Abstract
Poxviruses are notorious for encoding multiple proteins that regulate cellular signaling pathways, including the ubiquitin-proteasome system. Bioinformatics indicated that ectromelia virus, the causative agent of lethal mousepox, encoded four proteins, EVM002, EVM005, EVM154, and EVM165, containing putative F-box domains. In contrast to cellular F-box proteins, the ectromelia virus proteins contain C-terminal F-box domains in conjunction with N-terminal ankyrin repeats, a combination that has not been previously reported for cellular proteins. These observations suggested that the ectromelia virus F-box proteins interact with SCF (Skp1, cullin-1, and F-box) ubiquitin ligases. We focused our studies on EVM005, since this protein had only one ortholog in cowpox virus. Using mass spectrometry, we identified cullin-1 as a binding partner for EVM005, and this interaction was confirmed by overexpression of hemagglutinin (HA)-cullin-1. During infection, Flag-EVM005 and HA-cullin-1 colocalized to distinct cellular bodies. Significantly, EVM005 coprecipitated with endogenous Skp1, cullin-1, and Roc1 and associated with conjugated ubiquitin, suggesting that EVM005 interacted with the components of a functional ubiquitin ligase. Interaction of EVM005 with cullin-1 and Skp1 was abolished upon deletion of the F-box, indicating that the F-box played a crucial role in interaction with the SCF complex. Additionally, EVM002 and EVM154 interacted with Skp1 and conjugated ubiquitin, suggesting that ectromelia virus encodes multiple F-box-containing proteins that regulate the SCF complex. Our results indicate that ectromelia virus has evolved multiple proteins that interact with the SCF complex.
Ubiquitin is a 76-amino-acid protein that plays a crucial role in protein regulation (25, 57). The covalent attachment of ubiquitin to target substrates results in protein degradation or dramatic alterations in protein function (25, 57). The process of ubiquitination is essential for cellular homeostasis and tightly regulates a wide range of cellular functions, such as the cell cycle, signal transduction, transcription, and DNA repair (25, 46, 57).
A large family of ubiquitin ligases is responsible for transferring ubiquitin to specific substrates. Ubiquitin ligases exist either as single subunit ligases or as part of a multisubunit ubiquitin ligase complex (3). Multisubunit ubiquitin ligases are composed of a scaffold protein, a linker protein, and a substrate adaptor protein that is responsible for recruiting substrates to the complex for ubiquitination (45, 59). Multiprotein ubiquitin ligases incorporate a member of the cullin family to act as the molecular scaffold for the ubiquitin ligase and support the recruitment of substrates to the complex. Seven cullin family members have been identified, and each contains a cullin homology domain at the C terminus that binds Roc1, which is responsible for conferring the ubiquitin ligase activity (45). Substrate adaptor proteins contain protein-protein interaction domains that recruit the substrate to the complex and bind either directly to the cullin protein or through a linker protein (45, 59).
The cellular SCF (Skp1, cullin-1, and F-box) complex is the best characterized of the multiprotein ubiquitin ligases and regulates a wide range of cellular processes (4, 32, 38, 40, 41). Substrate adaptors are recruited to the SCF complex through interactions with the linker protein Skp1 (14, 37, 63). Substrate recruitment relies upon a highly conserved F-box domain that is typically found at the N terminus of substrate adaptor proteins and is required for interaction with the linker protein Skp1 (15, 29, 53, 61). The majority of cellular F-box proteins also possess C-terminal leucine-rich repeats or WD40 repeats that are responsible for recognizing and recruiting substrates (15, 29, 53, 61). Over 70 genes encoding F-box proteins exist in the human genome, indicating that a wide range of substrates are recruited to the SCF complex for ubiquitination (14, 15, 22, 29, 33, 53, 61).
Recently, it has become apparent that many viruses exploit the ubiquitination machinery, including human immunodeficiency virus (9, 10, 19, 48, 62), adenoviruses (17, 47), herpesviruses (11, 12), and poxviruses (7, 31, 36, 42). The Poxviridae are a large family of DNA viruses that encode an array of proteins that interfere with cellular signaling pathways (30, 50). Several proteins encoded by strain Moscow of ectromelia virus, the causative agent of mousepox, have been shown to specifically regulate the ubiquitin-proteasome system. These include p28, a RING domain-containing protein that functions as a ubiquitin ligase (28, 42), and EVM150 and EVM167, which contain bric-a-brac, tramtrack, broad complex (BTB) and kelch domains and interact with cullin-3, likely regulating ubiquitination of currently unknown substrates (60). More recently, it has been suggested that poxviruses also regulate the SCF complex through the expression of multiple proteins that contain N-terminal ankyrin repeat domains and putative C-terminal F-box domains (36, 54). Intriguingly, this combination is unique to poxviruses and to date has not been found within cellular proteins. Using bioinformatics, we identified seven ectromelia virus genes predicted to encode ankyrin repeats: the EVM002, EVM005, EVM010, EVM021, EVM022, EVM154, and EVM165 genes. Of the products of these seven genes, four contain putative C-terminal F-box domains. Here, we report that EVM005 contains a C-terminal F-box domain that is necessary for interaction with components of the SCF complex. Significantly, EVM002 and EVM154, which contain putative F-box domains, also interact with the SCF complex component Skp1. These results suggest that ectromelia virus has evolved multiple proteins that function to modulate the activity of SCF complex ubiquitin ligases upon infection.
MATERIALS AND METHODS
Cell culture and viruses.
CV-1, HeLa, and HEK293T cells were obtained from the American Type Culture Collection and cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 50 U/ml of penicillin, 50 μg/ml of streptomycin, and 200 μM glutamine (Invitrogen Corporation). Baby green monkey kidney cells were cultured in DMEM supplemented with 10% newborn calf serum, 50 U/ml of penicillin, 50 μg/ml of streptomycin, and 200 μM glutamine. HuTk−143B cells were cultured in DMEM supplemented with 10% fetal bovine serum, 50 U/ml of penicillin, 50 μg/ml of streptomycin, 200 μM glutamine, and 25 μg/ml bromodeoxyuridine. Vaccinia virus (VV) strain Copenhagen was propagated in baby green monkey kidney cells and harvested as previously described (55). The constructions of VV-Flag-EVM004 (60), VV-Flag-EVM150 (60), and VV-Flag-FPV039 (5) have been previously described.
Alignments.
Protein alignments for the ectromelia virus proteins were created with the AlignX program (Invitrogen Corporation). Skp2, a known cellular F-box-containing protein, was included in the alignments (48). Predicted secondary structures included in the alignment were previously described for the Skp1/Skp2 interface (48).
Plasmid constructs.
pcDNA3 plasmids with hemagglutinin (HA)-cullin-1, HA-cullin-1 with amino acids 610 to 615 deleted (cullin-1Δ610-615), HA-cullin-1ΔN53, Myc-cullin-1, and T7-Skp1 were previously described (21, 37). The genes encoding HA-cullin-1, HA-cullin-1Δ610-615, and HA-cullin-1ΔN53 were amplified by PCR using the primers 5′-(NotI)GCGGCCGCAATACGACTCACTATAGGGA-3′ (forward) and 5′-(XmaI)CCCGGGTTAAGCCAAGTAACTGTAGGTGTC-3′ (reverse). PCR products were subcloned into pGEM-T (Promega) and subsequently subcloned into pSC66 via NotI and XmaI (New England Biolabs). The EVM005 gene was amplified from ectromelia virus DNA and Flag tagged by PCR using the primers 5′-(SalI)GTCGACATGGACTACAAAGACGATGACGACAAGGAAAGATATTCATTACATA-3′ (forward) and 5′-(NotI)GCGGCCGCTCATTCATGTGTCTGTGTTTGGAC-3′ (reverse). The EVM002 gene was amplified from ectromelia virus DNA and Flag tagged by using the primers 5′-(KpnI)GGTACCAT GGACTACAAAGACGATGACGACAAGGGCGAGATGGACGAGATT-3′ (forward) and 5′-(NheI)GCTAGCTTATGAATA ATATTTGTA-3′ (reverse). The EVM154 gene was amplified from ectromelia virus DNA and Flag tagged by using the primers 5′-(SalI)GTCGACATGGACTACAAAGACGATGACGACAAGGATTTTTTTAAAAAGGAA-3′ (forward) and 5′-(NotI)GCGGCCGCTTATATTTTAATAGTGTT-3′ (reverse). The Flag-EVM005, Flag-EVM002, and Flag-EVM154 PCR products were cloned into pGemT and subcloned into pSC66 by digestion with either NotI and SalI for EVM005 and EVM154 or KpnI and NheI for EVM002 (Invitrogen Corporation). The EVM005 gene encoding amino acids 1 to 380 [the EVM005(1-380) gene] (Flag tagged) was amplified from pSC66-Flag-EVM005 DNA by using the forward primer previously described for EVM005 and the reverse primer 5′-(NotI)GCGGCCGCTCAATTATTATAAGTTCGTAACGT-3′. The gene encoding EVM005(381-650) (Flag tagged) was also amplified from pSC66-Flag-EVM005 DNA with the previously described reverse primer and the Flag-tagged forward primer 5′-(SalI)GTCGACATGGACTACAAAGACGATGACGACAAGAAAACTATTTTCTATTTGGA. Both Flag-EVM005(1-380) and Flag-EVM005(381-650) PCR products were cloned into pGEM-T and subcloned into pSC66 by digestion with NotI and SalI (Invitrogen Corporation). For transient expression of Flag-EVM005, the EVM005 gene was codon optimized (GeneArt) and subcloned into pcDNA3 for expression in HEK293T cells. The gene encoding EVM005(1-593) (Flag tagged) was amplified by PCR using the primers 5′-(BamHI)GGATCCATGGACTACAAAGACGATGACGACAAGGAGCGGTACAGCCTGCAC-3′ (forward) and 5′-(NotI)-GCGGCCGCTCACAGGTAGGTGGGCTGGTC-3′ (reverse). The Flag-EVM005(1-593) PCR product was cloned into pGEM-T and subcloned into pcDNA3 by digestion with BamHI and NotI.
Generation of recombinant VVs.
VV-Flag-EVM005, VV-Flag-EVM005(1-380), VV-Flag-EVM005(381-650), VV-Flag-EVM002, and VV-Flag-EVM154 were created by infecting CV-1 cells with VV Copenhagen at a multiplicity of infection (MOI) of 0.05 and transfecting 5 μg of either pSC66-Flag-EVM005, pSC66-Flag-EVM005(1-380), or pSC66-Flag-EVM005(381-650) by using Lipofectin reagent (Invitrogen Corporation). The VV Copenhagen recombinants were selected by growth on HuTk−143B cells in the presence of 25 μg/ml bromodeoxyuridine, and recombinant plaques were purified using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Rose Scientific, Ltd.) (18).
Antibodies.
Mouse anti-Flag (M2) was purchased from Sigma-Aldrich. Mouse anti-HA (clone 12CA5) was purchased from Roche Applied Science. Mouse anti-T7 was purchased from Novagen. Mouse anti-Myc (clone 9E10) was a gift from T. Hobman, University of Alberta. Antiubiquitin (clone FK2) was purchased from Biomol International (20). Antibodies specific for cullin-1, Skp1, and Roc1 were previously described (37, 44). An antibody specific to I5L was generated by immunizing rabbits with 500 μg of peptide (TYVKSLLMKS) conjugated to KLH as previously described (8).
Immunoprecipitations and Western blot analysis.
HEK293T cells were infected with VV Copenhagen or recombinant VV expressing Flag-EVM005, Flag-EVM005(1-380), Flag-EVM005(381-650), Flag-EVM004, Flag-EVM150, Flag-FPV039, Flag-EVM002, or Flag-EVM154 at an MOI of 5 for 12 h. All infected HEK293T cells were lysed in buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris (pH 8.0), and Complete mini-protease inhibitors (Roche Diagnostics). Following lysis, samples were incubated with anti-Flag (M2) and protein G-Sepharose beads (GE Healthcare).
To express HA-cullin-1, HA-cullin-1Δ610-615, and HA-cullin-1ΔN53 during infection, HEK293T cells were transfected with the corresponding pSC66 constructs and infected with VV Copenhagen or recombinant VV expressing either Flag-EVM005 or Flag-EVM004. HEK293T cells were infected at an MOI of 5 and transfected with 2 μg of the cullin-1 constructs by using the Lipofectamine 2000 reagent (Invitrogen Corporation). Infected 293T cells were harvested with NP-40 lysis buffer and subjected to immunoprecipitation at 12 h postinfection. Cell lysates were incubated with anti-HA (clone 12CA5) and protein G beads (GE Healthcare).
To transiently express T7-Skp1, Myc-cullin-1, Flag-EVM005, and Flag-EVM005(1-593), HEK293T cells were transfected with 3 μg of pcDNA3-T7-Skp1 or pcDNA3-Myc-cullin-1 and 2 μg of pcDNA3-Flag-EVM005 or pcDNA3-Flag-EVM005(1-593). Cells were harvested at 18 h posttransfection, lysed with 1% NP-40 lysis buffer, and incubated with either anti-T7 or anti-Myc (clone 9E10) and protein G beads (GE Healthcare).
Protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by transfer to nitrocellulose membrane (Fisher Scientific). To detect conjugated ubiquitin, membranes were blocked overnight in 1% bovine serum albumin (Roche Diagnostics) containing 50 mM Tris (pH 7.6) and 0.1% Tween 20. All other membranes were blocked overnight in 5% skim milk containing 50 mM Tris (pH 7.6) and 0.1% Tween 20. Anti-HA (clone 12CA5), anti-Myc (9E10), and anti-FK2 were used at a dilution of 1:2,000, and anti-Flag (M2), anti-T7, anti-cullin-1, anti-Skp1, anti-Roc1, and anti-I5L were all used at a dilution of 1:5,000. Membranes were treated with anti-mouse antibody conjugated to horseradish peroxidase (Bio-Rad or Jackson Laboratories) or anti-rabbit antibody-horseradish peroxidase (Jackson Laboratories), and protein bands were visualized using enhanced chemiluminescence (GE Healthcare).
Mass spectrometry.
HEK293T cells (2 × 107) were infected with VV Copenhagen or VV-Flag-EVM005 at an MOI of 5 for 12 h. Following infection, cells were harvested and subjected to immunoprecipitation using the anti-Flag (M2) antibody. Samples were separated by SDS-PAGE, and proteins were visualized by silver staining (51). Bands were excised, digested with trypsin, and analyzed by mass spectrometry at the Institute for Biomolecular Design, University of Alberta, by using a Fourier transform ion cyclotron resonance Apex Q mass spectrometer (Bruker).
Confocal microscopy.
HeLa cells (2 × 105) were plated on coverslips and infected with either VV Copenhagen or a recombinant VV expressing Flag-EVM005 or Flag-EVM004 at an MOI of 5 for 12 h. For expression of HA-cullin-1, infected HeLa cells were transfected with 2 μg of pSC66-HA-cullin-1. Alternatively, HeLa cells were transiently transfected with pcDNA3-HA-cullin-1 or pcDNA3-Flag-EVM005. Following transfection or infection, cells were fixed with 2% paraformaldehyde, permeabilized using 1% NP-40 (Sigma Aldrich), and blocked with 30% goat serum (Invitrogen Corporation). The cells were incubated with antiubiquitin (FK2) at a dilution of 1:200 and stained with anti-mouse conjugated with Alexa Fluor 488 (Invitrogen Corporation) at a dilution of 1:400. The cells were then fixed with 2% paraformaldehyde and further stained with anti-Flag M2 directly conjugated to Cy3 (Sigma-Aldrich) at a dilution of 1:200. Cells that were transfected with HA-cullin-1 were incubated with anti-cullin-1 at a dilution of 1:200 and anti-Flag (M2) at a dilution of 1:200. The secondary antibodies used were anti-rabbit conjugated with Alexa Fluor 546 (Invitrogen Corporation) and anti-mouse conjugated with Alexa Fluor 488, both at a dilution of 1:400. Coverslips were mounted in 50% glycerol containing 4 mg/ml N-propyl-gallate (Sigma-Aldrich) and 250 μg/ml 4′,6-diamino-2-phenylindole (DAPI) (Invitrogen Corporation) for visualization of nuclei and viral factories. Cells were visualized by using a Zeiss Axiovert laser scanning confocal microscope. To quantify the percentage of cells expressing EVM005, EVM002, or EVM154 that colocalized with cullin-1 or conjugated ubiquitin, >100 cells were counted.
RESULTS
Ectromelia virus encodes four ankyrin repeat/F-box proteins.
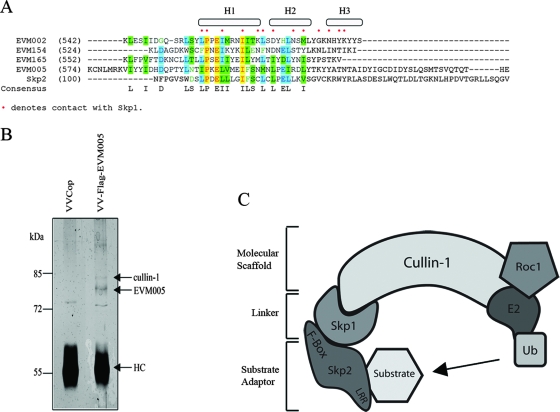
Members of the poxvirus family have recently been shown to modulate the ubiquitin-proteasome system (7, 28, 31, 42, 60). Prompted by these recent findings and the identification of putative F-box domain-containing proteins in Orf virus, a poxvirus that infects sheep (36), we used bioinformatics to identify potential F-box proteins in ectromelia virus strain Moscow. This approach identified seven ectromelia virus ankyrin repeat-containing proteins. Four of these, EVM002, EVM005, EVM154, and EVM165, contained putative C-terminal F-box-like domains. An alignment of EVM002, EVM005, EVM154, and EVM165 with the F-box from the cellular protein Skp2 demonstrated the conservation of key residues. Moreover, many of the contact points between the F-box of Skp2 and the linker protein Skp1 were maintained in the ectromelia virus Moscow F-box domains (Fig. 1A) (48, 63). In contrast to cellular F-box proteins, the ectromelia virus F-boxes were located at the C terminus in combination with a series of N-terminal ankyrin repeats. To date, no cellular F-box proteins have been found in conjunction with ankyrin repeats, suggesting that ectromelia virus has evolved a novel set of genes to regulate the ubiquitin-proteasome pathway. Bioinformatics further indicated that multiple orthologs for EVM002, EVM154, and EVM165 were present in a variety of poxviruses. Notably, bioinformatics indicated that EVM005 had only one ortholog, CPVBR011, in cowpox virus strain Brighton Red, suggesting that EVM005 and CPVBR011 may play a role specific to ectromelia virus and cowpox virus.
FIG. 1.
Ectromelia virus encodes four ankyrin repeat/F-box proteins. (A) AlignX was used to align the C termini of EVM002, EVM005, EVM154, and EVM165 with the F-box domain of Skp2, a cellular F-box protein. The dots indicate known contact points between Skp2 and the linker protein Skp1 (48). H1, H2, and H3 represent helical secondary structures (48). (B) HEK293T cells were infected with VV Copenhagen (VVCop) or VV-Flag-EVM005 at an MOI of 5 for 12 h. Cell lysates were subjected to immunoprecipitation with anti-Flag, and samples were separated by SDS-PAGE and silver stained. Bands were excised and analyzed by mass spectrometry. Peptides that corresponded to cullin-1 and EVM005 were identified. HC, antibody heavy chain. (C) Cullin-1 is a scaffold protein for a cellular SCF ubiquitin (Ub) ligase complex. Roc1, which supplies the ubiquitin ligase activity, binds to the C terminus of cullin-1, and the linker protein Skp1 interacts with the N terminus of cullin-1. Substrates are recruited for ubiquitination through substrate adaptors containing F-box domains. LRR, leucine-rich repeat.
EVM005 interacts with HA-cullin-1 during infection.
The presence of a putative F-box domain in EVM005 suggested that EVM005 interacted with components of the SCF ubiquitin ligase complex. To test this possibility, we generated a Flag-tagged version of EVM005 and constructed a recombinant VV, VV-Flag-EVM005, that expresses EVM005 during infection. To identify cellular binding partners for EVM005, HEK293T cells were infected with wild-type VV Copenhagen or VV-Flag-EVM005, and protein samples were subjected to immunoprecipitation with the anti-Flag antibody followed by mass spectrometry. Mass spectrometry identified both EVM005 and cullin-1 in VV-Flag-EVM005-infected cells, but not in VV Copenhagen-infected cells, by immunoprecipitation (Fig. 1B). The interaction between EVM005 and cullin-1, an essential component of the SCF complex ubiquitin ligase, suggested that EVM005 modulates the activity of the SCF complex in a manner similar to that of cellular F-box proteins (Fig. 1C) (14, 63).
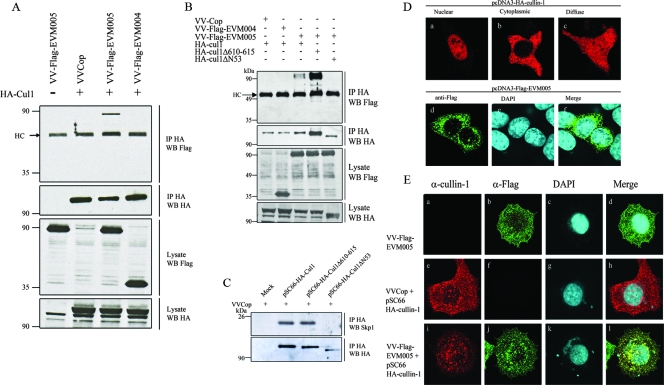
To confirm the mass spectrometry results showing an interaction between Flag-EVM005 and cullin-1, HEK293T cells were infected with VV-Flag-EVM005, VV Copenhagen, or VV-Flag-EVM004, which expresses a Flag-tagged BTB-only protein, as a negative control (60). To express HA-cullin-1 during infection, cells were infected and subsequently transfected with pSC66-HA-cullin-1 to express cullin-1 from a poxvirus-specific promoter (16, 34). Immunoprecipitation with anti-HA clearly demonstrated that overexpressed HA-cullin-1 interacted with Flag-EVM005 during infection but not Flag-EVM004 (Fig. 2A). Western blotting lysates with anti-Flag and anti-HA indicated that HA-cullin-1, Flag-tagged EVM005, and Flag-tagged EVM004 were expressed. To further elucidate the region of cullin-1 necessary for interaction with EVM005, we used two cullin-1 mutant constructs, HA-cullin-1Δ610-615 and HA-cullin-1ΔN53, which are missing the C-terminal Roc1 binding domain and the N-terminal Skp1 binding domain, respectively (Fig. 1C) (21). The mutant with a deletion of amino acids 610 to 615 in cullin-1, which are critical for Roc1 interaction with cullin-1, retained the ability to interact with EVM005, as expected (Fig. 2B). In fact, more EVM005 was immunoprecipitated with HA-cullin-1Δ610-615, due to increased immunoprecipitation of HA-cullin-1Δ610-615 in this particular assay, further supporting the notion of interaction between cullin-1Δ610-615 and EVM005 (Fig. 2B). Deletion of the N-terminal 53 amino acids of cullin-1, which have previously been shown to be necessary for Skp1 interaction, dramatically affected the ability of EVM005 to interact with cullin-1 (Fig. 2B). Additionally, Skp1 failed to interact with HA-cullin-1ΔN53, indicating the possibility that Skp1 acts as a linker between cullin-1 and EVM005 (Fig. 2C).
FIG. 2.
EVM005 interacts with HA-cullin-1 during infection. (A) HEK293T cells were infected with VV Copenhagen (VVCop), VV-Flag-EVM005, or VV-Flag-EVM004 and mock transfected or transfected with pSC66-HA-cullin-1. At 12 h postinfection, cells were lysed in NP-40 lysis buffer, subjected to immunoprecipitation (IP) with anti-HA to precipitate cullin-1 (Cul1), and Western blotted (WB) with anti-Flag for the detection of EVM005. (B) EVM005 fails to interact with HA-cullin-1ΔN53. HEK293T cells were infected with VV Copenhagen, VV-Flag-EVM004, or VV-Flag-EVM005 and transfected with HA-cullin-1, HA-cullin-1Δ610-615, or HA-cullin-1ΔN53. Cellular lysates were immunoprecipitated with anti-HA to precipitate cullin-1 and Western blotted with anti-HA or anti-Flag for the detection of cullin-1 or EVM005, respectively. (C) Endogenous Skp1 fails to interact with HA-cullin-1ΔN53 during infection. HEK293T cells were mock transfected or transfected with pSC66-HA-cullin-1, pSC66-HA-cullin-1Δ610-615, or pSC66-HA-cullin-1ΔN53. Cellular lysates were immunoprecipitated with anti-HA to precipitate cullin-1 and Western blotted with anti-HA or anti-Skp1 to detect cullin-1 or endogenous Skp1, respectively. (D) Localization of ectopically expressed cullin-1 and EVM005. HeLa cells (2 × 105) were transfected with HA-cullin-1 or Flag-EVM005 for 16 h, cullin-1 was visualized by anti-HA (a to c), and EVM005 was visualized with anti-Flag (d to f). (E) Flag-EVM005 colocalizes with HA-cullin-1 during infection. HeLa cells (2 × 105) were infected and transfected with pSC66-HA-cullin-1. Cells were costained with anti-cullin-1, anti-Flag, and DAPI to detect viral factories and the nucleus. (a to d) HeLa cells infected with VV-Flag-EVM005. (e to h) HeLa cells infected with VV Copenhagen and cotransfected with pSC66-HA-cullin-1. (i to l) HeLa cells infected with VV-Flag-EVM005 and cotransfected with pSC66-HA-cullin-1. A total of 77% of infected/transfected cells displayed colocalization between Flag-EVM005 and HA-cullin-1. α, anti.
Since coimmunoprecipitation data indicated that cullin-1 interacted with EVM005, we next investigated the subcellular localization of Flag-EVM005 and cullin-1 during infection. As previously described, ectopic expression of HA-cullin-1 in uninfected HeLa cells produced three different expression patterns, with the most prominent being nuclear accumulation (Fig. 2D, panels a to c) (21). Transient expression of Flag-EVM005 demonstrated a unique punctate staining pattern throughout the cytoplasm of the cell (Fig. 2D, panels d to f). HeLa cells infected with VV-Flag-EVM005 and stained with anti-Flag displayed similar punctate staining patterns for EVM005 in the absence and presence of infection (compare Fig. 2D, panels d to f, and E, panels b to d). Expression of HA-cullin-1 during infection was detected by using an antibody specific for cullin-1. Although the cullin-1 antibody detected ectopically expressed HA-cullin-1, the antibody failed to detect endogenous cullin-1 in this assay, likely due to low levels of cullin-1 (Fig. 2E, panel a). The cullin-1 antibody, however, clearly demonstrated that HA-cullin-1 localized throughout the infected cell to discrete punctate structures (Fig. 2E, panels e to h). Furthermore, coexpression of Flag-EVM005 and HA-cullin-1 indicated colocalization (Fig. 2E, panels i to l), supporting our previous results demonstrating that HA-cullin-1 and Flag-EVM005 interact during infection (Fig. 2A).
EVM005 interacts with components of an active SCF complex.
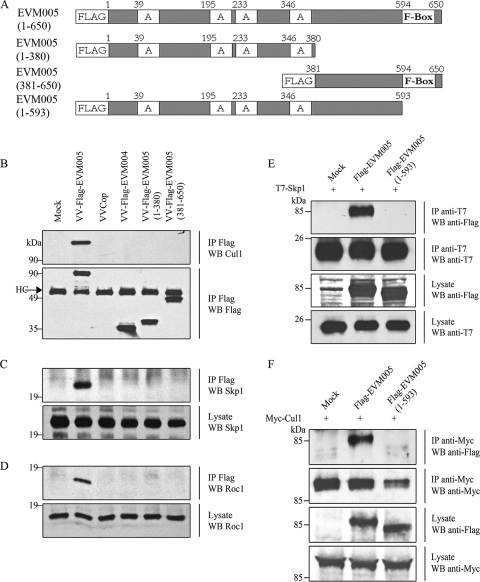
In order to determine whether the F-box domain of EVM005 was responsible for the observed interaction between Flag-EVM005 and cullin-1, three truncation mutants of Flag-EVM005 were created (Fig. 3A). Flag-EVM005(1-380) was composed of amino acids 1 to 380 of EVM005 and contained all four ankyrin repeats, whereas Flag-EVM005(381-650) lacked the ankyrin domains and contained the C-terminal F-box domain of EVM005. Flag-EVM005(1-593) lacked only the C-terminal 57 amino acids comprising the F-box domain. HEK293T cells were mock infected or infected with VV Copenhagen, VV-Flag-EVM004 as a negative control, or recombinant VV expressing full-length EVM005, EVM005(1-380), or EVM005(381-650). Cell lysates were subjected to immunoprecipitation with anti-Flag and Western blotted with anti-cullin-1 in order to detect an interaction with endogenous cullin-1 (Fig. 3B). The data demonstrated that only full-length Flag-EVM005 interacted with endogenous cullin-1, reaffirming our initial mass spectrometry data (Fig. 1B). Neither Flag-EVM005(1-380), which lacks the F-box domain, nor Flag-EVM005(381-650), which lacks the ankyrin domains but contains the F-box domain, interacted with cullin-1, suggesting that the C-terminal F-box was necessary but not sufficient for interaction with cullin-1 (Fig. 3B). Samples were additionally subjected to Western blotting with antibodies specific for the linker protein Skp1 and the ubiquitin ligase Roc1 to determine if EVM005 coprecipitated with endogenous Skp1 or Roc1 (Fig. 3C and D). Full-length Flag-EVM005 interacted with both endogenous Skp1 and Roc1, suggesting that EVM005 interacted with an intact ubiquitin ligase and may serve as a unique substrate adaptor for the SCF complex (Fig. 3C and D).
FIG. 3.
EVM005 interacts with endogenous components of the SCF complex ubiquitin ligase. (A) Full-length and truncated versions of Flag-EVM005 were constructed. (B) Mutant versions of EVM005 fail to interact with cullin-1. HEK293T cells were mock infected or infected with VV Copenhagen (VVCop), VV-Flag-EVM005, VV-Flag-EVM004, VV-Flag-EVM005(1-380), and VV-Flag-EVM005(381-650) at an MOI of 5. At 12 h postinfection, cells were lysed in NP-40 lysis buffer, immunoprecipitated (IP) with anti-Flag, and Western blotted (WB) with anti-Flag or anti-cullin-1 (Cul1). HC, antibody heavy chain. (C) Full-length EVM005 interacted with endogenous Skp1. HEK293T cells were infected and immunoprecipitated with anti-Flag to detect EVM005 and Western blotted with anti-Skp1. (D) EVM005 coprecipitates with endogenous Roc-1. HEK293T cells were infected and immunoprecipitated with anti-Flag to detect EVM005 and Western blotted with anti-Roc1. (E) HEK293T cells were cotransfected with pcDNA3-T7-Skp1 and either pcDNA3-Flag-EVM005 or pcDNA3-Flag-EVM005(1-593). Cellular lysates were immunoprecipitated with anti-T7 and Western blotted with anti-T7 to detect Skp1 or anti-Flag to detect EVM005. (F) HEK293T cells were cotransfected with pcDNA3-Myc-cullin-1 and either pcDNA3-Flag-EVM005 or pcDNA3-Flag-EVM005(1-593). Cellular lysates were immunoprecipitated with anti-Myc and Western blotted with anti-Myc to detect cullin-1 or anti-Flag to detect EVM005.
To further demonstrate that the F-box domain of EVM005 was necessary for interaction with Skp1 and cullin-1, we generated a third EVM005 mutant, Flag-EVM005(1-593), which lacked the C-terminal 57 amino acids comprising the F-box domain. HEK293T cells were cotransfected with pcDNA3-Flag-EVM005, pcDNA3-Flag-EVM005(1-593), and pcDNA3-T7-Skp1. Immunoprecipitation with anti-T7 to pull down Skp1 demonstrated a clear interaction with EVM005 but not with EVM005(1-593), which lacked the F-box domain (Fig. 3E). Additionally, cells cotransfected displayed interaction between cullin-1 and EVM005 but not EVM005(1-593), reaffirming that the F-box domain was necessary for interaction with the SCF complex (Fig. 3F).
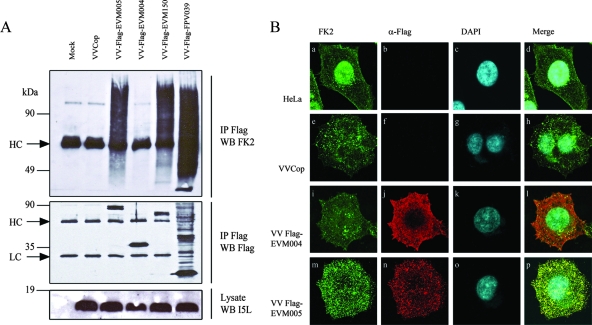
Given that Flag-EVM005 interacted with components of an active SCF complex, this observation suggested that EVM005 interacted with a functional SCF complex. To test this possibility, we used the antiubiquitin antibody clone FK2, which recognizes conjugated ubiquitin but not free ubiquitin, in order to determine an association between EVM005 and conjugated ubiquitin (20). HEK293T cells were mock infected or infected with VV Copenhagen, VV-Flag-EVM005, or VV-Flag-EVM004. We also used two viruses that served as positive controls, VV-Flag-EVM150, which expresses a BTB/kelch protein that we previously showed to interact with cullin-3 and conjugated ubiquitin, and VV-Flag-FPV039, a Bcl-2 homolog which is encoded by fowlpox virus and is regulated by ubiquitination (Fig. 4A) (5, 60; L. Banadyga and M. Barry, unpublished data). Infected cells were lysed and subjected to immunoprecipitation with anti-Flag followed by Western blotting with antiubiquitin (FK2), anti-Flag, or anti-I5L, which detects a late protein expressed by VV (Fig. 4A) (60). The expression of I5L indicated that all recombinant VVs replicated equally in HEK293T cells (Fig. 4A). Western blotting the immunoprecipitations with anti-Flag clearly indicated that EVM005, EVM004, EVM150, and FPV039 were expressed and that Flag-FPV039 was subjected to heavy ubiquitination, resulting in the presence of multiple Flag-positive bands (Fig. 4A). Western blotting of the Flag immunoprecipitations with anti-FK2 confirmed that both Flag-EVM150 and Flag-FPV039 associated with conjugated ubiquitin due to the presence of high-molecular-weight ubiquitin adducts, whereas Flag-EVM004 did not, as previously described (Fig. 4A) (60). Significantly, Flag-EVM005 also associated with conjugated ubiquitin, suggesting that Flag-EVM005 interacted with an active SCF complex (Fig. 4A).
FIG. 4.
EVM005 associates with conjugated ubiquitin. (A) HEK293T cells were mock infected or infected with VV Copenhagen (VVCop), VV-Flag-EVM005, VV-Flag-EVM004, VV-Flag-EVM150, or VV-Flag-FPV039 at an MOI of 5. At 12 h postinfection, cells were lysed in NP-40 lysis buffer, immunoprecipitated (IP) with anti-Flag, and Western blotted (WB) with antiubiquitin (clone FK2) to detect conjugated ubiquitin, anti-Flag to detect Flag-tagged proteins, or anti-I5L to detect the VV protein I5L. HC and LC represent antibody heavy and light chains, respectively. (B) EVM005 colocalizes with conjugated ubiquitin. HeLa cells (2 × 105) were mock infected or infected with VV Copenhagen, VV-Flag-EVM004, or VV-Flag-EVM005 at an MOI of 5, and colocalization with conjugated ubiquitin was visualized by confocal microscopy. Twelve hours postinfection, cells were fixed and costained with antiubiquitin (FK2) and anti-Flag-Cy3 (α-Flag) to detect the virus proteins and with DAPI to visualize the nucleus and viral factories. (a to d) Mock-infected cells. (e to h) Cells infected with VV Copenhagen. (i to l) Cells infected with VV-Flag-EVM004. (m to p) Cells infected with VV-Flag-EVM005. Eighty percent of HeLa cells infected with VV-Flag-EVM005 displayed colocalization with conjugated ubiquitin.
To confirm the coimmunoprecipitation results, we used confocal microscopy. HeLa cells were mock infected or infected with VV Copenhagen, VV-Flag-EVM005, or VV-Flag-EVM004 and costained with anti-Flag in order to detect EVM005 and EVM004 and with antiubiquitin (FK2) to detect conjugated ubiquitin. In uninfected HeLa cells, the majority of conjugated ubiquitin was located in the nucleus (Fig. 4B, panels a to d). Upon infection of cells with VV Copenhagen, the subcellular localization of conjugated ubiquitin changed and the majority of conjugated ubiquitin was not found in the nucleus but instead localized to punctate structures throughout the infected cell, indicating that conjugated ubiquitin was present in infected cells (Fig. 4B, panels e to h). Infection with VV-Flag-EVM004 demonstrated that Flag-EVM004 localized diffusely throughout the infected cell and did not colocalize with conjugated ubiquitin (Fig. 4B, panels i to l). In contrast, HeLa cells infected with VV-Flag-EVM005 demonstrated that Flag-EVM005 localized to punctate structures throughout the infected cell, and staining with antiubiquitin (FK2) determined that the punctate EVM005 structures colocalized with conjugated ubiquitin (Fig. 4B, panels m to p). The colocalization of Flag-EVM005 with conjugated ubiquitin lends further support to our hypothesis that EVM005 interacts with a functional SCF ubiquitin ligase complex.
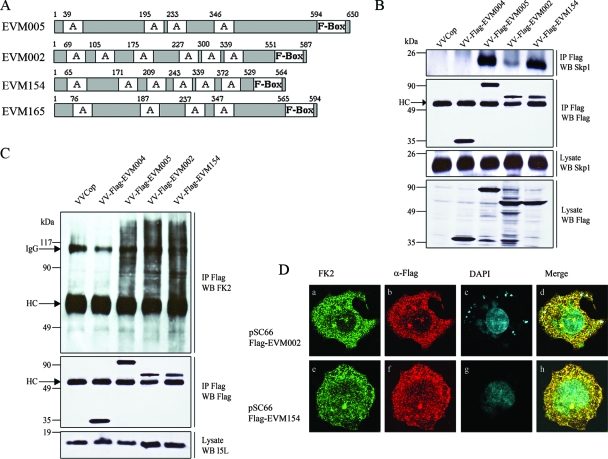
EVM002 and EVM154 contain F-box domains and interact with Skp1.
Bioinformatics indicated that ectromelia virus encoded three proteins besides EVM005 that contain putative F-box domains (Fig. 1A and 5A), namely, EVM002, EVM154, and EVM165, all of which contain N-terminal ankyrin domains in combination with C-terminal F-box domains (Fig. 5A). The recent demonstration that a modified VV Ankara ortholog of EVM165 interacts with Skp1 clearly suggests that EVM165 also interacts with the SCF complex (54). Prompted by this observation and our bioinformatics data, we sought to determine if EVM002 and EVM154 also interacted with the SCF complex. HeLa cells were infected with VV Copenhagen, VV-Flag-EVM004, VV-Flag-EVM005, VV-Flag-EVM002, or VV-Flag-EVM154. Cellular lysates were immunoprecipitated with anti-Flag and Western blotted with anti-Skp1 to determine an interaction with endogenous Skp1 (Fig. 5B). Similar to EVM005, both EVM154 and EVM002 interacted with Skp1 (Fig. 5B). Interestingly, EVM002 reproducibly demonstrated a reduced ability to interact with Skp1, suggesting that perhaps differences in amino acid composition within the F-box domain may account for this observation. Despite this difference, both EVM002 and EVM154 interacted with conjugated ubiquitin (Fig. 5C) and colocalized with conjugated ubiquitin, as assessed by confocal microscopy (Fig. 5D).
FIG. 5.
EVM002 and EVM154 interact with Skp1 and conjugated ubiquitin. VVCop, VV Copenhagen. (A) Schematic representation of EVM005, EVM002, EVM154, and EVM165 containing C-terminal F-boxes and N-terminal ankyrin domains (indicated with an “A” in the diagram). (B) EVM002 and EVM154 interact with Skp1. HEK293T cells were infected with VV Copenhagen, VV-Flag-EVM004, VV-Flag-EVM005, VV-Flag-EVM002, or VV-Flag-EVM154. Lysates were immunoprecipitated (IP) with anti-Flag and Western blotted (WB) with anti-Flag to detect the viral proteins or Western blotted with anti-Skp1 to detect endogenous Skp1. HC, antibody heavy chain. (C) EVM002 and EVM154 interact with conjugated ubiquitin. HEK293T cells were mock infected or infected with VV Copenhagen, VV-Flag-EVM004, VV-Flag-EVM005, VV-Flag-EVM002, or VV-Flag-EVM154 at an MOI of 5. At 12 h postinfection, cells were lysed in NP-40 lysis buffer and immunoprecipitated with anti-Flag and Western blotted with antiubiquitin (clone FK2) to detect conjugated ubiquitin, anti-Flag to detect Flag-tagged viral proteins, or anti-I5L to detect the VV protein I5L. IgG, immunoglobulin G antibody; HC, antibody heavy chain. (D) EVM002 and EVM154 colocalize with conjugated ubiquitin. HeLa cells were infected with VV Copenhagen and cotransfected with pSC66-Flag-EVM002 or pSC66-Flag-EVM154. Colocalization with conjugated ubiquitin was visualized by confocal microscopy. Twelve hours postinfection, coverslips were fixed and costained with antiubiquitin (FK2), anti-Flag-Cy3 (α-Flag), and DAPI to visualize the nucleus and viral factories. (a to d) VV Copenhagen infected and transfected with pSC66-Flag-EVM002. (e to h) Cells infected with VV Copenhagen and transfected with pSC66-Flag-EVM154. Ninety-five percent of cells transfected with pSC66-Flag-EVM002 or pSC66-EVM154 demonstrated colocalization between conjugated ubiquitin and EVM002 or EVM154.
DISCUSSION
Previous studies have shown an important role for F-box proteins in substrate recruitment to the SCF complex (14, 29, 45, 59). More than 70 cellular proteins containing F-box domains have been identified, many of which function as substrate adaptor molecules for the cellular SCF complex ubiquitin ligase (14, 29, 45, 59, 63). Using a bioinformatics approach, we identified four ectromelia virus genes, EVM002, EVM005, EVM154, and EVM165 genes, which encode N-terminal ankyrin repeats in combination with C-terminal F-box domains (Fig. 1A and 5A). Significantly, the combination of ankyrin repeats and F-box domains is unique to poxviruses and not found within cellular F-box proteins (36). The presence of F-box domains led us to speculate that these four ectromelia virus proteins would play a role in regulating the ubiquitin-proteasome system via interaction with the cellular SCF complex. To perform these studies, we used an overexpression strategy in which EVM002, EVM005, and EVM154 were expressed during virus infection. This approach allowed us to determine interaction with components of the SCF complex during infection. This approach was favored due to the inability to express EVM002, EVM005, and EVM154 in the absence of infection, and codon optimization was necessary to express EVM005 in the absence of infection (Fig. 2D, panels d to f) (6). The use of recombinant VVs demonstrated that EVM005 interacted with components of the SCF complex, including Skp1, cullin-1, and Roc1 (Fig. 1B, 2, and 3). Transient expression of EVM005 also demonstrated interaction with Skp1 and cullin-1 (Fig. 3E and F). EVM005 also colocalized and associated with conjugated ubiquitin molecules during infection further providing a link between EVM005 and the ubiquitin proteasome system (Fig. 4). Moreover, EVM002 and EVM154 also interacted with Skp1 during infection (Fig. 5). Together, our data indicate that ectromelia virus encodes multiple proteins that interact with components of the SCF complex.
In contrast to the majority of cellular SCF substrate adaptor proteins, the ectromelia virus F-boxes are located at the C terminus, in combination with a series of N-terminal ankyrin repeats. An alignment of EVM002, EVM005, EVM154, and EVM165 with the F-box from the cellular protein Skp2 demonstrated that key residues were maintained in the ectromelia virus proteins (Fig. 1A) (48, 63). The cocrystal structure of Skp1 and the cellular substrate adaptor protein Skp2 demonstrates that the F-box domain of Skp2 is composed of three α-helices containing residues important for contact with Skp1 (Fig. 1A) (48, 63). The ectromelia virus F-box domains appear to have conserved key amino acids within helices H1 and H2, but little homology was maintained within α-helix H3 (Fig. 1A). We have shown that, despite the lack of obvious homology of the F-box domains to the H3 α-helix, three of the ectromelia virus proteins, EVM002, EVM005, and EVM154, interact with Skp1 (Fig. 3C and 5B). Additionally, data from Sperling and colleagues confirm that the VV ortholog of EVM165 also interacts with Skp1 (54). Significantly, deletion of the F-box domain in EVM005 resulted in a protein that no longer interacted with cullin-1, Skp1, and Roc1, confirming that the F-box domain was necessary for interaction with the SCF complex (Fig. 3B to F). However, we were unable to observe an interaction between Skp1 and a mutant of EVM005, EVM005(381-650), that contains the F-box domain but lacks the ankyrin repeat domains, suggesting that the ankyrin domains may also play a role in interaction (Fig. 3A to D). The cellular substrate adaptor protein Skp2 contains 10 leucine-rich repeats in conjunction with an N-terminal F-box domain. The first three leucine-rich repeats in Skp2 serve a structural linker role, suggesting that one or more of the ankyrin repeats in EVM005 may provide similar functions (Fig. 4) (48, 63).
The combination of N-terminal ankyrin repeats and C-terminal F-box domains appears to be unique to poxviruses (36). Bioinformatics has identified the presence of multiple ankyrin/F-box proteins in every genus of vertebrate poxvirus, with the exception of Molluscipox (36). Significantly, fowlpox virus and canarypox virus, members of the Avipoxvirus genus, encode the largest family of ankyrin/F-box proteins, and these are likely to interact with avian SCF complexes (2, 56). The initial identification of the putative F-box domains in various poxviruses prompted our screening of the ectromelia virus genome (36). Using bioinformatics, we identified seven ankyrin-containing proteins in ectromelia virus, and significantly, four of these contained F-box domains. Of the four ankyrin/F-box proteins that we identified in ectromelia virus, EVM005 appears to be unique. Whereas multiple orthologs of EVM002, EVM154, and EVM165 can be found in other members of the poxvirus family, only one ortholog, CPVBR011, encoded by cowpox virus, exists for EVM005 (23). We therefore focused our studies on EVM005, since it likely plays an important role during ectromelia virus and cowpox virus infections.
In all cases, the ectromelia virus F-box domains were found in combination with N-terminal ankyrin domains. Ankyrin-repeat domains are composed of a 30- to 34-amino-acid repeat important for protein-protein interactions and found in a wide range of proteins (39, 49). Interestingly, the majority of cellular SCF complex substrate adaptors contain F-box domains in combination with either leucine-rich repeats or WD repeats in order to recruit substrates to the SCF complex (14, 15, 29, 45, 61). Although cellular F-boxes are not found in combination with ankyrin domains, ankyrin/suppressors of cytokine signaling-box proteins, which recruit substrates to cullin-2 ubiquitin ligases, are common (43), suggesting that the poxvirus ankyrin/F-box proteins may have evolved through recombination. Given that a large number of cellular F-box proteins function as substrate adaptors for the SCF complex (14, 15, 29, 45, 61), we speculate that the ectromelia virus ankyrin/F-box proteins may also function as substrate adaptors for the SCF complex. The interaction of EVM002, EVM005, EVM154, and EVM165 with components of the SCF complex strongly supports this possibility. Several cellular F-box proteins, however, serve functions other than substrate adaptors (14, 15, 29, 45, 61). The best characterized of these is cyclin F, which contains a cyclin domain and an F-box to regulate levels of itself, ultimately controlling the cell cycle (4). Accordingly, it is possible that the ectromelia virus ankyrin/F-box proteins may be important for regulating the levels of EVM002, EVM005, EVM154, and EVM165 during infection. Alternatively, the ectromelia virus ankyrin/F-box proteins could simply bind and sequester cullin-1 in order to inhibit the SCF complex. At the present, our data do not distinguish between these possibilities; however, the presence of conjugated ubiquitin upon expression of EVM002, EVM005, and EVM154 suggested that active ubiquitination was occurring (Fig. 4 and 5C and D). Undoubtedly, the identification of additional binding partners for the ectromelia virus ankyrin/F-box proteins will lead to further insights regarding their function.
Although members of the poxvirus family encode multiple proteins with ankyrin repeats, few have been studied extensively. One of the best characterized poxviral ankyrin repeat proteins is CP77, which plays a role in host-range determination of cowpox virus (26, 27). Like CP77, the ankyrin repeat protein K1L, encoded by VV, also mediates host-range function, and it has more recently been shown to inhibit NF-κB activation (13, 52). MT-5, an ankyrin/F-box protein encoded by myxoma virus, a member of the Leporipoxvirus genus, interacts with cullin-1, resulting in degradation of p27Kip and cell cycle arrest (31, 58). Additionally, the ectromelia virus ankyrin repeat proteins EVM010, EVM021, and EVM022, which do not contain putative C-terminal F-box domains, are likely to serve important, but as yet unknown, functions during ectromelia virus infection.
In addition to encoding ankyrin/F-box proteins, poxviruses have multiple strategies to exploit the ubiquitin-proteasome system. The avipoxviruses encode an extended family of RING finger proteins that are predicted to function as ubiquitin ligases (56, 60). Additionally, canarypox virus encodes its own molecule of ubiquitin (56), and crocodilepox virus encodes a homolog to APC11, a member of the anaphase-promoting complex/cyclosome ubiquitin ligase proteins (1). Myxoma virus regulates the cell surface expression of major histocompatibility complex class I and CD4 molecules via expression of the ubiquitin ligase M153R (24, 35). Moreover, members of the Orthopoxvirus and Leporipoxvirus genus encode a unique ubiquitin ligase, p28, that localizes to the virus factory (28, 42). Several members of the poxvirus family encode multiple BTB/kelch proteins, two of which we have shown to interact with cullin-3 ubiquitin ligases (60). Thus, it is clear that poxviruses have evolved a wide variety of strategies to exploit the ubiquitin-proteasome system, and the presence of multiple ankyrin/F-box proteins suggests that poxviruses have also evolved a unique mechanism to exploit the SCF complex. Identifying the substrates that are degraded by the SCF complex during poxvirus infection remains the goal of future studies.
Acknowledgments
We thank R. Garneau for technical assistance and L. Banadyga, S. Campbell, and K. Veugelers for critical reviews of the manuscript.
Work in our laboratories is supported by grants to M.B. from the Canadian Institutes of Health Research (CIHR), the Howard Hughes Medical Institute (HHMI), and an NIH grant (GM067113) to Y.X. M.B. is a CIHR New Investigator, an Alberta Heritage Foundation for Medical Research Senior Scholar, and a HHMI International Research Scholar.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, G. Delhon, Z. Lu, G. J. Viljoen, D. B. Wallace, G. F. Kutish, and D. L. Rock. 2006. Genome of crocodilepox virus. J. Virol. 804978-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 743815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ardley, H. C., and P. A. Robinson. 2005. E3 ubiquitin ligases. Essays Biochem. 4115-30. [DOI] [PubMed] [Google Scholar]
- 4.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86263-274. [DOI] [PubMed] [Google Scholar]
- 5.Banadyga, L., J. Gerig, T. Stewart, and M. Barry. 2007. Fowlpox virus encodes a Bcl-2 homologue that protects cells from apoptotic death through interaction with the proapoptotic protein Bak. J. Virol. 8111032-11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett, J. W., Y. Sun, S. H. Nazarian, T. A. Belsito, C. R. Brunetti, and G. McFadden. 2006. Optimization of codon usage of poxvirus genes allows for improved transient expression in mammalian cells. Virus Genes 3315-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barry, M., and K. Fruh. 2006. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE 2006pe21. [DOI] [PubMed] [Google Scholar]
- 8.Barry, M., S. Hnatiuk, K. Mossman, S. F. Lee, L. Boshkov, and G. McFadden. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239360-377. [DOI] [PubMed] [Google Scholar]
- 9.Besnard-Guerin, C., N. Belaidouni, I. Lassot, E. Segeral, A. Jobart, C. Marchal, and R. Benarous. 2004. HIV-1 Vpu sequesters beta-transducin repeat-containing protein (betaTrCP) in the cytoplasm and provokes the accumulation of beta-catenin and other SCFbetaTrCP substrates. J. Biol. Chem. 279788-795. [DOI] [PubMed] [Google Scholar]
- 10.Bour, S., C. Perrin, H. Akari, and K. Strebel. 2001. The human immunodeficiency virus type 1 Vpu protein inhibits NF-kappa B activation by interfering with beta TrCP-mediated degradation of Ikappa B. J. Biol. Chem. 27615920-15928. [DOI] [PubMed] [Google Scholar]
- 11.Boutell, C., A. Orr, and R. D. Everett. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 778686-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradley, R. R., and M. Terajima. 2005. Vaccinia virus K1L protein mediates host-range function in RK-13 cells via ankyrin repeat and may interact with a cellular GTPase-activating protein. Virus Res. 114104-112. [DOI] [PubMed] [Google Scholar]
- 14.Cardozo, T., and M. Pagano. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5739-751. [DOI] [PubMed] [Google Scholar]
- 15.Cenciarelli, C., D. S. Chiaur, D. Guardavaccaro, W. Parks, M. Vidal, and M. Pagano. 1999. Identification of a family of human F-box proteins. Curr. Biol. 91177-1179. [DOI] [PubMed] [Google Scholar]
- 16.Davison, A. J., and B. Moss. 1990. New vaccinia virus recombination plasmids incorporating a synthetic late promoter for high level expression of foreign proteins. Nucleic Acids Res. 184285-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 2721470-1473. [DOI] [PubMed] [Google Scholar]
- 18.Earl, P. L., B. Moss, L. S. Wyatt, and M. W. Carroll. 2001. Generation of recombinant vaccinia viruses, chapter 16, unit 16-17. In Current protocols in molecular biology. John Wiley and Sons, Hoboken, NJ. [DOI] [PubMed]
- 19.Estrabaud, E., E. Le Rouzic, S. Lopez-Verges, M. Morel, N. Belaidouni, R. Benarous, C. Transy, C. Berlioz-Torrent, and F. Margottin-Goguet. 2007. Regulated degradation of the HIV-1 Vpu protein through a betaTrCP-independent pathway limits the release of viral particles. PLoS Pathog. 3e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimuro, M., and H. Yokosawa. 2005. Production of antipolyubiquitin monoclonal antibodies and their use for characterization and isolation of polyubiquitinated proteins. Methods Enzymol. 39975-86. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa, M., Y. Zhang, J. McCarville, T. Ohta, and Y. Xiong. 2000. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 208185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagne, J. M., B. P. Downes, S. H. Shiu, A. M. Durski, and R. D. Vierstra. 2002. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 9911519-11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gubser, C., S. Hue, P. Kellam, and G. L. Smith. 2004. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 85105-117. [DOI] [PubMed] [Google Scholar]
- 24.Guerin, J. L., J. Gelfi, S. Boullier, M. Delverdier, F. A. Bellanger, S. Bertagnoli, I. Drexler, G. Sutter, and F. Messud-Petit. 2002. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J. Virol. 762912-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2195-201. [DOI] [PubMed] [Google Scholar]
- 26.Hsiao, J. C., C. C. Chao, M. J. Young, Y. T. Chang, E. C. Cho, and W. Chang. 2006. A poxvirus host range protein, CP77, binds to a cellular protein, HMG20A, and regulates its dissociation from the vaccinia virus genome in CHO-K1 cells. J. Virol. 807714-7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao, J. C., C. S. Chung, R. Drillien, and W. Chang. 2004. The cowpox virus host range gene, CP77, affects phosphorylation of eIF2 alpha and vaccinia viral translation in apoptotic HeLa cells. Virology 329199-212. [DOI] [PubMed] [Google Scholar]
- 28.Huang, J., Q. Huang, X. Zhou, M. M. Shen, A. Yen, S. X. Yu, G. Dong, K. Qu, P. Huang, E. M. Anderson, S. Daniel-Issakani, R. M. Buller, D. G. Payan, and H. H. Lu. 2004. The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J. Biol. Chem. 27954110-54116. [DOI] [PubMed] [Google Scholar]
- 29.Jin, J., T. Cardozo, R. C. Lovering, S. J. Elledge, M. Pagano, and J. W. Harper. 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 182573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston, J. B., and G. McFadden. 2003. Poxvirus immunomodulatory strategies: current perspectives. J. Virol. 776093-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston, J. B., G. Wang, J. W. Barrett, S. H. Nazarian, K. Colwill, M. Moran, and G. McFadden. 2005. Myxoma virus M-T5 protects infected cells from the stress of cell cycle arrest through its interaction with host cell cullin-1. J. Virol. 7910750-10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latres, E., D. S. Chiaur, and M. Pagano. 1999. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of beta-catenin. Oncogene 18849-854. [DOI] [PubMed] [Google Scholar]
- 33.Lechner, E., P. Achard, A. Vansiri, T. Potuschak, and P. Genschik. 2006. F-box proteins everywhere. Curr. Opin. Plant Biol. 9631-638. [DOI] [PubMed] [Google Scholar]
- 34.Mackett, M., G. L. Smith, and B. Moss. 1992. Vaccinia virus: a selectable eukaryotic cloning and expression vector. 1982. Bio/Technology 24495-499. [PubMed] [Google Scholar]
- 35.Mansouri, M., E. Bartee, K. Gouveia, B. T. Hovey Nerenberg, J. Barrett, L. Thomas, G. Thomas, G. McFadden, and K. Fruh. 2003. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 771427-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercer, A. A., S. B. Fleming, and N. Ueda. 2005. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 31127-133. [DOI] [PubMed] [Google Scholar]
- 37.Michel, J. J., and Y. Xiong. 1998. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 9435-449. [PubMed] [Google Scholar]
- 38.Minella, A. C., and B. E. Clurman. 2005. Mechanisms of tumor suppression by the SCF(Fbw7). Cell Cycle 41356-1359. [DOI] [PubMed] [Google Scholar]
- 39.Mosavi, L. K., T. J. Cammett, D. C. Desrosiers, and Z. Y. Peng. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 131435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama, K. I., S. Hatakeyama, and K. Nakayama. 2001. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem. Biophys. Res. Commun. 282853-860. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama, K. I., and K. Nakayama. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 6369-381. [DOI] [PubMed] [Google Scholar]
- 42.Nerenberg, B. T., J. Taylor, E. Bartee, K. Gouveia, M. Barry, and K. Fruh. 2005. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J. Virol. 79597-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholson, S. E., and D. J. Hilton. 1998. The SOCS proteins: a new family of negative regulators of signal transduction. J. Leukoc. Biol. 63665-668. [DOI] [PubMed] [Google Scholar]
- 44.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3535-541. [DOI] [PubMed] [Google Scholar]
- 45.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 69-20. [DOI] [PubMed] [Google Scholar]
- 46.Pickart, C. M., and D. Fushman. 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8610-616. [DOI] [PubMed] [Google Scholar]
- 47.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 713788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408381-386. [DOI] [PubMed] [Google Scholar]
- 49.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24311-316. [DOI] [PubMed] [Google Scholar]
- 50.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21377-423. [DOI] [PubMed] [Google Scholar]
- 51.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 52.Shisler, J. L., and X. L. Jin. 2004. The vaccinia virus K1L gene product inhibits host NF-κB activation by preventing IκBα degradation. J. Virol. 783553-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91209-219. [DOI] [PubMed] [Google Scholar]
- 54.Sperling, K. M., A. Schwantes, B. S. Schnierle, and G. Sutter. 2008. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology 374234-239. [DOI] [PubMed] [Google Scholar]
- 55.Stuart, D., K. Graham, M. Schreiber, C. Macaulay, and G. McFadden. 1991. The target DNA sequence for resolution of poxvirus replicative intermediates is an active late promoter. J. Virol. 6561-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2004. The genome of canarypox virus. J. Virol. 78353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2169-178. [DOI] [PubMed] [Google Scholar]
- 58.Werden, S. J., J. W. Barrett, G. Wang, M. M. Stanford, and G. McFadden. 2007. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J. Virol. 812340-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willems, A. R., M. Schwab, and M. Tyers. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695133-170. [DOI] [PubMed] [Google Scholar]
- 60.Wilton, B. A., S. Campbell, N. Van Buuren, R. Garneau, M. Furukawa, Y. Xiong, and M. Barry. 2008. Ectromelia virus BTB/kelch proteins, EVM150 and EVM167, interact with cullin-3-based ubiquitin ligases. Virology 37482-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winston, J. T., D. M. Koepp, C. Zhu, S. J. Elledge, and J. W. Harper. 1999. A family of mammalian F-box proteins. Curr. Biol. 91180-1182. [DOI] [PubMed] [Google Scholar]
- 62.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 63.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416703-709. [DOI] [PubMed] [Google Scholar]