Abstract
Whole-genome sequencing has been skewed toward bacterial pathogens as a consequence of the prioritization of medical and veterinary diseases. However, it is becoming clear that in order to accurately measure genetic variation within and between pathogenic groups, multiple isolates, as well as commensal species, must be sequenced. This study examined the pangenomic content of Escherichia coli. Six distinct E. coli pathovars can be distinguished using molecular or phenotypic markers, but only two of the six pathovars have been subjected to any genome sequencing previously. Thus, this report provides a seminal description of the genomic contents and unique features of three unsequenced pathovars, enterotoxigenic E. coli, enteropathogenic E. coli, and enteroaggregative E. coli. We also determined the first genome sequence of a human commensal E. coli isolate, E. coli HS, which will undoubtedly provide a new baseline from which workers can examine the evolution of pathogenic E. coli. Comparison of 17 E. coli genomes, 8 of which are new, resulted in identification of ∼2,200 genes conserved in all isolates. We were also able to identify genes that were isolate and pathovar specific. Fewer pathovar-specific genes were identified than anticipated, suggesting that each isolate may have independently developed virulence capabilities. Pangenome calculations indicate that E. coli genomic diversity represents an open pangenome model containing a reservoir of more than 13,000 genes, many of which may be uncharacterized but important virulence factors. This comparative study of the species E. coli, while descriptive, should provide the basis for future functional work on this important group of pathogens.
Escherichia coli is commonly found in the normal microflora in the human gastrointestinal tract (16) and is intricately involved in the lives of humans. This bacterium can be grown readily, and its genetics are easily manipulated in the laboratory, making it a common workhorse and one of the best-studied prokaryotic model organisms. However, E. coli isolates can cause serious illness in humans and are associated with at least six distinct disease presentations (29) that result in billions of dollars in lost work time and doctor and hospital visits each year (56). Diarrheagenic E. coli strains are well known from recent outbreaks in the United States (http://www.cdc.gov/ecoli/); however, a significant proportion of E. coli isolates cause disease outside the intestinal tract, and these isolates are known as extraintestinal pathogenic E. coli (ExPEC) (54, 56). The ExPEC isolates cause a range of diseases in humans, including urinary tract infections and neonatal meningitis. The diarrheagenic pathogenic variants (pathovars) of E. coli are also diverse in terms of their clinical presentation, age groups affected, and associated virulence factors. Five distinct clinical groups of diarrheagenic isolates have been identified: enteroaggregative E. coli (EAEC), enterohemorrhagic E. coli (EHEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), and enterotoxigenic E. coli (ETEC). Other pathogenic groups have been proposed; however, these five groups are distinct and can be agreed upon by the community at large (29). EAEC is considered by many to be an emerging pathogen that can infect a wide range of age groups (18, 23). The molecular basis of the virulence of EAEC is not well defined, suggesting that further molecular characterization of the pathovar is required (18, 23). Prototypical EAEC infection is characterized by the formation of a biofilm in the colon, followed by secretion of toxins and cytolytic factors (18, 23). EHEC isolates are often associated with serotype O157:H7, because this serotype has been responsible for recent outbreaks associated with spinach in the United States (10). There are many serotypes that are associated with EHEC, but almost all isolates contain a type III secretion system (TTSS) involved in the direct secretion of bacterial factors into the epithelium, resulting in destruction of the colonic architecture and leading to bloody diarrhea (3). Additionally, EHEC isolates produce a Shiga-like toxin responsible for systemic clinical symptoms such as hemolytic-uremic syndrome and secondary neuronal and kidney sequelae (30). EPEC isolates also use a TTSS to disrupt the epithelial layer; however, a tropism for the small intestine and the lack of Shiga toxin genes distinguish this pathovar from EHEC (5, 29). ETEC isolates, as the name implies, secrete a number of toxins, including stable toxin and labile toxin (49). The clinical presentation of ETEC infection is often referred to as traveler's diarrhea and occurs predominantly in developing countries (49). Members of this pathovar bind to the epithelial layer in the small intestine and secrete toxins that cause an altered water balance in the intestinal lumen, resulting in watery diarrhea (49). While none of the other E. coli pathovars routinely invades the epithelial cells, members of the EIEC group readily enter the enterocytes of the colon and move laterally, thus avoiding many of the host innate immune responses (12, 44). Clinically and diagnostically, EIEC is similar to Shigella species, sharing many of the same virulence factors (12, 44). Overall, E. coli isolates and the diseases that they cause are diverse, and while many virulence factors are known for each of the different pathovars, the genetic content of these pathogens has not been examined previously on a genomic level.
The microbial species that have been investigated using whole-genome sequencing have been markedly skewed toward pathogens as a result of microbe-associated human and veterinary diseases. In addition, and despite the power inherent in whole-genome analyses, the inability to reliably assign a function to the predicted products of uncharacterized genes, such as virulence factors, remains a major limitation. Functional gene characterization following genome sequencing relies on wet lab bench work to validate bioinformatic predictions. Genomic comparisons can rapidly narrow, by at least a factor of 10 (38), the number of targets or virulence factors that need to be investigated using further classic molecular biology techniques. Comparative genomic analysis of carefully selected isolates with well-defined phenotypic characteristics maximizes the insights available through whole-genome sequencing. To this end, the current study was aimed at comparing the genomic contents of multiple pathogenic strains and one commensal isolate of E. coli in an effort to identify unique virulence determinants in each pathovar, as well as common features shared by members of the species. We used the comparisons to predict the pangenome (60) of the species E. coli and to identify potential novel virulence factors in a wide range of isolates.
In this paper, we also describe a genomic comparison of a true human commensal E. coli isolate, E. coli HS (34), to previously determined and publicly available E. coli genome sequences (Table 1). Previous genome sequencing efforts with this species have focused on laboratory-adapted E. coli K-12 strains (1, 19, 53) or a limited number of pathogenic isolates (20, 22, 46, 52, 67). The use of the genome of a gastrointestinal tract-colonizing commensal as a reference genome should provide a more accurate account of which features may be associated with colonization and which features may be disease associated. Additionally, comparison of multiple isolates of the same pathovar should allow examination within the groups to determine conserved or convergent methods of virulence. This data set provides a novel genomic perspective on the evolution of E. coli as a pathogen and a species.
TABLE 1.
Characteristics of isolates sequenced
| Pathovar | Strain | Serotype | Virulence factorsa | Genome status | No. of contigs | Plasmid | GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| Laboratory adapted | K-12 | Complete | 1 | No | U00096.2 | 1 | ||
| Laboratory adapted | W3110 | Complete | 1 | No | AP009048 | 19, 53 | ||
| Commensal | HS | O9 | Complete | 1 | No | CP000802 | This study | |
| ETEC | E24377A | O139:H28 | LT+ ST+; CFA CS1 and CS3 | Complete | 1 | Yes | CP000800 | This study |
| ETEC | B7A | O148:H28 | LT+ ST+; CFA CS6 | Draft | 198 | Yesb | AAJT00000000 | This study |
| EHEC | EDL933 | O157:H7 | LEE, Shiga toxin | Complete | 1 | Yesc | AE005174.2 | 46 |
| EHEC | Sakai | O157:H7 | LEE, Shiga toxin | Complete | 1 | Yesc | BA000007.2 | 20 |
| ExPEC/UPEC | CFT073 | O6:K2:H1 | MRHA/S, MRHA/H, hemolysin, aerobactin, pap pili (X2) | Complete | 1 | No | AE014075.1 | 67 |
| ExPEC/UPEC | F11 | O6:H31 | MRHA/S, MRHA/H, hemolysin, aerobactin, pap pili, and PapG III allele | Draft | 88 | No | AAJU00000000 | This study |
| ExPEC/UPEC | UTI89 | Cysitis isolation; pap pili, PapG III allele, iron-regulated genes | Complete | 1 | Yesc | CP000243.1 | 6 | |
| ExPEC/UPEC | 536 | O6:K15:H31 | MRHA; hemolysin, aerobactin, pap pili | Complete | 1 | No | CP000247.1 | 22 |
| EPEC | E22 | O103:H2 | Rabbit pathogen, EAF plasmid, LEE | Draft | 109 | Unknown | AAJV00000000 | This study |
| EPEC | E110019 | O111:H9 | Atypical EPEC, LEE, EAF plasmid not present | Draft | 115 | Unknown | AAJW00000000 | This study |
| EPEC | B171 | O111:NM | EAF plasmid, LEE | Draft | 159 | Unknownb | AAJX00000000 | This study |
| EAEC | Ec042 | O44:H18 | enterotoxin (Pet and EAST), pAAF plasmid | Incomplete | 7 | Yes | NAd | Sanger |
| EAEC | 101-1 | untypable:H10 | Heat-stable enterotoxin production, lacks pAAF plasmid | Draft | 70 | Yesb | AAMK00000000 | This study |
| ExPEC/avian | APEC01 | O1 | Avian disease-causing isolate, pAPEC01 | Complete | 1 | Yes | CP000468.1 | 27 |
LT, labile toxin; ST, stable toxin; MRHA, mannose-resistant hemagglutinin.
The draft sequences contain contigs that appear to be plasmid in origin; however, no plasmids have been closed for the projects.
Strain EDL933 was sequenced with a plasmid designated pO157 (accession number AF074613) that is 92.1 kb long; there is no smaller plasmid in this isolate. Strain Sakai was sequenced with two plasmids, pO157 (accession number AB011549) and pOSAK1 (accession number AB011548), which were 92.7 and 3.3 kb long, respectively. Strain UTI89 contains a 114-kb plasmid (accession number CP000244), and strain B171 contains a 68.8-kb plasmid that has been sequenced, pB171 (EAF plasmid; accession number AB024946).
NA, not applicable.
MATERIALS AND METHODS
Isolate selection.
The characteristics of the strains sequenced in the present study and the publicly available genomes used for comparison are summarized in Table 1.
(i) Commensal isolate.
The E. coli HS culture used for sequencing was obtained from the Center for Vaccine Development at the University of Maryland School of Medicine as a frozen stock early in the history of this isolate. This isolate was obtained from a healthy human with no disease and has been shown to colonize the human gastrointestinal tract without any apparent clinical symptoms (37).
(ii) ETEC isolates.
ETEC isolates E24377A and B7A are verified human pathogens (36, 59), and stocks were prepared from a master GMP bank at Walter Reed Army Institute of Research (Silver Spring, MD). Both strains produce labile toxin and stable toxin. E24377A is a serotype O139:H28 strain and produces the CS1 and CS3 colonization factor antigens (CFA) (4), whereas strain B7A is a serotype O148:H28 strain and produces only the CS6 CFA.
(iii) ExPEC isolate.
ExPEC isolate F11 is a uropathogenic isolate obtained from a 20-year-old patient with cystitis (with only bladder involvement [57]). The ability of this strain to colonize mice has been experimentally demonstrated, and a number of virulence factors have been functionally characterized (26). This strain is a serotype O6:H31 strain, encodes pap pili, contains the papG III allele, two distinct mannose-resistant hemagglutinins (MRHA/S and MRHA/H), and hemolysin, and has been shown to produce aerobactin (38).
(iv) EPEC isolates.
E. coli E22 is a model strain used in rabbit model experiments (rabbit EPEC) (7). This strain is a serotype O103:H2 strain and contains the locus of enterocyte effacement (LEE) characteristic of strains causing attaching and effacing lesions (40). E. coli E110019 was obtained from an outbreak in Finland in 1987 (66) and is notable because it lacks many of the previously identified virulence factors for the EPEC group, including the EAF plasmid (63). This isolate is considered an atypical EPEC isolate and is serotype O111:H9. E. coli B171 is the prototypic member of the EPEC2 group and encodes potential virulence factors, such as LEE and the bundle-forming pilus-encoding plasmid EAF (GenBank accession number AB024946) (63). Additionally, this isolate is serotype O111:NM.
(v) EAEC isolate.
E. coli 101-1 was characterized as an atypical EAEC isolate and was responsible for a large outbreak in Japan in the late 1990s (8, 24). This strain contains few classic EAEC virulence factors, as determined by DNA hybridization (8); however, the nature of this isolate suggests that it has a higher level of virulence than other EAEC isolates.
Genomic DNA genomic preparation.
Precautions were taken to minimize the possibility of genomic mutation via an increased number of passages and virulence attenuation due to laboratory growth. Genomic DNA was obtained from each E. coli isolate using the method of Ge and Taylor (15), with the following modification. E. coli isolates were grown in Luria broth for 16 h, and the cell biomass was used for genomic DNA isolation. The genomic DNA was quantified by spectrophotometer and Nanodrop methods; ∼30 to 40 μg was used for library construction and closure reactions.
Sequencing of the E. coli genomes.
Library construction, random shotgun sequencing, and genome assembly were performed as described previously (51). One large-insert random plasmid sequencing library (10 to 12 kb) and one small-insert random plasmid sequencing library (4 to 5 kb) were sequenced for each of the genome projects; the sequencing success rates were greater than 85%, and the average high-quality read lengths were greater than 800 nucleotides. The completed genome sequences contained reads from the large- and small-insert libraries that resulted in an average of >10-fold sequence coverage per nucleotide for closed genomes and >7.5-fold coverage for draft genomes. For the complete genome projects, editing, walking library clones, and linking assemblies were used to close the remaining unsequenced regions. The Glimmer Gene Finder (55) was utilized to identify potential coding regions, and annotation and assignment of role categories were performed as described previously (61). GenBank accession numbers for all projects are shown in Table 1.
BSR analysis.
BLAST score ratio (BSR) analyses were performed as previously described by Rasko et al. (50). Briefly, for each of the predicted proteins of a selected E. coli reference strain, we obtained a raw BLASTP score for the alignment against itself (REF_SCORE) and the most similar protein (QUE_SCORE) for each of the other E. coli genomes listed in Table 1. Dividing the QUE_SCORE obtained for each query genome protein by REF_SCORE normalized the scores. Peptides with a normalized ratio of ≤0.4 were considered nonhomologous. A normalized BSR of 0.4 is similar to two proteins being ∼30% identical over their entire lengths (50). Normalized scores were used as an indication of conservation (BSR, ≥0.8), divergence (0.4 < BSR < 0.8), or uniqueness (BSR, ≤0.4) (50).
Pangenome calculations.
Tables containing the complete data set for the BSRs were compiled, and then the unique and conserved gene sets were determined as previously described (60); however, instead of using the BLAST P values, the BSR scores with a stringent threshold of inclusion of ∼>80% over the length of the protein were used. We felt that the stringent threshold applied in this study more accurately predicted the similarities and differences in this group of pathogens. The complete tables for BSR data for each genome can be obtained from the corresponding author.
RESULTS AND DISCUSSION
E. coli HS: What makes a commensal?
E. coli HS was originally isolated from a stool of a healthy laboratory scientist and has been used as a nondomesticated E. coli isolate in a number of human colonization studies (34, 35). In all cases, E. coli HS has been shown to stably colonize the human gastrointestinal tract but cause no detectable adverse effects at doses greater than 1010 CFU (34). The utility of using E. coli HS as the reference genome and isolate is that it provides a colonization potential background that most laboratory-adapted isolates no longer possess. Additionally, inclusion of this isolate provided the opportunity to examine evolution of pathogenic E. coli on a genomic scale by comparing its sequence to the sequences of pathogens that were isolated from various clinical presentations. We believe that the HS genome sequence will become the model for comparative studies in E. coli.
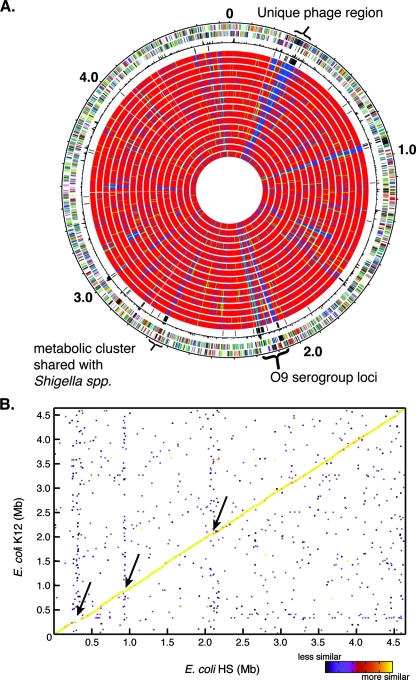
There are relatively few genes (94 genes) that were found only in the HS genome and not in the other E. coli genomes (Table 2). Sixty-four of these genes have no functional annotation and are annotated as hypothetical. The unique genes with functional annotations are either related to production of the serogroup O9 lipopolysaccharide, a serotype not represented in the other genomes, or prophage remnants. The commensal genome does not contain or lack any one specific region that we can conclusively attribute to the colonization phenotype compared to the laboratory-adapted isolates (Fig. 1A; see Fig. S1 in the supplemental material). The E. coli HS genome is highly syntenic with the other sequenced E. coli genomes (Fig. 1B). While it possesses a rather small number of unique genes, it does contain features that are shared with each of the compared pathogenic isolates. Detailed comparative examination of the genomes indicated the mosaic genome structure of the commensal species. Welch et al. first described the mosaic E. coli genome structure based on a comparison of three isolates (67). The inclusion of 14 additional strains in this study further supports this conclusion. Using E. coli HS as a reference, it is possible to rapidly identify clusters of genes that are shared by specific isolates or pathovars and HS (see Fig. S2 in the supplemental material). Multiple examples of this mosaicism were found throughout the genome comparisons.
TABLE 2.
Identification of conserved, unique, and group-specific genes in E. coli
| E. coli strain | Pathovara | No. of genes | No. of conserved genesb | No. of unique genesc | No. of group-specific genesd |
|---|---|---|---|---|---|
| K-12 | Commensal | 4,238 | 2,312 | 21 | 11 |
| W3110 | Commensal | 4,384 | 2,324 | 31 | 11 |
| HS | Commensal | 4,433 | 2,321 | 94 | 11 |
| E24377A | ETEC | 5,111 | 2,318 | 246 | 5 |
| B7A | ETEC | 5,357 | 2,446 | 256 | 4 |
| EDL933 | EHEC | 4,863 | 2,327 | 27 | 122 |
| Sakai | EHEC | 5,497 | 2,337 | 208 | 126 |
| CFT073 | ExPEC/UPEC | 5,589 | 2,341 | 308 | 56 |
| F11 | ExPEC/UPEC | 5,198 | 2,412 | 203 | 46 |
| UTI89 | ExPEC/UPEC | 5,176 | 2,288 | 109 | 45 |
| 536 | ExPEC/UPEC | 4,734 | 2,347 | 134 | 46 |
| APEC01 | ExPEC/Avian | 4,561 | 2,295 | 158 | 3d |
| E22 | EPEC | 5,575 | 2,343 | 274 | 5 |
| E110019 | EPEC | 5,586 | 2,397 | 219 | 4 |
| B171 | EPEC | 5,330 | 2,382 | 234 | 6 |
| Ec042 | EAEC | 4,899 | 2,305 | 308 | 3 |
| 101-1 | EAEC | 4,812 | 2,356 | 155 | 4 |
Pathovar assignment is based on previous identification of the strain (see references).
Conserved genes are genes whose BSR is ≥0.8 in all isolates.
Unique genes are genes whose BSR is <0.4 in all other isolates tested.
Group-specific genes are genes whose BSR is >0.8 in the members of the group but <0.4 in all other isolates.
FIG. 1.
Gene content and synteny of the commensal isolate E. coli HS. (A) Gene conservation using E. coli HS as the reference strain. Starting from the outside, the first circle shows the genes in the forward orientation. The second circle shows the genes in the reverse orientation relative to the origin. The third circle shows the chi-square values, representing differences in the local G+C content. The fourth circle shows all of the genes that are unique to E. coli HS. Circles 5 to 20 show the gene conservation in all of the other E. coli genomes compared in the following order: MG1655, W3110, E24377A, B7A, EDL933, Sakai, CFT073, F11, UTI89, 536, E22, E110019, B171, Ec042, 101-1, and APEC01 (Table 1). The color indicates that a gene is present (red), divergent (green), or absent (blue). Three additional regions which are unique to E. coli HS are indicated: one phage region (∼0.3 Mb) that is not shared with any of the other sequenced strains, the serogroup O9-specific region (∼2.1 Mb), and one additional cluster that is shared only with Shigella species (∼2.6 Mb). (B) Gene synteny (conserved gene order) for E. coli HS and E. coli K-12. The color indicates the level of similarity between regions, as shown by the scale on the lower right. The arrows indicate three regions of diversity for these genomes. The upper and lower arrows indicate the unique phage and the O9 serogroup cluster. Overall, there is a great deal of synteny between the two genomes. A similar pattern was observed for most other complete genomes; the exception was the EHEC strain EDL933 genome, which contains a single large inversion.
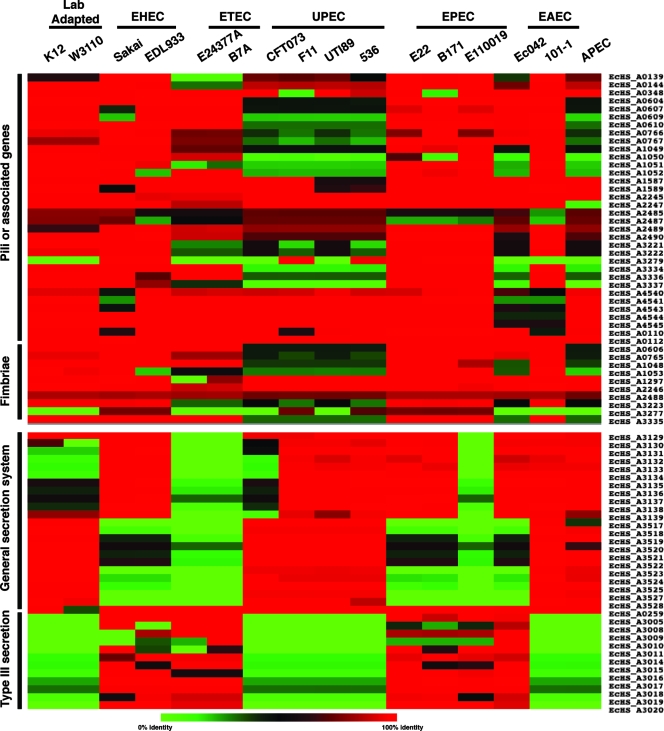
We examined a number of features that are shared by HS and one or more of the pathogens that are thought to be involved in colonization or virulence. Pili and fimbriae are two mechanisms by which bacteria adhere to biotic and abiotic surfaces. Comparisons of the genes annotated as genes encoding pilus or fimbrial components in E. coli HS to genes of the other pathogens revealed that these genes are conserved in the genomes compared and that none of these genes are unique to HS (Fig. 2). Conversely, when the secretion systems were examined, the results showed that E. coli HS contains two divergent copies of a general secretory pathway, also known as the type II secretion system (43). While both copies are absent in the ETEC isolates and one EPEC isolate (E110019), one version is conserved in the EHEC and remaining EPEC isolates, whereas ExPEC isolates contain both copies. In E. coli HS, this type II secretion system is not flanked by any mobile elements, further supporting the possibility that the presence of these pathways in certain isolates represents niche specialization and not a random horizontal transfer event. Additionally, a nonfunctional TTSS lacking all components of a functional TTSS, designated ETT2, is present in E. coli HS (Fig. 2) (39). This cluster is conserved in E. coli HS and the EHEC and EPEC isolates but in no other pathovars. ETT2 has been found in EPEC strains and also in some non-O157 Shiga toxin-producing E. coli strains (39). It was originally thought that ETT2 was a pathogenic marker for particular isolates and that the locus may be used for identification of pathogenic isolates of human and animal epidemic strains (39); however, the presence of ETT2 in E. coli HS suggests that it is not a marker of pathogenesis. These are only a few examples of the mosaic nature of the E. coli HS genome.
FIG. 2.
Commensal features are often shared with one or more pathogens. Using E. coli HS as the reference, we identified regions, based on the annotation or similarity to known features, that could be associated with colonization of the human gastrointestinal tract. These regions were grouped into four general categories: pili or pilus-associated genes, fimbriae, general secretion, and type III secretion. Isolates are arranged in vertical lines, and each horizontal group is based on a single gene or peptide. The color indicates the level of similarity; red indicates the most similar (∼100% identical), green indicates little or no similarity, and black indicates ∼50% identity over the length of the sequence queried. It is clear that some pathogenic groups have features more or less similar to features of E. coli HS. Notably, the general secretion system genes for two separate systems are absent in both ETEC strains; however, they are present in three of four UPEC strains. The presence of one of the secretion systems is variable in EHEC, EAEC, and EPEC isolates, and the opposite phenotype is present in the laboratory-adapted strains, suggesting that this system may play a role in colonization.
The mosaic structure suggests that the commensal strains do have pathogenic potential and may also act as genetic repositories for virulence factors. These factors include many factors that were previously thought to be pathogen specific, suggesting that they are (i) not strictly pathogenic features, (ii) utilized by the commensal species for colonization, or (iii) “in transit” through the commensal species en route to another pathogenic isolate. Pupo et al. suggested that pathogenic E. coli lineages may have evolved multiple times (48), and it is possible that commensal E. coli strains act as genetic repositories with the ability to acquire DNA from multiple sources, as well as the ability to act as DNA donors. Acquisition of the appropriate pathogenic features may result in transfer of the ability to cause disease to a commensal isolate; conversely, pathogens may be able to revert to a commensal state by loss or donation of DNA.
ETEC.
This study provides the first description of the complete genome sequence of an ETEC isolate. The genome of isolate E24377A was sequenced to completion, and enough of the isolate B7A genome was sequenced to obtain a high-coverage draft. Both isolates are verified human pathogens (Carl Brinkley, Walter Reed Army Institute of Research, personal communication) and contain stable and labile toxins characteristic of ETEC isolates (65). One feature of the E24377A genome is that the chromosome of this isolate is smaller than those of other pathogenic E. coli isolates examined to date (∼4.9 Mb versus >5 Mb for EHEC and ExPEC isolates). A smaller genome may be a characteristic of ETEC and may be a result of the high percentage of identified insertion (IS) elements and mobile features. The frequency of insertion and repeated mobile features made this genome difficult to complete. The presence of these elements in associated plasmids may also provide a mechanism by which the plasmids can integrate into or excise from the chromosome. The IS elements that are readily identifiable and occur in multiple copies in the E24377A genome include IS1 to IS4, IS605, IS66, IS91, IS605, IS621, IS629, IS630, and IS911 (http://www-is.biotoul.fr/). Approximately 4% of the genome is composed of these mobile elements. The B7A genome does not appear to contain as many mobile elements as the E24377A genome; however, the high frequency of repeated elements impaired assembly and resulted in the B7A genome having the greatest number of contigs of any draft genome sequence (Table 1).
The chromosome of ETEC contains a unique gene cluster, EcE24377A_2278 to EcE24377A_2297, that is responsible for the utilization of propanediol. Propanediol can be obtained from catalysis of fucose under anaerobic conditions, leading to the production of ATP, an electron sink, and multiple compounds diverted into central metabolism (2, 64). The use of propanediol as a sole carbon source in Salmonella enterica serovar Typhimurium is linked to cobalamin cofactor production via the cobalamin-dependent propanediol dehydratase enzyme pathway (2, 25); however, this is a rare phenotype in E. coli (17). It is noteworthy that in E24377A the cobalamin biosynthetic pathway genes are adjacent to the propanediol utilization gene cluster, and pathway reconstruction suggested that a fully functional pathway is present. While propanediol utilization genes are limited to E23477A, cobalamin biosynthesis is not. Lawrence and Roth have proposed that the propanediol utilization pathway was lost in an ancestor of E. coli and Salmonella and that the reintroduction of this pathway into these organisms was the result of a relatively recent horizontal gene transfer event resulting in altered metabolism (33). The role that this gene cluster plays in virulence, if any, is unclear; it is possible that fucose is obtained from the surface of epithelial cells and catalyzed.
One interesting aspect of the E24377A genome is the presence of multiple plasmids. No complete closed plasmids were identified in the B7A genome sequence. We could identify some plasmid contig sequences, but the high level of repeated sequences prevented rapid closure. The only other large ETEC plasmid whose sequence has been reported is designated pCOO (13). Accurate closure of the E24377A plasmids was difficult due to the presence of IS elements and repeated elements both within other plasmids and in the chromosome. The three smallest plasmids of E24377A do not contain any IS elements, whereas the three largest plasmids contain at least five IS elements and multiple identical copies of some of these plasmids (Table 3). We completed analysis of six plasmids from E24377A whose sizes ranged from 5,033 to 79,237 bp, for a total of 269,669 bp of extrachromosomal DNA (Table 3). The G+C contents of the E24377A plasmids range from 47.3 to 51.6%; in contrast, the chromosome has a G+C content of 50%. Interestingly, the largest plasmid in this isolate has the lowest G+C content (Table 3), whereas the highest G+C content was found in the small plasmid pE24377A_6. If pE24377A_5 is disregarded as an outlier or neutral plasmid, as it has no identifiable phenotype and can be considered cryptic, the G+C content is inversely correlated to plasmid size. Additionally, the only other sequenced ETEC plasmid, pCOO (∼98 kb), exhibits a similar trend and has an even lower G+C content, 46.74% (13). The inverse correlation of size and G+C content may be a result of the introduction foreign DNA in the form of more AT-rich IS or repeated elements.
TABLE 3.
Features of the plasmids from E24377A
| Plasmid | Size (nucleotides) | G+C content (%) | IS elementsa | Virulence factors |
|---|---|---|---|---|
| pE24377A_5 | 5,033 | 49.63 | NP | None, cryptic, pColE1 replicon |
| pE24377A_6 | 6,199 | 52.57 | NP | Streptomycin and sulfonamide resistance |
| pE24377A_35 | 34,367 | 51.62 | NP | Transfer capabilities |
| pE24377A_70 | 70,609 | 50.21 | IS1 (1*), IS2 (1), IS66 (2), IS629 (1*) | CS fimbriae |
| pE24377A_74 | 74,224 | 49.85 | IS1 (1), IS2 (2*), IS66 (4), IS91 (1*) IS629 (2*) | EatA autotransporter |
| pE24377A_79 | 79,237 | 47.27 | IS1 (2*), IS2 (2*), IS66 (1*), IS91 (4*) IS629 (1*) | Enterotoxin A, CS3 fimbriae |
| pCOO | 98,396 | 46.74 | NDb | CS1 fimbriae, EatA serine protease |
The number of copies of each IS element is indicated in parentheses. An asterisk indicates that the element contains a frameshift and/or a premature stop. NP, no IS elements present; ND, not determined (multiple IS elements have been identified in pCOO, but most of them are not grouped with the pE24377A plasmids).
The gene contents of the smaller plasmids of E24377A, pE24377A_5 and pE24377A_6, are similar to the gene contents of previously identified plasmids, and their role in pathogenesis is questionable. Plasmid pE24377A_35 does not resemble other known plasmids, with the exception of a ∼5-kb region that is shared with pE24377A_73 and other ColIb-P9-based plasmids (GenBank accession number AB021078). Putative components of a plasmid transfer system are present on pE24377A_35 (EcE24377A_C0001 to EcE24377A_C0005 and EcE24377A_C0028), but the complete transfer system is not present. Plasmid transfer genes are present on all of the large E24377A plasmids, suggesting that all these plasmids may be required to form a functional transfer apparatus. Alternatively, it is possible that these plasmids constitute one larger cointegrated plasmid that undergoes rearrangement and that the forms that we observed in the sequencing project represented a snapshot of the plasmid(s) in flux. The concept of a large cointegrated plasmid in ETEC was suggested during the characterization of pCOO (13) and requires further functional examination with E24377A.
The E24377A plasmids each contain virulence factors known to be essential to ETEC pathogenesis (65). Plasmid pE24377A_70 encodes the fimbrial subunit required for the CFA CS1 with only a single amino acid change compared to the CooD precursor protein (13). pE24377A_74 encodes the EatA autotransporter protein (45). EatA is a member of the type V autotransporter family of secretion systems, and a single peptide contains all of the functional information necessary for it to cross the inner and outer membranes of gram-negative bacteria (21). In this case, EatA is a serine protease that plays a significant role in the virulence of ETEC. It has also been shown by hybridization that a panel of ETEC isolates contain plasmid-borne copies of the EatA gene (45). The final large plasmid, pE24377A_79, encodes an additional subunit of the CFAs, CS3, as well as heat-labile enterotoxin A (EcE24377A_F0020). The toxin is directly responsible for the diarrhea seen with this pathovar because it alters the ionic balance within the intestinal epithelium and causes water to enter the lumen, resulting in diarrhea. While the pCOO plasmid contained these virulence factors on a single plasmid, in E24377A these factors are on separate plasmids.
Examination of the chromosome and plasmids of E24377A revealed a genome that is undergoing rapid change through gene rearrangement and loss compared to the other E. coli strains in this study. A comparison of the draft and complete ETEC genome sequences demonstrated that regions not inundated with IS elements are stable and exhibit conserved synteny with other E. coli strains; however, it must be acknowledged that ETEC appears to be a pathovar in genetic flux.
EHEC and EPEC.
EHEC and EPEC both cause attaching and effacing lesions in the gastrointestinal tract; the former has a tropism for the colon, while the latter resides in the small intestine (29). In both pathovars virulence is driven mainly by the genes in the LEE (40). The LEE is composed of ∼41 genes, which are distributed in five distinct transcriptional units designated LEE1 to LEE5 (see Fig. S4 in the supplemental material). The gene contents at this locus are almost identical in these two pathovars, and the majority of the diversity is in LEE5, specifically in the genes encoding the Tir (translocated intimin receptor) and intimin proteins, which function as a receptor-ligand pair. LEE1 to LEE4 contain the structural and regulatory genes involved in the synthesis of a TTSS which secretes Tir into eukaryotic cells. Tir then inserts into the eukaryotic membrane and acts as the receptor for intimin, which then mediates the “intimate attachment” (31). Since this locus is highly conserved in these pathovars, we expected and found that all the EHEC and EPEC isolates contained the LEE genes (see Fig. S4 in the supplemental material).
The number of unique genes found in each of the EHEC isolates analyzed mainly reflects a difference in the annotation strategies that were employed for them (20, 46). For example, the Sakai genome contains approximately 10 times more unique genes than the EDL933 genome (Table 2); however, many of the genes are small (<100 nucleotides) and are not present in the EDL933 genome annotation, suggesting that the difference is not biological but rather is a result of the gene-finding algorithm used. In contrast, genes shared by the two EHEC isolates in this study exhibited the greatest level of genomic similarity with the greatest number of shared genes, which suggests that there is a common evolutionary history (Table 2) (58, 71). Interestingly, ∼43% of the genes are prophage or phage-related genes (see Table S1 in the supplemental material). The presence of such a high proportion of phage genes confirms the important role that phages have played in the evolution of this pathogen (32, 47).
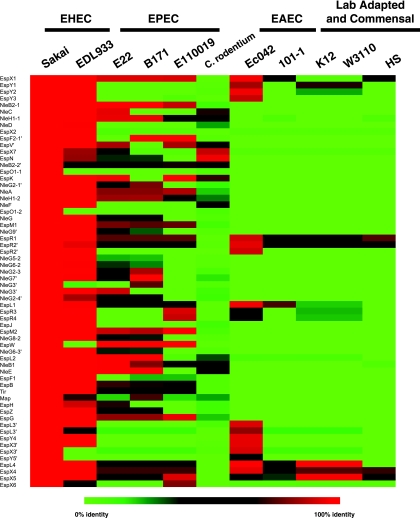
The TTSS utilized by EHEC and EPEC has been shown to secrete a number of effectors not encoded in the LEE (70). A recent study by Tobe et al. (62) identified a number of secreted EHEC effectors, and many of these non-LEE-encoded secreted effectors were associated with phage and prophage. The identification of conserved phage genes in the EHEC isolates (see Table S1 in the supplemental material) with no other functional annotation may suggest that these genes are being maintained in EHEC and in some cases encode secreted effectors required for pathogenesis. Because EHEC, EPEC, and Citrobacter rodentium (a model organism that causes attaching and effacing lesions in mice) have similar TTSS, we wanted to determine if the secreted effectors identified in EHEC were being conserved (9, 62). Our analysis demonstrated that the EHEC, EPEC, and C. rodentium isolates do not exhibit a consistent profile with respect to conservation of the secreted effectors (Fig. 3). Thus, it is not clear if each isolate contains a distinct repertoire of effectors and how this may affect pathogenesis. ExPEC isolates did not contain any of the secreted effectors (not shown), whereas a small but distinct group of effectors were conserved in the commensal and laboratory-adapted isolates. In the commensal and laboratory-adapted strains these effectors may represent distant homologs of the secreted effectors retained in phage and phage remnants. The two EAEC isolates exhibited different profiles for the secreted effectors (Fig. 3), which may reflect the presence or absence of a functional TTSS in these isolates, as suggested by Harrington et al. (18). It appears that there is some conservation of secreted effectors in isolates that maintain a TTSS, but these secreted effectors are not maintained in strains unable to secrete them. A significant number of secreted effectors are present in the other pathogenic E. coli strains that contain a functional TTSS (i.e., EPEC isolates), whereas strains thought to lack the TTSS (uropathogenic E. coli [UPEC], commensal, and laboratory-adapted strains) do not contain as many similar peptides, suggesting that the genes encoding these peptides are not maintained in the genomes when the peptides are not being secreted. Interestingly, while the EAEC strains are thought to contain an active TTSS, they do not seem to contain the same repertoire of secreted effectors.
FIG. 3.
Secreted effector molecules identified by Tobe et al. (62) identified in other E. coli genomes. The BLAST identity is shown as a heat map constructed using the functionally and bioinformatically identified secreted effector molecules from the EHEC isolate E. coli Sakai. Red indicates a higher level of similarity, and green indicates a lower level of similarity.
No complete closed EPEC genome is available; however, the sequence of isolate E2348/69 should be released in the near future (http://www.sanger.ac.uk/Projects). Analysis of the sequences in this study revealed that the draft genomes of the EPEC isolates contained more than 200 unique genes compared to the current data set (Table 2). In contrast to EHEC, relatively few pathovar-specific genes were identified in the EPEC isolates (Table 2), suggesting that the EPEC isolates, while having similar clinical definitions, harbor diverse molecular mechanisms. Many of the pathogenesis-related genes (i.e., the LEE region) are shared with EHEC; however, the EHEC pathovar appears to be younger, either from an evolutionary standpoint or in terms of clinical identification (68).
Analysis of unique genes in each of the two pathovars revealed that while the pathogenesis is similar for the two groups, their evolutionary paths may not be similar (Table 2). EHEC isolates appear to be evolving along a similar evolutionary path, as indicated by the greatest number of group-specific genes and a relatively low number of unique genes in each genome, when annotation strategy is accounted for. In contrast, EPEC may be utilizing multiple molecular mechanisms to achieve similar pathogenic outcomes, as the numbers of group-specific genes are among the lowest in genomes and yet the numbers of unique genes are relatively high. Alternatively, it is possible that the EHEC isolates are defined by very restrictive criteria, whereas the EPEC criteria are encompassing and thus the diversity observed is a result of the diffuse classification of this group of pathogens.
EAEC.
Comparison of the EAEC genomes revealed a pattern similar to that observed with EPEC; there are a significant number of unique genes in each genome and very few conserved pathovar-specific genes (Table 2). This was expected as Ec042 is the EAEC type strain and causes disease in the majority of volunteers (41), whereas isolate 101-1 is labeled “atypical” as it does not possess the pAA plasmids or the aggregation adherence fimbriae that are used to type EAEC isolates (24). A number of toxins have been implicated in the pathogenesis of EAEC, but most of these toxins have homologs in other pathogenic E. coli strains (18). We identified a 25-kb region of the 101-1 genome sequence that was not present in Ec042 but contained a bundle-forming pilus-like locus similar to that identified by Tobe et al. in the EAF plasmid from B171 (63) (see Fig. S5 in the supplemental material). Western blots of whole-cell lysates grown under various medium conditions and probed with anti-bundle-forming pilus antibodies did not detect an antigenically similar pilus (D. A. Rasko, unpublished data). This is not surprising considering the low level of conservation between the EPEC and EAEC gene clusters and the lack of any similarity between the fimbrial subunits. The novel fimbriae may act as the initial adhesin in the absence of the aggregation adherence fimbriae; however, this hypothesis should be functionally examined. A region adjacent to the novel fimbriae contains the AatBA gene cluster (see Fig. S5 in the supplemental material), which encodes gene products responsible for the secretion of dispersin, a small peptide required for transit of EAEC across the epithelial layer during infection (42). While there are intriguing hints and a large number of unique genes to functionally characterize, it is not immediately obvious from the genome data why the 101-1 isolate caused an outbreak.
E. coli conserved core genome.
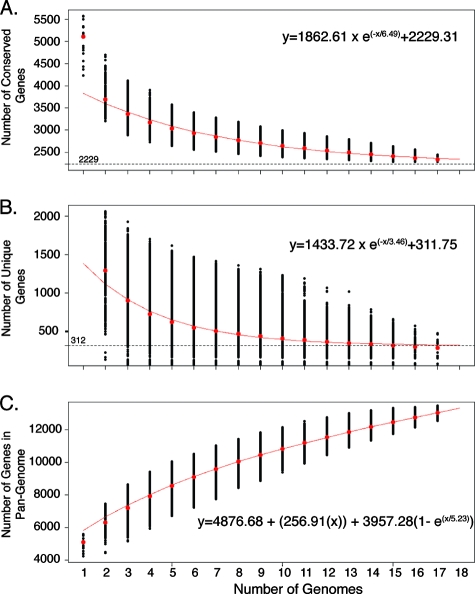
Comparative genome analysis performed with the 17 genomes provided a glimpse of the genetic diversity within the species E. coli. Examination of the number of genes in each of the 17 E. coli genomes revealed that the isolates have a genome size of 5,020 ± 446 genes (mean ± standard deviation). Using the BSR analyses (50), we calculated that the “conserved core” genome size (the genes that are highly conserved in all 17 isolates) is 2,344 ± 43 genes (mean ± standard deviation) (Table 2). The exponential decay model shown in Fig. 4A suggests that the number of conserved core genes is approaching an asymptote with the comparison of 17 genomes. The model is based on the median number of conserved genes in each of the permutations of genome comparisons and predicts that the number of core genes in E. coli is approximately 2,200 genes (Fig. 4A). Examination of the functional annotation of these genes suggests that the conserved core genes encode mostly core metabolic processes. Previous comparative genome microarray hybridization-based analysis using 22 isolates with an incomplete array data set indicated that the “conserved core” of the E. coli genome included approximately 2,800 genes (14). Additionally, Chen et al. used bioinformatics methods to determine that the core genome size of E. coli was 2,865 genes (6). This number was derived from a data set consisting of the genomic contents of seven isolates, two of which were Shigella flexneri isolates (6). While the core genome values obtained in previous studies correlate well with our current model, the smallest number of genes in the conserved core was identified by the current analysis. It must also be noted that in the current study also the most stringent threshold for inclusion was used.
FIG. 4.
Conserved core, unique, and pangenome calculations for E. coli. (A) Each point indicates the number of genes that are conserved in genomes. The red line shows the exponential decay model based on the median value for conserved genes when increasing numbers of genomes were compared. (B) Decreasing number of unique genes in a genome with increasing number of genomes compared. The red line shows the exponential decay model based on the median value for unique genes when increasing numbers of genomes were compared. (C) Pangenome of the species E. coli. The extrapolated curve continues to increase, and thus E. coli has an open pangenome.
Unique gene identification.
The pangenome analysis also identified truly unique genes (TUG), i.e., the genes present only in the reference genome and not in any of the other genomes (Table 2). The number of TUG ranges from ∼20 in the laboratory-adapted isolates to more than 300 depending on the genome used as the reference. The mean number of TUG found in our E. coli genome data set is 176 ± 95. The large deviation from the mean is indicative of the high degree of variation within E. coli, as well as the fact that not all isolates have similar clinical presentations. We also applied the exponential decay model to the identification of unique genes using the median value (Fig. 4B). Application of this model resulted in a value of ∼300 TUG per new genome sequenced and more accurately represented the data set. The use of the median for the exponential decay model minimizes the effect of comparing the genomes of isolates with similar origins. As expected, the majority of the TUG do not have a functional annotation, and thus they may represent novel biosynthetic or pathogenic features.
Two isolates, CFT073 and Ec042, contain the greatest number of unique genes (308 TUG) (Table 2). This finding was anticipated, as UPEC (ExPEC) isolates have long been known to have greater genetic diversity, mostly due to multiple pathogenicity and/or genomic islands (11, 38, 67). The identification of EAEC as a group with significant diversity was unexpected. Our understanding of the genetic diversity within the EAEC group is limited. Most typical EAEC isolates are known to contain a large virulence plasmid (18, 23). However, the large number of TUG in each EAEC isolate in the present study (308 TUG in isolate Ec042 and 155 TUG in isolate 101-1) suggests that chromosomal differences may also play a role in pathogenesis. Identification of the unique genes in EAEC may provide targets for further functional studies on the pathogenesis of this emerging group of pathogens. The unique genes identified in each isolate may not always represent the virulence factors of the group, as these genes could be shared with isolates having a similar clinical origin.
Pathovar shared gene content.
The current data set for 17 E. coli genomes contains at least one genome sequence from each of the E. coli pathovars identified, providing a unique opportunity to identify pathovar-specific genes (Table 2). Surprisingly, there are fewer pathovar-specific genes (genes conserved within pathovar isolates that are not found in any other isolate) than we anticipated. Only the EHEC genomes contain a significant proportion of pathovar-specific genes, more than 120 genes (see Table S1 in the supplemental material). Of the genes that are unique to EHEC, 43% are associated with prophage and phage elements, confirming that phages have played a significant role in the evolution of this pathovar (58, 69, 71). The phage regions may carry genes encoding as-yet-unidentified toxins or virulence factors. In contrast, 11 or fewer pathovar-specific genes could be identified in the isolates belonging to the commensal or laboratory-adapted ETEC, EPEC, and EAEC groups. This suggests that while these isolates are grouped as members of pathovars, there is significant genetic heterogeneity within the groups, which is most likely a reflection on how these isolates cause disease.
The ExPEC genomes share a significant level of similarity, suggesting that outside the gastrointestinal tract E. coli utilizes common molecular mechanisms. One exception to this observation is the avian E. coli isolate, which shares three genes with the other ExPEC isolates (27, 28). This indicates that the avian pathogenic E. coli strain has a distinct genetic repertoire for colonization and infection of avian species that is not required for infection of humans. In general, these comparisons suggest that E. coli pathovars are not distinct on the molecular level, with the exception of EHEC and (to a lesser extent) ExPEC, or that E. coli, as a pathogen, is a “generalist” having the genetic potential to colonize and infect humans. Conversely, the genomic data set has outliers such as EAEC isolate 101-1 and EPEC isolate E110019, both of which are considered “atypical.” Thus, identification of pathovar-specific genes using an atypical isolate and a typical isolate may represent a biased comparison. More typical and atypical isolates need to be examined on the molecular level to accurately determine the genomic boundaries of the pathovar designations.
Pangenome of E. coli.
In contrast to previous pangenome studies, which examined closely related clinical groups of Streptococcus and found a limited number of novel unique genes in each genome (60), we observed a much broader range for the number of unique genes when we examined a group of clinically unrelated isolates. The E. coli isolates included in this study represent a broad sample of the diverse pathogens that comprise this species and are not a narrowly defined group of clinical isolates. We used a methodology similar to that of Tettelin et al. for determination of the pangenome (60). The trend for the data is continual addition of new genes with each newly sequenced genome (Fig. 4C), and thus, the pangenome of E. coli is considered open. An open species pangenome indicates that the species is still evolving by gene acquisition and diversification. Our calculations suggest that the E. coli pangenome has a reservoir consisting of more than 13,000 genes. This number has tremendous implications in terms of the diversity and pathogenesis of the species E. coli and its ability to colonize and cause disease in the human host.
In this study we analyzed the genome contents of 17 E. coli isolates, including 8 new isolates which represent three pathovars that had not been sequenced previously in addition to a true commensal, E. coli HS. This study identified novel genome features and potentially novel pathogenic mechanisms by using comparative genomics. Approximately one-half of the genome content of any E. coli isolate represents the “core conserved” genome. The open pangenome of the species E. coli indicates that continued sequencing should result in identification of ∼300 novel genes per genome. The complete sequence of a commensal, E. coli HS, revealed significant genome mosaicism between a pathogen and a commensal, as features previously thought to be pathogen associated or restricted to pathogens were identified in the commensal genome. This has significant implications for the evolution of pathogenic E. coli, in that members of the commensal microflora may act as genetic sinks and the interaction of “precursor” pathogen species with commensals may allow development of fully pathogenic isolates via horizontal gene transfer. The E. coli HS genome represents the new reference sequence for pathogen comparison, and data for this genome will be the basis for functional work for years to come.
Supplementary Material
Acknowledgments
This work was supported by federal funds from the National Institute of Allergy and Infectious Diseases (contract no. N01-AI-30071).
The data for each genome can be obtained by contacting the corresponding author. The sequence data for the E. coli 042 and C. rodentium genomes were produced by the Pathogen Genome Sequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens. Additional thanks to investigators who provided isolates for sequencing: Carl Brinkley, Edward Boedeker, James B. Kaper, Harry L. T. Mobley, James P. Nataro, and Stephen J. Savarino.
Footnotes
Published ahead of print on 1 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 2.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 1815967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caron, E., V. F. Crepin, N. Simpson, S. Knutton, J. Garmendia, and G. Frankel. 2006. Subversion of actin dynamics by EPEC and EHEC. Curr. Opin. Microbiol. 940-45. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter, C. M., E. R. Hall, R. Randall, R. McKenzie, F. Cassels, N. Diaz, N. Thomas, P. Bedford, M. Darsley, C. Gewert, C. Howard, R. B. Sack, D. A. Sack, H. S. Chang, G. Gomes, and A. L. Bourgeois. 2006. Comparison of the antibody in lymphocyte supernatant (ALS) and ELISPOT assays for detection of mucosal immune responses to antigens of enterotoxigenic Escherichia coli in challenged and vaccinated volunteers. Vaccine 243709-3718. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H. D., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 2983-98. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 1035977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheney, C. P., P. A. Schad, S. B. Formal, and E. C. Boedeker. 1980. Species specificity of in vitro Escherichia coli adherence to host intestinal cell membranes and its correlation with in vivo colonization and infectivity. Infect. Immun. 281019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 672692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 696323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Departments of State and Local Health, E. coli O157:H7 Investigation Team, and Centers for Disease Control and Prevention. 2006. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb. Mortal. Wkly. Rep. 551045-1046. [PubMed] [Google Scholar]
- 11.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 1851831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escobar-Paramo, P., C. Giudicelli, C. Parsot, and E. Denamur. 2003. The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J. Mol. Evol. 57140-148. [DOI] [PubMed] [Google Scholar]
- 13.Froehlich, B., J. Parkhill, M. Sanders, M. A. Quail, and J. R. Scott. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J. Bacteriol. 1876509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukiya, S., H. Mizoguchi, T. Tobe, and H. Mori. 2004. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 1863911-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge, Z., and D. E. Taylor. 1992. H. pylori DNA transformation by natural competence and electroporation, p. 145-152. In C. L. Clayton and H. L. T. Mobley (ed.), Helicobacter pylori protocols. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- 16.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 3121355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacking, A. J., J. Aguilar, and E. C. Lin. 1978. Evolution of propanediol utilization in Escherichia coli: mutant with improved substrate-scavenging power. J. Bacteriol. 136522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington, S. M., E. G. Dudley, and J. P. Nataro. 2006. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 25412-18. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi, K., N. Morooka, Y. Yamamoto, K. Fujita, K. Isono, S. Choi, E. Ohtsubo, T. Baba, B. L. Wanner, H. Mori, and T. Horiuchi. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 22006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 811-22. [DOI] [PubMed] [Google Scholar]
- 21.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochhut, B., C. Wilde, G. Balling, B. Middendorf, U. Dobrindt, E. Brzuszkiewicz, G. Gottschalk, E. Carniel, and J. Hacker. 2006. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol. Microbiol. 61584-595. [DOI] [PubMed] [Google Scholar]
- 23.Huang, D. B., A. Mohanty, H. L. DuPont, P. C. Okhuysen, and T. Chiang. 2006. A review of an emerging enteric pathogen: enteroaggregative Escherichia coli. J. Med. Microbiol. 551303-1311. [DOI] [PubMed] [Google Scholar]
- 24.Itoh, Y., I. Nagano, M. Kunishima, and T. Ezaki. 1997. Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J. Clin. Microbiol. 352546-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeter, R. M. 1990. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J. Gen. Microbiol. 136887-896. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D. E., C. V. Lockatell, R. G. Russell, J. R. Hebel, M. D. Island, A. Stapleton, W. E. Stamm, and J. W. Warren. 1998. Comparison of Escherichia coli strains recovered from human cystitis and pyelonephritis infections in transurethrally challenged mice. Infect. Immun. 663059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 1893228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, T. J., K. E. Siek, S. J. Johnson, and L. K. Nolan. 2005. DNA sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid. Antimicrob. Agents Chemother. 494681-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 30.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295405-418. [DOI] [PubMed] [Google Scholar]
- 31.Kenny, B., and B. B. Finlay. 1995. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc. Natl. Acad. Sci. USA 927991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotewicz, M. L., S. A. Jackson, J. E. LeClerc, and T. A. Cebula. 2007. Optical maps distinguish individual strains of Escherichia coli O157:H7. Microbiology 1531720-1733. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence, J. G., and J. R. Roth. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 14211-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i1119-1122. [DOI] [PubMed] [Google Scholar]
- 35.Levine, M. M., E. S. Caplan, D. Waterman, R. A. Cash, R. B. Hornick, and M. J. Snyder. 1977. Diarrhea caused by Escherichia coli that produce only heat-stable enterotoxin. Infect. Immun. 1778-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, M. M., D. R. Nalin, D. L. Hoover, E. J. Bergquist, R. B. Hornick, and C. R. Young. 1979. Immunity to enterotoxigenic Escherichia coli. Infect. Immun. 23729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine, M. M., and M. B. Rennels. 1978. E. coli colonization factor antigen in diarrhoea. Lancet ii534. [DOI] [PubMed] [Google Scholar]
- 38.Lloyd, A. L., D. A. Rasko, and H. L. Mobley. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 1893532-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makino, S., T. Tobe, H. Asakura, M. Watarai, T. Ikeda, K. Takeshi, and C. Sasakawa. 2003. Distribution of the secondary type III secretion system locus found in enterohemorrhagic Escherichia coli O157:H7 isolates among Shiga toxin-producing E. coli strains. J. Clin. Microbiol. 412341-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 921664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nataro, J. P., Y. Deng, S. Cookson, A. Cravioto, S. J. Savarino, L. D. Guers, M. M. Levine, and C. O. Tacket. 1995. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J. Infect. Dis. 171465-468. [DOI] [PubMed] [Google Scholar]
- 42.Nishi, J., J. Sheikh, K. Mizuguchi, B. Luisi, V. Burland, A. Boutin, D. J. Rose, F. R. Blattner, and J. P. Nataro. 2003. The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J. Biol. Chem. 27845680-45689. [DOI] [PubMed] [Google Scholar]
- 43.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6519-527. [DOI] [PubMed] [Google Scholar]
- 44.Parsot, C. 2005. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 25211-18. [DOI] [PubMed] [Google Scholar]
- 45.Patel, S. K., J. Dotson, K. P. Allen, and J. M. Fleckenstein. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 721786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 47.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 1811767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 9710567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasko, D. A., G. S. Myers, and J. Ravel. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 42381-86. [DOI] [PubMed] [Google Scholar]
- 52.Reigstad, C. S., S. J. Hultgren, and J. I. Gordon. 2007. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 28221259-21267. [DOI] [PubMed] [Google Scholar]
- 53.Riley, M., T. Abe, M. B. Arnaud, M. K. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett III, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo, T. A., and J. R. Johnson. 2006. Extraintestinal isolates of Escherichia coli: identification and prospects for vaccine development. Expert Rev. Vaccines 545-54. [DOI] [PubMed] [Google Scholar]
- 55.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, J. L., P. M. Fratamico, and N. W. Gunther. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4134-163. [DOI] [PubMed] [Google Scholar]
- 57.Stapleton, A., S. Moseley, and W. E. Stamm. 1991. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J. Infect. Dis. 163773-779. [DOI] [PubMed] [Google Scholar]
- 58.Steele, M., K. Ziebell, Y. Zhang, A. Benson, P. Konczy, R. Johnson, and V. Gannon. 2007. Identification of Escherichia coli O157:H7 genomic regions conserved in strains with a genotype associated with human infection. Appl. Environ. Microbiol. 7322-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tacket, C. O., G. Losonsky, S. Livio, R. Edelman, J. Crabb, and D. Freedman. 1999. Lack of prophylactic efficacy of an enteric-coated bovine hyperimmune milk product against enterotoxigenic Escherichia coli challenge administered during a standard meal. J. Infect. Dis. 1802056-2059. [DOI] [PubMed] [Google Scholar]
- 60.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 10213950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 62.Tobe, T., S. A. Beatson, H. Taniguchi, H. Abe, C. M. Bailey, A. Fivian, R. Younis, S. Matthews, O. Marches, G. Frankel, T. Hayashi, and M. J. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 10314941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tobe, T., T. Hayashi, C. G. Han, G. K. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 675455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsang, A. W., A. R. Horswill, and J. C. Escalante-Semerena. 1998. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J. Bacteriol. 1806511-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner, S. M., A. Scott-Tucker, L. M. Cooper, and I. R. Henderson. 2006. Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 26310-20. [DOI] [PubMed] [Google Scholar]
- 66.Viljanen, M. K., T. Peltola, S. Y. Junnila, L. Olkkonen, H. Jarvinen, M. Kuistila, and P. Huovinen. 1990. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet 336831-834. [DOI] [PubMed] [Google Scholar]
- 67.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 611619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 1871783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaharik, M. L., S. Gruenheid, A. J. Perrin, and B. B. Finlay. 2002. Delivery of dangerous goods: type III secretion in enteric pathogens. Int. J. Med. Microbiol. 291593-603. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, Y., C. Laing, M. Steele, K. Ziebell, R. Johnson, A. K. Benson, E. Taboada, and V. P. Gannon. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.