Abstract
The pvc gene cluster from Pseudomonas aeruginosa has been linked to the biosynthesis of both the pyoverdine chromophore and pseudoverdine. Our reinvestigation of the role this gene cluster plays in P. aeruginosa secondary metabolite biosynthesis shows that its major product is actually paerucumarin, a novel isonitrile functionalized cumarin.
One of the most extensively studied families of secondary metabolites in pseudomonads is the pyoverdines, cyclic peptide siderophores that are important virulence factors in Pseudomonas aeruginosa (16, 19, 24, 25). Pyoverdines contain both a variable cyclic peptide moiety and a conserved tricyclic chromophore. In P. aeruginosa, the biosynthesis of the pyoverdine chromophore has been attributed to two different loci: a nonribosomal peptide synthase known as pvdL and a four-gene operon known as the pvc gene cluster (Fig. 1) (18, 22, 23). The pvc gene cluster is unique in that it is also reported to be involved in the biosynthesis of pseudoverdine, a fluorescent bicyclic metabolite that resembles the pyoverdine chromophore (15). Our recent observation that many homologs of pvcA are actually isonitrile synthases led us to reinvestigate the role that the pvc gene cluster plays in P. aeruginosa secondary metabolite biosynthesis (1, 2). Here we report the characterization of paerucumarin, an isonitrile functionalized cumarin that is produced by the pvc gene cluster, and demonstrate that the pvc genes are not required for pyoverdine production.
FIG. 1.
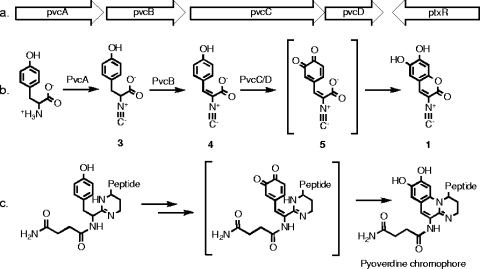
(a) pvc gene cluster. (b) Proposed biosynthetic scheme for paerucumarin. (c) Proposed biosynthetic scheme for the pyoverdine chromophore (6).
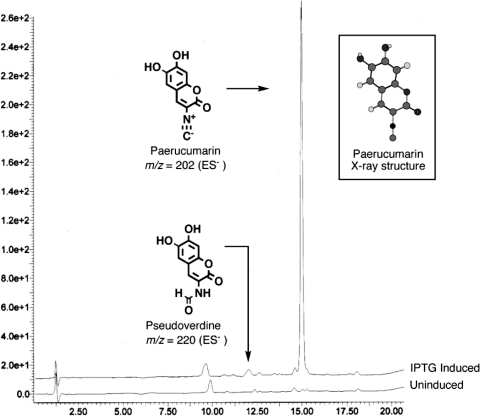
To better understand the role of the pvc cluster in P. aeruginosa secondary metabolite production, the pvc operon was PCR amplified and cloned into the broad-host-range expression vector pMMB67, and the resulting construct, pPVCAD, was conjugated into P. aeruginosa PAK (8, 13). A comparison of the ethyl acetate extracts derived from IPTG (isopropyl-β-d-thiogalactopyranoside)-induced and uninduced cultures by silica gel thin-layer chromatography indicated that the overexpression of the pvc gene cluster led to the production of a single organic extractable metabolite. This compound was purified by silica gel flash chromatography, and the structure was elucidated by using a combination of mass spectrometry and nuclear magnetic resonance experiments (1). The final structure was confirmed by single crystal X-ray diffraction (Fig. 2) and found to be a novel isonitrile-functionalized dihydroxycumarin, which we have given the trivial name paerucumarin (for P. aeruginosa cumarin).
FIG. 2.
LC-MS (negative-ion electrospray ionization) traces of the ethyl acetate extracts derived from IPTG-induced and uninduced cultures of P. aeruginosa transformed with pPVCAD are shown.
The biosynthesis of paerucumarin can be easily rationalized from the general function predictions for each of the enzymes found in the pvc gene cluster (Fig. 1). PvcA is related to known isonitrile synthases (2), PvcB is related to amino acid oxidizing enzymes (2), and PvcC and D are related to HpaB- and HpaC-like two-component flavin adenine dinucleotide-dependent monooxygenases that have been shown to oxidize phenols to both dihydroxy phenols and catechols (9, 26). In the biosynthetic scheme proposed in Fig. 1b, PvcA generates an isonitrile-functionalized tyrosine (compound 3) that is oxidized by PvcB to give the unsaturated intermediate compound 4. Oxidation of intermediate compound 4 to a catechol (compound 5) by PvcC/D, followed by intermolecular cyclization of the catechol and the carboxylate, would give paerucumarin (compound 1).
In our initial thin-layer chromatography analyses of the extracts derived from P. aeruginosa cultures that overexpress the pvc gene cluster, we did not detect pseudoverdine, the N-formyl adduct of paerucumarin that was previously reported to arise from the overexpression of the pvc gene cluster (15). However, upon closer inspection of these extracts by liquid chromatography-mass spectrometry (LC-MS), we were able to identify a minor IPTG-inducible peak that corresponds to the predicted mass for pseudoverdine (molecular weight of 221) (Fig. 2). In Escherichia coli-based studies with other pvcA containing gene clusters, both isonitrile and N-formyl functionalized metabolites have also been observed, and in at least one case a pvcA homolog containing gene cluster was found to produce exclusively an N-formyl adduct (1). Isonitrile and N-formyl functional groups are very closely related, differing by only the addition or loss of water. The isolation of both isonitrile and N-formyl functionalized metabolites from cultures that express different pvcA containing gene clusters suggests that individual members of this family of enzymes may actually produce mixtures of these two functionalities. Although it is possible that paerucumarin was not found in earlier studies on the pvc gene cluster because isonitrile functionalized metabolites are unstable and often rapidly decompose during purification, it is also possible that different pvc gene clusters produce different ratios of isonitrile and N-formyl functionalized metabolites.
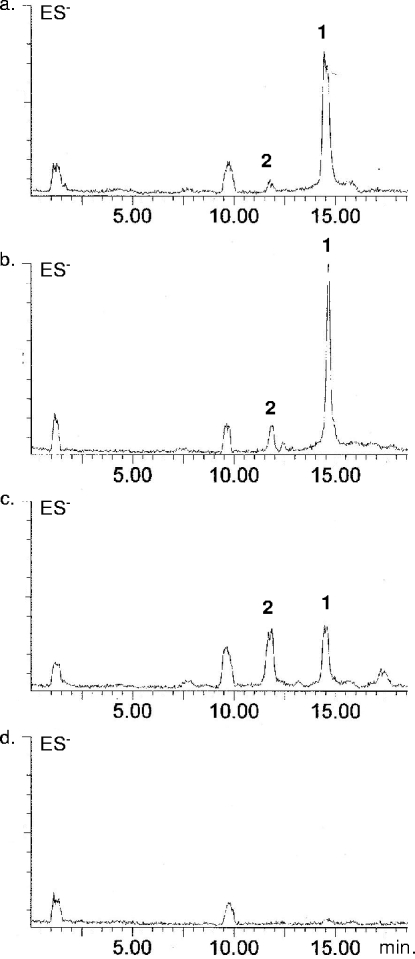
It is known that the pvc gene cluster is positively regulated by PtxR, a LysR transcription factor that also controls exotoxin A production through regA and homoserine lactone production through lasI (3-5, 7, 10, 21, 23). To investigate the products of pvc gene clusters from different P. aeruginosa strains, we overexpressed PtxR in PAO1, PAK, and PA14 strains. Culture broth extracts from each of these strains were then examined by LC-MS (for methods, see the supplemental material). The LC-MS analysis showed that each strain produced a different ratio of paerucumarin to pseudoverdine (Fig. 3). PAK and PAO1 produce almost exclusively paerucumarin, while PA14 produces approximately a one-to-one mixture of paerucumarin and pseudoverdine. Neither paerucumarin nor pseudoverdine was detected in extracts from strains that did not overexpress PtxR, nor was either compound seen in extracts from pvcA knockouts that overexpressed PtxR. pvcA genes from different sequenced P. aeruginosa strains contain a small number of point mutations. Whether these mutations result in the strain-to-strain difference we observed, or whether other factors contribute to the observed differences will likely require a detailed structural analysis of at least one PvcA homolog.
FIG. 3.
LC-MS (negative-ion electrospray ionization) traces of ethyl acetate extracts from cultures of PAK (a), PAO1 (b), and PA14 (c) strains transformed with pPTXR and strain PAK (d) transformed with a vector control. Peaks for paerucumarin (peak 1) and pseudoverdine (peak 2) are marked.
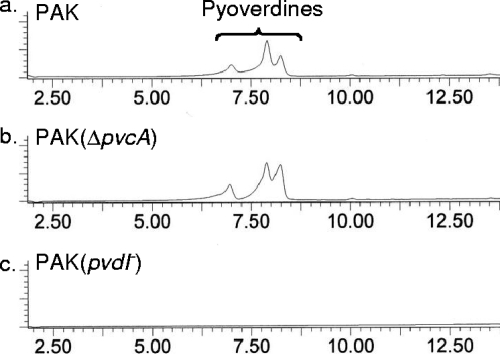
In addition to its role in the biosynthesis of pseudoverdine and now paerucumarin the pvc gene cluster has been reported to be involved in the biosynthesis of pyoverdine (22, 23). The catechol formation and intermolecular cyclization steps in the proposed biosynthetic scheme for paerucumarin are very similar to the biosynthetic steps that have been proposed for the formation of the pyoverdine chromophore (Fig. 2c) (6). To explore the possibility that the pvc gene cluster might play a role in pyoverdine biosynthesis, we generated both pvc gene cluster deletion mutants (ΔpvcA and ΔpvcA-ptxR) and pyoverdine biosynthesis transposon knockouts (12, 20). Under the culture conditions we examined (Luria-Bertani broth, succinate minimal medium [SM] [17], and SM supplemented with 1% Casamino Acids) pvc gene cluster knockouts continued to produced pyoverdines (Table 1 and Fig. 4) (14). Although similar reactions may be used in the biosynthesis of both paerucumarin and the pyoverdine chromophore, it appears that only paerucumarin and pseudoverdine are produced by the pvc gene cluster.
TABLE 1.
Pyoverdine and paerucumarin production by PAK strains
| Strain (genotype)/plasmid | Productiona of:
|
|
|---|---|---|
| Pyoverdine | Paerucumarin | |
| PAK | + | - |
| PAK/pPVCAD | + | + |
| PAK/pPTXR | + | + |
| PAK (ΔpvcA) | + | - |
| PAK (ΔpvcA-ptxR) | + | - |
| PAK (ΔpvcA)/pPTXR | + | - |
| PAK2 (ΔpvdI) | - | - |
| PAK2 (ΔpvdI)/pPTXR | - | + |
| PAK2 (ΔpvdI)/pPVCAD | - | + |
Production of pyoverdine was monitored in SM medium. Production of paerucumarin was monitored in LB medium.
FIG. 4.
High-pressure liquid chromatography (UV) traces of the material extracted by solid-phase extraction from cultures of strains PAK (a), PAK (ΔpvcA) (b), and PAK2 (ΔpvdI) (c).
PvcA and PvcB homologs are found in a number of different bacteria, including Frankia sp. strain CcI3, Erwinia carotovora, Photorhabdus luminescens, Burkholderia mallei, Bdellovibrio bacteriovorus, Legionella pneumophila, and Vibrio cholerae. Even though many of these bacteria are well-studied pathogens, no functions have yet been assigned to any of the metabolites that are produced by pvcA/B-containing gene clusters. In L. pneumophila the pvcA/B homologs are regulated in sessile (biofilm) cells with respect to iron, and in P. aeruginosa the pvc gene cluster expression is controlled by the same transcription factor that controls at least two other important pathogenicity determinants (3, 10, 11). Although the functional significance of the strain-to-strain differences in paerucumarin and pseudoverdine production by P. aeruginosa is still unknown, the assignment of a well-defined function to the pvc gene cluster should make it possible to more easily decipher the various roles that pvc-like biosynthetic clusters play in bacteria.
Supplementary Material
Acknowledgments
This study was supported by NIH grant GM077516.
Footnotes
Published ahead of print on 8 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Brady, S. F., J. D. Bauer, M. F. Clarke-Pearson, and R. Daniels. 2007. Natural products from isnA-containing biosynthetic gene clusters recovered from the genomes of cultured and uncultured bacteria. J. Am. Chem. Soc. 12912102-12103. [DOI] [PubMed] [Google Scholar]
- 2.Brady, S. F., and J. Clardy. 2005. Cloning and heterologous expression of isocyanide biosynthetic genes from environmental DNA. Angew. Chem. Int. Ed. Engl. 447063-7065. [DOI] [PubMed] [Google Scholar]
- 3.Carty, N. L., N. Layland, J. A. Colmer-Hamood, M. W. Calfee, E. C. Pesci, and A. N. Hamood. 2006. PtxR modulates the expression of QS-controlled virulence factors in the Pseudomonas aeruginosa strain PAO1. Mol. Microbiol. 61782-794. [DOI] [PubMed] [Google Scholar]
- 4.Colmer, J. A., and A. N. Hamood. 1999. Expression of ptxR and its effect on toxA and regA expression during the growth cycle of Pseudomonas aeruginosa strain PAO1. Can. J. Microbiol. 451008-1016. [PubMed] [Google Scholar]
- 5.Colmer-Hamood, J. A., H. Aramaki, J. M. Gaines, and A. N. Hamood. 2006. Transcriptional analysis of the Pseudomonas aeruginosa toxA regulatory gene ptxR. Can. J. Microbiol. 52343-356. [DOI] [PubMed] [Google Scholar]
- 6.Dorrestein, P. C., K. Poole, and T. P. Begley. 2003. Formation of the chromophore of the pyoverdine siderophores by an oxidative cascade. Org. Lett. 52215-2217. [DOI] [PubMed] [Google Scholar]
- 7.Ferrell, E., N. L. Carty, J. A. Colmer-Hamood, A. N. Hamood, and S. E. West. 2008. Regulation of Pseudomonas aeruginosa ptxR by Vfr. Microbiology 154431-439. [DOI] [PubMed] [Google Scholar]
- 8.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48119-131. [DOI] [PubMed] [Google Scholar]
- 9.Gibello, A., M. Suarez, J. L. Allende, and M. Martin. 1997. Molecular cloning and analysis of the genes encoding the 4-hydroxyphenylacetate hydroxylase from Klebsiella pneumoniae. Arch. Microbiol. 167160-166. [PubMed] [Google Scholar]
- 10.Hamood, A. N., J. A. Colmer, U. A. Ochsner, and M. L. Vasil. 1996. Isolation and characterization of a Pseudomonas aeruginosa gene, ptxR, which positively regulates exotoxin A production. Mol. Microbiol. 2197-110. [DOI] [PubMed] [Google Scholar]
- 11.Hindre, T., H. Bruggemann, C. Buchrieser, and Y. Hechard. 2008. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 15430-41. [DOI] [PubMed] [Google Scholar]
- 12.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 13.Ishimoto, K. S., and S. Lory. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J. Bacteriol. 1743514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilz, S., C. Lenz, R. Fuchs, and H. Budzikiewicz. 1999. A fast screening method for the identification of siderophores from fluorescent Pseudomonas spp. by liquid chromatography/electrospray mass spectrometry. J. Mass Spectrom. 34281-290. [DOI] [PubMed] [Google Scholar]
- 15.Longerich, I., K. Taraz, H. Budzikiewicz, L. Tsai, and J. M. Meyer. 1993. Pseudoverdin, a compound related to the pyoverdin chromophore from a Pseudomonas aeruginosa strain incapable to produce pyoverdins. Zentralbl. Naturforsch. C 48425-429. [DOI] [PubMed] [Google Scholar]
- 16.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, J. M. a. M. A. A. 1978. The Fluorescent Pigment of Pseudomonas fluorescens: biosynthesis, purification, and physiocochemical properties. J. Gen. Microbiol. 107319-328. [Google Scholar]
- 18.Mossialos, D., U. Ochsner, C. Baysse, P. Chablain, J. P. Pirnay, N. Koedam, H. Budzikiewicz, D. U. Fernandez, M. Schafer, J. Ravel, and P. Cornelis. 2002. Identification of new, conserved, nonribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 451673-1685. [DOI] [PubMed] [Google Scholar]
- 19.Ravel, J., and P. Cornelis. 2003. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 11195-200. [DOI] [PubMed] [Google Scholar]
- 20.Rietsch, A., I. Vallet-Gely, S. L. Dove, and J. J. Mekalanos. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1028006-8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47597-626. [DOI] [PubMed] [Google Scholar]
- 22.Stintzi, A., P. Cornelis, D. Hohnadel, J. M. Meyer, C. Dean, K. Poole, S. Kourambas, and V. Krishnapillai. 1996. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiology 142(Pt. 5)1181-1190. [DOI] [PubMed] [Google Scholar]
- 23.Stintzi, A., Z. Johnson, M. Stonehouse, U. Ochsner, J. M. Meyer, M. L. Vasil, and K. Poole. 1999. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS. J. Bacteriol. 1814118-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 681834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visca, P., F. Imperi, and I. L. Lamont. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 1522-30. [DOI] [PubMed] [Google Scholar]
- 26.Xun, L., and E. R. Sandvik. 2000. Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl. Environ. Microbiol. 66481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.