. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides cytochrome c2 gene. J. Bacteriol. 168962-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eraso, J. M., and S. Kaplan. 1995. Oxygen-insensitive synthesis of the photosynthetic membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J. Bacteriol. 1772695-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eraso, J. M., and S. Kaplan. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 17632-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eraso, J. M., J. H. Roh, X. Zeng, S. J. Callister, M. S. Lipton, and S. Kaplan. 2008. Role of the global transcriptional regulator PrrA in Rhodobacter sphaeroides 2.4.1: combined transcriptome and proteome analysis. J. Bacteriol. 1904831-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Fales, L., L. Kryszak, and J. Zeilstra-Ryalls. 2001. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: effect of a transposon insertion in the hbdA gene. J. Bacteriol. 1831568-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomelsky, L., J. Sram, O. V. Moskvin, I. M. Horne, H. N. Dodd, J. M. Pemberton, A. G. McEwan, S. Kaplan, and M. Gomelsky. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149377-388. [DOI] [PubMed] [Google Scholar]
- 13.Gomelsky, M., I. M. Horne, H. J. Lee, J. M. Pemberton, A. G. McEwan, and S. Kaplan. 2000. Domain structure, oligomeric state, and mutational analysis of PpsR, the Rhodobacter sphaeroides repressor of photosystem gene expression. J. Bacteriol. 1822253-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomelsky, M., and S. Kaplan. 1998. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel fad binding domain. J. Biol. Chem. 27335319-35325. [DOI] [PubMed] [Google Scholar]
- 15.Gomelsky, M., and S. Kaplan. 1995. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J. Bacteriol. 1771634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27497-500. [DOI] [PubMed] [Google Scholar]
- 18.Grainger, D. C., H. Aiba, D. Hurd, D. F. Browning, and S. J. Busby. 2007. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 35269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, Y., M. H. Meyer, M. Keusgen, and G. Klug. 2007. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol. Microbiol. 641090-1104. [DOI] [PubMed] [Google Scholar]
- 20.Jaubert, M., S. Zappa, J. Fardoux, J. M. Adriano, L. Hannibal, S. Elsen, J. Lavergne, A. Vermeglio, E. Giraud, and D. Pignol. 2004. Light and redox control of photosynthesis gene expression in Bradyrhizobium: dual roles of two PpsR. J. Biol. Chem. 27944407-44416. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, S., J. Eraso, and J. H. Roh. 2005. Interacting regulatory networks in the facultative photosynthetic bacterium, Rhodobacter sphaeroides 2.4.1. Biochem. Soc. Trans. 3351-55. [DOI] [PubMed] [Google Scholar]
- 22.Kiley, P. J., and S. Kaplan. 1988. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol. Rev. 5250-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, D. Y., and K. K. Kim. 2005. Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38266-274. [DOI] [PubMed] [Google Scholar]
- 24.Kim, Y. J., I. J. Ko, J. M. Lee, H. Y. Kang, Y. M. Kim, S. Kaplan, and J. I. Oh. 2007. Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1895617-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laguri, C., R. A. Stenzel, T. J. Donohue, M. K. Phillips-Jones, and M. P. Williamson. 2006. Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides. Biochemistry 457872-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 994632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. E. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 7019-41. [DOI] [PubMed] [Google Scholar]
- 28.Mackenzie, C., J. M. Eraso, M. Choudhary, J. H. Roh, X. Zeng, P. Bruscella, A. Puskas, and S. Kaplan. 2007. Postgenomic adventures with Rhodobacter sphaeroides. Annu. Rev. Microbiol. 61283-307. [DOI] [PubMed] [Google Scholar]
- 29.Mao, L., C. Mackenzie, J. H. Roh, J. M. Eraso, S. Kaplan, and H. Resat. 2005. Combining microarray and genomic data to predict DNA binding motifs. Microbiology 1513197-3213. [DOI] [PubMed] [Google Scholar]
- 30.Masuda, S., and C. E. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110613-623. [DOI] [PubMed] [Google Scholar]
- 31.Masuda, S., C. Dong, D. Swem, A. T. Setterdahl, D. B. Knaff, and C. E. Bauer. 2002. Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc. Natl. Acad. Sci. USA 997078-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuda, S., K. Hasegawa, and T. A. Ono. 2005. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry 441215-1224. [DOI] [PubMed] [Google Scholar]
- 33.Moskvin, O. V., L. Gomelsky, and M. Gomelsky. 2005. Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development. J. Bacteriol. 1872148-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moskvin, O. V., S. Kaplan, M. A. Gilles-Gonzalez, and M. Gomelsky. 2007. Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 28228740-28748. [DOI] [PubMed] [Google Scholar]
- 35.O'Gara, J. P., J. M. Eraso, and S. Kaplan. 1998. A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1804044-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Gara, J. P., and S. Kaplan. 1997. Evidence for the role of redox carriers in photosynthesis gene expression and carotenoid biosynthesis in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1791951-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh, J. I., and S. Kaplan. 2001. Generalized approach to the regulation and integration of gene expression. Mol. Microbiol. 391116-1123. [DOI] [PubMed] [Google Scholar]
- 38.Oh, J. I., and S. Kaplan. 2002. Oxygen adaptation. The role of the CcoQ subunit of the cbb3 cytochrome c oxidase of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 27716220-16228. [DOI] [PubMed] [Google Scholar]
- 39.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 194237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh, J. I., and S. Kaplan. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 382688-2696. [DOI] [PubMed] [Google Scholar]
- 41.Oh, J. I., I. J. Ko, and S. Kaplan. 2004. Reconstitution of the Rhodobacter sphaeroides cbb3-PrrBA signal transduction pathway in vitro. Biochemistry 437915-7923. [DOI] [PubMed] [Google Scholar]
- 42.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 1864748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Ranson-Olson, B., D. F. Jones, T. J. Donohue, and J. H. Zeilstra-Ryalls. 2006. In vitro and in vivo analysis of the role of PrrA in Rhodobacter sphaeroides 2.4.1 hemA gene expression. J. Bacteriol. 1883208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roh, J. H., and S. Kaplan. 2002. Interdependent expression of the ccoNOQP-rdxBHIS loci in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1845330-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roh, J. H., W. E. Smith, and S. Kaplan. 2004. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1. Redox active gene expression profile. J. Biol. Chem. 2799146-9155. [DOI] [PubMed] [Google Scholar]
- 45.Sistrom, W. R. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas spheroides. J. Gen. Microbiol. 28607-616. [DOI] [PubMed] [Google Scholar]
- 46.Sockett, R. E., T. J. Donohue, A. R. Varga, and S. Kaplan. 1989. Control of photosynthetic membrane assembly in Rhodobacter sphaeroides mediated by puhA and flanking sequences. J. Bacteriol. 171436-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wade, J. T., N. B. Reppas, G. M. Church, and K. Struhl. 2005. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 192619-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wade, J. T., K. Struhl, S. J. Busby, and D. C. Grainger. 2007. Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization. Mol. Microbiol. 6521-26. [DOI] [PubMed] [Google Scholar]
- 49.Wong, D. K., W. J. Collins, A. Harmer, T. G. Lilburn, and J. T. Beatty. 1996. Directed mutagenesis of the Rhodobacter capsulatus puhA gene and orf 214: pleiotropic effects on photosynthetic reaction center and light-harvesting 1 complexes. J. Bacteriol. 1782334-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Yang, M. K., Y. C. Lin, and C. H. Shen. 2006. Identification of two gene loci involved in poly-beta-hydroxy butyrate production in Rhodobacter sphaeroides FJ1. J. Microbiol. Immunol. Infect. 3918-27. [PubMed] [Google Scholar]
- 50.Zeilstra-Ryalls, J. H., and S. Kaplan. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell. Mol. Life Sci. 61417-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng, X., M. Choudhary, and S. Kaplan. 2003. A second and unusual pucBA operon of Rhodobacter sphaeroides 2.4.1: genetics and function of the encoded polypeptides. J. Bacteriol. 1856171-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng, X., J. H. Roh, S. J. Callister, C. L. Tavano, T. J. Donohue, M. S. Lipton, and S. Kaplan. 2007. Proteomic characterization of the Rhodobacter sphaeroides 2.4.1 photosynthetic membrane: identification of new proteins. J. Bacteriol. 1897464-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.