Abstract
Positive-strand RNA viruses use diverse mechanisms to regulate viral and host gene expression for ensuring their efficient proliferation or persistence in the host. We found that a small viral noncoding RNA (0.4 kb), named SR1f, accumulated in Red clover necrotic mosaic virus (RCNMV)-infected plants and protoplasts and was packaged into virions. The genome of RCNMV consists of two positive-strand RNAs, RNA1 and RNA2. SR1f was generated from the 3′ untranslated region (UTR) of RNA1, which contains RNA elements essential for both cap-independent translation and negative-strand RNA synthesis. A 58-nucleotide sequence in the 3′ UTR of RNA1 (Seq1f58) was necessary and sufficient for the generation of SR1f. SR1f was neither a subgenomic RNA nor a defective RNA replicon but a stable degradation product generated by Seq1f58-mediated protection against 5′→3′ decay. SR1f efficiently suppressed both cap-independent and cap-dependent translation both in vitro and in vivo. SR1f trans inhibited negative-strand RNA synthesis of RCNMV genomic RNAs via repression of replicase protein production but not via competition of replicase proteins in vitro. RCNMV seems to use cellular enzymes to generate SR1f that might play a regulatory role in RCNMV infection. Our results also suggest that Seq1f58 is an RNA element that protects the 3′-side RNA sequences against 5′→3′ decay in plant cells as reported for the poly(G) tract and stable stem-loop structure in Saccharomyces cerevisiae.
Many lines of recent evidence indicate that noncoding RNAs including microRNAs and small interfering RNAs play an important role in the control of gene expression in diverse cellular processes and in defense responses against molecular parasites such as viruses and transposons. Viruses also use many different types of RNAs in trans for regulating the expression of their own genomes or host genomes temporally and spatially to ensure efficient virus proliferation and/or latency in host cells. These RNAs include subgenomic RNAs (sgRNAs), viral genomic RNA itself, and many types of noncoding viral RNAs.
For example, the adenovirus virus-associated RNAs (VA RNAs) (23) are small noncoding RNA transcripts. They inhibit the activation of RNA-induced protein kinase and thereby interfere with the activation of the interferon-induced cellular antiviral defense systems (38). VA RNAs also interfere with RNA interference pathways by acting as substrates for Dicer and suppressing the activity of Dicer probably involved in cellular antiviral mechanisms (2, 55). Epstein-Barr virus-encoded RNAs (EBERs) (56) inhibit RNA-induced protein kinase as VA RNAs (38). They also are known to encode microRNAs, which are thought to work for persistent infection (28). On the other hand, recently, EBERs have been reported to be recognized by RIG-I, a cytosolic protein with a DexD/H box RNA helicase domain that recognizes viral RNA in mammalian cells, and to activate signaling to induce type I interferon (35). Thus, associations of viral small RNAs with virus infection are complicated.
sgRNAs also function as riboregulators in viral gene expression in addition to their original roles as mRNAs used for protein production. For example, a Flock house virus sgRNA trans activates the replication of genomic RNA (9). In Barley yellow dwarf virus (BYDV), sgRNA2 encoding a small open reading frame (ORF) of unknown function acts in trans to suppress the translation of replicase genes from genomic RNA but permits the translation of structural genes from sgRNA1 (40). Furthermore, Red clover necrotic mosaic virus (RCNMV) genomic RNA2 is required in trans for the synthesis of sgRNA from genomic RNA1 (42).
RCNMV is a member of the genus Dianthovirus in the family Tombusviridae. Its genome is divided into two RNA molecules, RNA1 and RNA2 (Fig. 1A) (11, 12, 30), each of which has no cap structure at the 5′ end and no poly(A) tail at the 3′ end (21, 25). RNA2 is a monocistronic RNA that encodes a 35-kDa protein required for viral cell-to-cell movement in plants (21, 51). RNA1 encodes putative RNA replicase components, a 27-kDa protein (p27), and an 88-kDa protein (p88). p88, containing an RNA-dependent RNA polymerase motif (19), is produced by programmed −1 ribosomal frameshifting (16, 17, 52) and is required in cis for the replication of RNA1 (29). RNA1 also encodes a 37-kDa coat protein (CP) that is expressed from CP sgRNA (57). The transcription of CP sgRNA requires intermolecular interaction between RNA1 and RNA2 as described above. The 3′ UTR of RNA1 contains RNA elements (3′TE-DR1) essential for cap-independent translation (27) and also contains the promoter for negative-strand RNA synthesis (14).
FIG. 1.
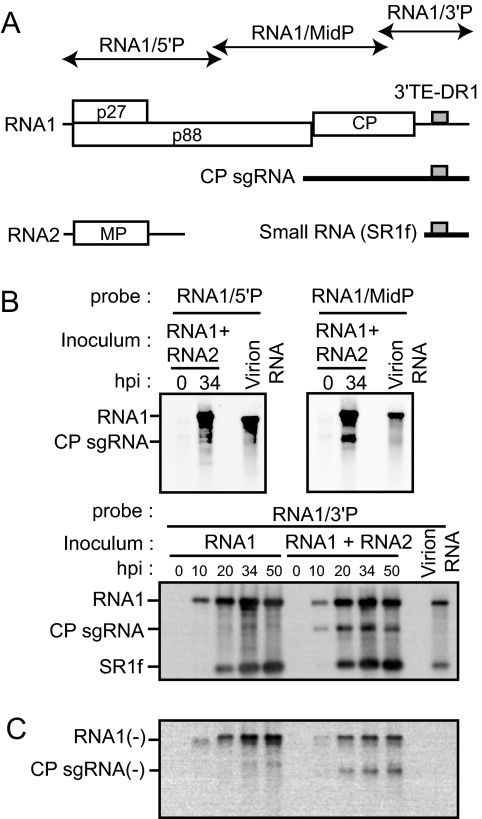
A small RNA is generated from the 3′-proximal region of RNA1 and packaged into virions in RCNMV-infected cells. (A) Schematic representation of viral RNAs of RCNMV. The genome is shown as horizontal lines with coding regions depicted as open boxes with the assigned viral protein designated. CP sgRNA and a small RNA (SR1f) are shown as thick horizontal lines below the genome diagram. The gray boxes indicate 3′TE-DR1. The regions covered by RNA probes are shown as thin arrows. MP, movement protein. (B) Accumulations of RNA1, CP sgRNA, and a small RNA (SR1f) in BY-2 protoplasts. Inoculated protoplasts were incubated at 17°C for the indicated times. Total RNAs extracted from protoplasts and purified virions were separated by gel electrophoresis and blotted onto a membrane. The top left, top right, and bottom panels show membranes hybridized with the DIG-labeled RNA probes specific for the 5′ UTR, middle region, and 3′ region, respectively. hpi, hours postinfection. (C) Accumulations of negative-strand RNA1 and CP sgRNA in BY-2 protoplasts. The same set of membranes shown in the bottom panel in panel B was hybridized with the DIG-labeled RNA probe specific for negative-strand RNA1.
We noticed that a large amount of small viral RNAs (0.4 kb) accumulated in RCNMV-infected plants and protoplasts during our RCNMV study. Here, we analyzed the origin, generation mechanisms, and possible functions of this small RNA in vivo and in vitro. Our results show that (i) the small RNA (defined here as SR1f) consists of nearly the entire 3′ UTR of RNA1; (ii) SR1f is not an sgRNA but a degradation intermediate generated by cis-RNA element-mediated protection against 5′→3′ decay, and a 58-nucleotide (nt) sequence (Seq1f58) is necessary and sufficient for protection against 5′→3′ decay; and (iii) SR1f trans inhibits both cap/poly(A)-dependent and 3′TE-DR1-mediated cap-independent translation in vivo and in vitro, resulting in a decrease in negative-strand RNA synthesis of RCNMV genomic RNAs. Our results also suggest that Seq1f58 is a cis-acting RNA element that confers resistance to RNA against 5′→3′ decay in plant cells as the poly(G) tract and stable stem-loop structures reported in Saccharomyces cerevisiae RNAs.
MATERIALS AND METHODS
Plasmid constructions.
pUCR1 and pRC2|G are full-length cDNA clones of RNA1 and RNA2 of the RCNMV Australian strain, respectively (44, 54). pCR-R1 and pSC-R1 are full-length cDNA clones of RNA1 of Carnation ring spot virus (CRSV) (41) and Sweet clover necrotic mosaic virus (SCNMV) (10), respectively, which were kindly provided by S. A. Lommel (North Carolina State University) and C. Hiruki (University of Alberta), respectively. Constructs described previously that were used in this study include the following: pUCR1-ΔCP (44), pUCR1-d3′SLA (14), pUCR1-d3′SLB (14), pUCR1-d3′SLC (14), pUCR1-d3′SLD (14), pUCR1-d3′SLE (14), pUCR1-d3′SLF (14), pR1-5′-XbS (27), and pLUCpA60 (27). Escherichia coli DH5α was used for the construction of all plasmids, except for pSSL-luc. All constructs were verified by sequencing. The primers used are listed in Table 1.
TABLE 1.
List of primers and their sequences used for PCR to generate constructs described in the text
| Primer | Nucleotide positiona | Sequenceb |
|---|---|---|
| A1+3380 | 3361-3377 | TGCAGTTTTCAGGTTCC |
| d3′-ii− | 3469-3428 delta 3459-3447 | GAGGCTACACTTAACCAAGTATGAAAGTG |
| M4 | * | GTTTTCCCAGTCACGAC |
| d3′-ii+ | 3434-3477 delta 3447-3459 | CATACTTGGTTAAGTGTAGCCTCCACCCGAG |
| AC1+3036 | 3036-3055 | GAAGTCGAGTTCAAAGGACC |
| m1− | 3481-3446 | GCAACTCGGGTGGAGCAACGTCTTAAAAGAACCAAT |
| m2− | 3489-3455 | CCCCTCTTGCAACTCCCCACGAGGCTACACTTAAA |
| m3− | 3497-3463 | TGCGTGTTCCCCTCTACATCGTCGGGTGGAGGCTA |
| m4− | 3509-3472 | GGTCGGCGAGACTGCCACAAGGGCTCTTGCAACTCGGG |
| m5− | 3516-3480 | CCAACAGGGTCGGCGTCTGACGGTGTTCCCCTCTTGC |
| m6− | 3522-3488 | TGTTTGCCAACAGGGAACCGGAGACTGCGTGTTCC |
| m7− | 3526-3494 | TTACTGTTTGCCAACTCCGTCGGCGAGACTGCG |
| m8− | 3532-3498 | GCAATTTTACTGTTTCGGCTCAGGGTCGGCGAGAC |
| m9− | 3538-3503 | TTTTTTGCAATTTTACACAAAGCCAACAGGGTCGGC |
| m10− | 3540-3508 | TATTTTTTGCAATTTACGTGTTTGCCAACAGGG |
| m11− | 3544-3511 | ACTCTATTTTTTGCATAAATACTGTTTGCCAACA |
| m12− | 3547-3535 | AGCACTCTATTTTTTCATATTTTACTGTTTGCC |
| m13− | 3554-3518 | TACTCCTAGCACTCTTAAAAAAGCAATTTTACTGTTT |
| m14− | 3561-3525 | CGGGAACTACTCCTACGTGAGAATTTTTTGCAATTTT |
| m1/m3− | 3491-3457 | CCTCTACATCGTCGGGTGGAGCAACGTCTTAAAAG |
| 3′R/C1 | ** | TACCCGGG GTACCTAGCCGTTATAC |
| m1+ | 3446-3481 | ATTGGTTCTTTTAAGACGTTGCTCCACCCGAGTTGC |
| m2+ | 3455-3489 | TTTAAGTGTAGCCTCGTGGGGAGTTGCAAGAGGGG |
| m3+ | 3462-3497 | GTAGCCTCCACCCGACGATGTAGAGGGGAACACGCA |
| m4+ | 3471-3509 | ACCCGAGTTGCAAGAGCCCTTGTGGCAGTCTCGCCGACC |
| m5+ | 3480-3516 | GCAAGAGGGGAACACCGTGAGACGCCGACCCTGTTGG |
| m6+ | 3488-3522 | GGAACACGCAGTCTCCGGTTCCCTGTTGGCAAACA |
| m7+ | 3494-3526 | CGCAGTCTCGCCGACGGAGTTGGCAAACAGTAA |
| m8+ | 3498-3532 | GTCTCGCCGACCCTGAGCCGAAACAGTAAAATTGC |
| m9+ | 3503-3537 | GCCGACCCTGTTGGCTTTGTGTAAAATTGCAAAAA |
| m10+ | 3508-3540 | CCCTGTTGGCAAACACGTAAATTGCAAAAAATA |
| m11+ | 3511-3544 | TGTTGGCAAACAGTATTTATGCAAAAAATAGAGT |
| m12+ | 3515-3547 | GGCAAACAGTAAAATATGAAAAAATAGAGTGCT |
| m13+ | 3518-3554 | AAACAGTAAAATTGCTTTTTTAAGAGTGCTAGGAGTA |
| m14+ | 3525-3561 | AAAATTGCAAAAAATTCTGACGTAGGAGTAGTTCCCG |
| m1/m3+ | 3456-3492 | TTAAGACGTTGCTCCACCCGACGATGTAGAGGGGAAC |
| Aus3115+ | 3096-3115 | CTCGGTGGGACACTCACTTC |
| Aus4291− | *** | ATTTAGAGCTTGACGGGGAAAG |
| SacI/T7 | GCGAGCTC TAATACGACTCACTATA | |
| 5′A1/R-Luc/L | CATAAACTTTCGAAGTCATGACTGGTACGAAAAGTAG | |
| 5′A1/R-Luc/R | CTACTTTTCGTACCAGTCATGACTTCGAAAGTTTATGAT | |
| R-Luc/3′A1/L | GCTACACTTAAAAGAACCAATTATTGTTCATTTTTG | |
| R-Luc/3′A1/R | CAAAAATGAACAATAATTGGTTCTTTTAAGTGTAGC | |
| 3′A1end/L | TCTAGAGGATCCCCGGG GTACCTAGCCGTTATAC | |
| R-1f− | GAGGCTACACGGATCCTTATTGTTCATTTTTGAGAAC | |
| R-m1− | GAGCAACGTCGGATCCTTATTGTTCATTTTTGAGAAC | |
| 1fNco1− | CATGCCATGG GCCAACAGGGTCGGCGAGAC | |
| R-1f+ | TGAACAATAAGGATCCGTGTAGCCTCCACCCGAGTTG | |
| R-m1+ | TGAACAATAAGGATCCGACGTTGCTCCACCCGAGTTGCAAGAGG | |
| pR/1f/F+ | CCCAAGCTT CCACCATGACTTCGAAAGTTTATGATC | |
| pR/1f/F− | TTTGTCTAC TTACACGGCGATCTTTCCGC | |
| T7/TC5′ | GCGAGCTC TAATACGACTCACTATAGTGTAGCCTCCACCCGAG | |
| 1f-m1(+) | GCGAGCTC TAATACGACTCACTATAGACGTTGCTCCACCCGAGTTGCAAG | |
| SSL/A1/R | GGTCCACCACGGCCGATATC ACGGCCGTGGTGGACCGCACAAACGTTTT | |
| T7/9/SSL/R | GCGAGCTC TAATACGACTCACTATAGCAACAACAAGCGGTCCACCACGG | |
| A1/SSL/2L | ACCACGGCCGTGATATC GGCCGTGGTGGACCGCGACTGGTACGAAAAGT | |
| SSL/14/Luc/2R | ATCACGGCCGTGGTGGACCGCAACAACAACAACAAATGGAAGACGCCAA | |
| T7NSR+ | GCGAGCTC TAATACGACTCACTATAGTTATTGTTCATTTTTGAGAAC | |
| RSma1− | TCCCCCGGG ATGACTTCGGTTCTTGATTC | |
| A5′RACE-3621 | 3621-3594 | CTGTTTCCCAGGGTCCGATGCCGGTTCG |
Nucleotide positions relative to the complete sequence of RCNMV RNA1. * and ***, primers specific for the vector sequences (pUC118). **, primer with an SmaI site and reverse-complement sequences of RNA1 from the 3′ end to nt 3871.
The altered sequences are underlined, and restriction enzyme sites are in bold and italics.
(i) pUCR1-p27fs.
pUCR1 was cut at an EcoRI site in the p27 ORF, end filled with T4 DNA polymerase, and relegated.
(ii) pUCR1-d3′SLA-L.
Two DNA fragments were amplified from pUCR1 using two primer pairs, A1+3380 plus d3′-2ii− and M4 plus d3′-2ii+. Then, a PCR fragment was amplified from a mixture of these two fragments using the primer pair A1+3380 plus M4, digested with MluI and SmaI, and inserted into the corresponding region in the 3′ UTR of pUCR1.
(iii) pUCR1-m1, pUCR1-m2, pUCR1-m3, pUCR1-m4, pUCR1-m5, pUCR1-m6, pUCR1-m7, pUCR1-m8, pUCR1-m9, pUCR1-m10, pUCR1-m11, pUCR1-m12, pUCR1-m13, pUCR1-m14, and pUCR1-m1/m3.
DNA fragments were amplified by PCR from pUCR1. The primer pairs used were AC1+3036 plus one each of the following: m1−, m2−, m3−, m4−, m5−, m6−, m7−, m8−, m9−, m10−, m11−, m12−, m13−, m14−, and m1/m3−. Another primer, 3′R/C1, was used together with one each of the following: m1+, m2+, m3+, m4+, m5+, m6+, m7+, m8+, m9+, m10+, m11+, m12+, m13+, m14+, and m1/m3+. Each recombinant PCR product was amplified with the primer pair AC1+3036 and 3′R/C1, digested with MluI and SmaI, and inserted into the corresponding region in the 3′ UTR of pUCR1.
(iv) pUCR1-m1/d3′SLF.
Two DNA fragments were amplified from pUCR1-d3′SLF using two primer pairs, AC1+3115 plus m1− and Aus4291− plus m1+. Then, a PCR fragment was amplified from a mixture of these two fragments using the primer pair AC1+3115 plus Aus4291−, digested with MluI and SmaI, and inserted into the corresponding region in the 3′ UTR of pUCR1.
(v) pA1-R-Luc-A1.
Two DNA fragments were amplified by PCR from pR1-5′-XbS using two primer pairs, SacI/T7 plus 5′A1/R-Luc/L and R-Luc/3′A1/R plus 3′A1end/L. Other DNA fragments were amplified from pSP64-RLUC (27) using the primer pair 5′A1/R-Luc/R and R-Luc/3′A1/L. These amplified DNA fragments were mixed and further amplified by PCR using the primer pair SacI/T7 plus 3′A1end/L. The amplified DNA fragment was digested with SacI and SacII and used to replace the corresponding region of pR1-5′-XbS.
(vi) pAR/1f/FA and pAR/m1/FA.
DNA fragments were amplified by PCR from pA1-RLuc-A1, and the primer pairs used were SacI/T7 plus one each of the following: R-1f− and R-m1−, respectively. Another primer, 1fNcoI−, was used together with one each of the following: R-1f+ and R-m1+, respectively. These recombinant PCR products were amplified with the primer pair SacI/T7/R plus 1fNco1−, digested with SacI and NcoI, and used to replace the corresponding region of pR1-5′-XbS, respectively.
(vii) pR/1f/F and pR/m1/F.
To construct pR/1f/F and pR/m1/F, DNA fragments were amplified by PCR using the primer pairs pR/1f/F+ plus pR/1f/F− from pAR/1f/FA and pAR/m1/FA, respectively. These fragments were digested with HindIII and AccI and used to replace the corresponding region of p3′-8 (27), respectively.
(viii) p1f-Luc and pm1-Luc.
To construct p1f-Luc and pm1-Luc, DNA fragments were amplified from pUCR1 using two primer pairs, 1fNco1− plus T7/TC5′ and 1fNco1− plus 1fm1+, respectively. These amplified fragments were digested with SacI and NcoI and used to replace the corresponding region of pR1-5′-XbS.
(ix) pSSL/R1-5′-XbS.
A DNA fragment was amplified from pR1-5′-XbS using the primer pair SSL/A1/R plus 3′R/C1. To complete the DNA fragments containing the T7 promoter and the SacI site, this DNA fragment was amplified by PCR again with the primer pair T7/9/SSL/R plus 3′R/C1. The final amplified product was digested with SacI and SacII and used to replace the corresponding region of pR1-5′-XbS.
(x) pSSL-Luc.
Two DNA fragments of the 5′ UTR of RCNMV RNA1 with a stable stem-loop structure and Luc ORF with the 3′UTR of RNA1 were amplified by PCR from pR1-5′-XbS using two primer pairs, SacI/T7 plus A1/SSL/2L and SSL/14/Luc/2R plus 3′R/C1, respectively. Both DNA fragments were used as a template for the PCR and were amplified using primers SacI/T7 and 3′R/C1. The final amplified product was digested by EcoRV and SacII and used to replace the corresponding region of pSSL/R1-5′-XbS. Because we failed to construct this plasmid with the expected sequences in the stable stem-loop site using the E. coli DH5α strain, this plasmid was amplified in the E. coli stbl4 strain (Invitrogen ElectroMAX Stbl4 cells). E. coli was cultured in a Tris-borate medium at 30°C.
(xi) pSR1f and pSR1f/TE-Lm1.
DNA fragments were amplified from pUCR1 and pRC1mL1 (27) by using the primer pair T7/TC5′ plus M4. The PCR products were digested with SacI and SmaI and used to replace the corresponding regions of pUC118.
(xii) pSR1f-m1.
A DNA fragment was amplified from pUCR1 by using the primer pair 1b-m1(+) plus M4. The PCR product was digested with SacI and SmaI and used to replace the corresponding region of pUC118.
(xiii) pBSRC1/5′P.
pBSRC1/5′P was generated by ligating the SacI/XhoI fragment of pUCR1 into EcoRV/XhoI-digested pBluescript II KS(+) (Stratagene, La Jolla, CA) in antisense orientation behind the T3 promoter. DNA fragments with incompatible ends were ligated after blunting with T4 DNA polymerase.
(xiv) pBSRC1/midP.
pBSRC1/midP was generated by ligating the PstI/XhoI fragment of pUCR1 into PstI/XhoI-digested pBluescript II KS(+) (Stratagene) in antisense orientation behind the T7 promoter.
(xv) pBSCR1P.
pBSCR1P was generated by ligating the HindIII/BamHI fragment of pCR-R1 into HindIII/BamHI-digested pBluescript II KS(+).
(xvi) pR-lucN.
A PCR fragment was amplified from pA1-R-luc-A1 by using the primer pair T7NSR+ plus RSmaI−. The PCR product was digested with SacI and SmaI and used to replace the corresponding region of pUC118.
RNA preparation.
All RNA transcripts except for CRSV RNA1, R/1f/F, and R/m1/F were synthesized in vitro from XmaI-linearized plasmids with T7 RNA polymerase. CRSV RNA1 was synthesized from HindIII-linearized pCR-R1 with T7 polymerase. R/1f/F and R/m1/F were synthesized from AccI-linearized plasmids with T7 polymerase. All transcripts were purified with a Sephadex G-50 fine column (GE Healthcare UK, Ltd., Buckinghamshire, United Kingdom). The RNA concentration was determined spectrophotometrically and its integrity verified by 1% agarose gel electrophoresis. All transcripts are named for their parent plasmids minus the “pUC,” “p,” or “pLUC” prefix. Capped transcripts were prepared using the ScriptCap m7G capping system (Epicentre Biotechnologies, Madison, WI) to achieve efficient capping rates.
Protoplast experiments.
Tobacco BY-2 protoplast experiments were performed as previously described (14). Briefly, RNA1 (1.1 pmol) with or without RNA2 (2.9 pmol) was suspended in 0.2 ml cold morpholineethanesulfonic acid buffer and mixed with 0.6 ml of BY-2 protoplast solution (1.67 × 106 cells/ml) before electroporation using a Pulse Controller Plus (Bio-Rad). Protoplasts were incubated at 17°C for 18 h in the dark. Total RNAs were subjected to Northern blot analysis, as previously described (14). In the SR1f competition assay, approximately 8 pmol of R1-5′-XbS or pA60(+) was electroporated with an eightfold molar excess of SR1f or SR1f/TE-Lm1 into BY-2 protoplasts. The protoplasts were incubated at 17°C in the dark for 6 h. The cells were lysed in passive lysis buffer (Promega) and subjected to one freeze-thaw cycle. Aliquots of the cell lysate were assayed with the luciferase reporter assay system (Promega).
Evacuolated BY-2 protoplast lysate (BYL) experiments.
Preparation of the cell extracts of evacuolated BY-2 protoplasts and the in vitro translation/replication reaction were described previously (14, 18). In the luciferase assays, aliquots of samples were diluted with 100-fold passive lysis buffer (Promega), and aliquots of these samples were assayed with the luciferase reporter assay system (Promega). See the figure legends for more information on each experiment.
Inoculation of Nicotiana benthamiana plants.
The inoculation of N. benthamiana plants was performed as previously described (25), except for using uncapped transcripts.
Northern and Western blot analysis.
Total RNAs extracted from N. benthamiana plants, BY-2 protoplasts, and BYL were subjected to Northern blot analysis, as previously described (14, 25). The digoxigenin (DIG)-labeled RNA probes specific to the 3′ UTRs of RCNMV RNA1 (RNA1/3′P), the negative-strand RNAs of RCNMV RNA1, the 3′ UTRs of RCNMV RNA2, the negative-strand RNAs of RCNMV RNA2, and the firefly luciferase ORF (F-Luc probe) have been described previously (14, 25-27). The DIG-labeled RNA probes specific to the 5′ region of RCNMV RNA1 (RNA1/5′P) and the middle region of RCNMV RNA1 (RNA1/MidP) were transcribed from SmaI-linearized pBSRC1/5′P with T3 RNA polymerase and from XhoI-linearized pBSRC1/midP with T7 RNA polymerase, respectively. The DIG-labeled RNA probe for the Renilla luciferase ORF (R-Luc probe) was transcribed from SmaI-linearized pR-lucN with T7 RNA polymerase. The DIG-labeled RNA probe for the region between nt 3673 and the 3′ end (nt 3890) of RCNMV RNA1 (3′ UTR short probe) was transcribed from BstPI-linearized pSRT1 (27) with T7 RNA polymerase. The DIG-labeled RNA probe for CRSV RNA1 was transcribed from BamHI-linearized pBSCR1P with T7 RNA polymerase. The RNA signals were detected with a luminescent image analyzer (LAS 1000 Plus; Fuji Photo Film, Japan).
Western blot analysis was performed as described previously (44). The signals were detected with a luminescent-image analyzer (LAS 1000 Plus; Fuji Photo Film, Japan), and the signal intensities were quantified with the Image Gauge program (Fuji Photo Film, Japan).
Determination of the 5′ and 3′ends of SR1f by primer extension and 3′-RACE.
The purification of RCNMV SR1f by electrophoresis through a denaturing polyacrylamide gel was performed as described previously (36). Oligonucleotide primer A5′RACE-3621 was used for 5′-end sequencing of SR1f. Sequencing reactions and treatment of the primer extension products with terminal deoxynucleotidyl transferase were performed as described previously (1), except for using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). The 3′-rapid amplification of cDNA ends (RACE) was done as described previously (25).
RESULTS
A small RNA is generated from the 3′-proximal region of RNA1 and packaged into virions in RCNMV-infected cells.
Northern blot analysis using a probe complementary to the 3′ region of RCNMV RNA1 revealed that infected plants and protoplasts contain a large amount of a 400-base RNA (termed SR1f) in addition to the viral genomic and sgRNAs (Fig. 1B). To identify SR1f and to study the biological roles of SR1f, we first investigated whether SR1f might contain any regions other than the 3′ UTR of RNA1. Tobacco BY-2 protoplasts were inoculated with in vitro transcripts of RCNMV RNA1 and RNA2, or RNA1 alone, and total RNA was analyzed by Northern blotting using RNA probes that specifically hybridized to the 5′-proximal (RNA1/5′P), the middle (RNA1/MidP), or the 3′-proximal regions (RNA1/3′P) of RNA1 (Fig. 1A). SR1f was detected only with RNA1/3′P (Fig. 1B), suggesting that SR1f consists mainly of the 3′-proximal region of RNA1. SR1f, unlike CP sgRNA, accumulated in protoplasts inoculated with RNA1 alone (Fig. 1B), indicating no requirement for RNA2 in its generation. This suggests that the mechanism that generates SR1f is different from the mechanism that generates CP sgRNA, which requires an intermolecular interaction between RNA1 and RNA2 (42, 46). Time course experiments consistently showed that SR1f started to accumulate after the initial detection of RNA1 or CP sgRNA. It is noteworthy that a small RNA similar in size to SR1f was detected in purified virions, whereas CP sgRNA was not (Fig. 1B), suggesting that SR1f is packaged into virions. To determine the precise 5′ and 3′ ends of SR1f, we used a primer extension analysis and a 3′-RACE analysis, respectively. Because purified RCNMV virions contain a small RNA that is probably identical to SR1f, as shown in Fig. 1B, we used virion RNAs directly or after separation on a 5% polyacrylamide gel containing 8 M urea as the template for the analysis. These analyses indicated that SR1f had a 3′ end coterminal with RNA1 consisting of 431 nt (data not shown). These results also suggested that SR1f is unlikely to be an RNA replicon.
Negative-strand RNAs of SR1f are not detected in BY-2 protoplasts.
Next, we analyzed the accumulation of negative-strand viral RNAs using full-length genomic RNA1 as a probe. Negative-strand RNAs corresponding to RNA1 or CP sgRNA were detected in protoplasts inoculated with RNA1 and RNA2 (Fig. 1C), supporting a model involving the premature termination mechanism that generates CP sgRNA (42). In contrast, no negative-strand RNA corresponding to the small RNA was detected. Thus, SR1f could be a transcript synthesized from negative-strand RNA1 by an internal initiation mechanism (24) or a degradation intermediate generated by self-cleavage or cleavage by endo- or exoribonucleases.
SR1f is not a transcript but a degradation intermediate of RNA1.
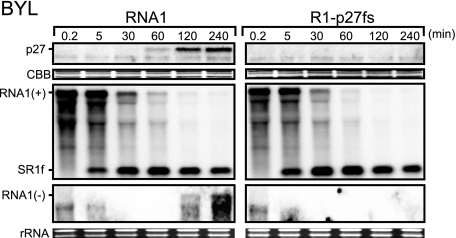
To investigate the mechanism that generates SR1f, we used BYL, which were initially developed for the study of tobamovirus translation and replication (18). BYL is a powerful tool for analysis of the translation and replication mechanisms of RCNMV (14, 26, 29). BYL reflects the cap-independent translation activity of RCNMV RNA1 with a series of mutations in the 3′ UTR in BY-2 protoplasts (14, 26). Furthermore, in BYL, several RNA viruses can replicate (18), and negative-strand RNAs of RCNMV are easily detected following the translation of viral replication proteins (14, 29). Therefore, we used BYL to investigate the timing of the accumulation of SR1f and the generation mechanism of SR1f. First, we incubated uncapped RNA1 in BYL and analyzed the accumulations of p27, positive- and negative-strand RNA1, and SR1f at different time points after incubation, using Western and Northern blotting, respectively. p27 started to be detected 30 min after incubation, and negative-strand RNA1 started to be detected at 120 min (Fig. 2). Surprisingly, SR1f was detected 5 min after incubation or earlier (Fig. 2; data not shown). This suggested no link between viral RNA replication and the generation of SR1f. To test this, we used an RNA1 mutant (R1-p27fs) that does not produce functional replication proteins. SR1f accumulated with the incubation of R1-p27fs to a level similar to that after the incubation of wild-type RNA1 in BYL (Fig. 2). The stability of R1-p27fs was not significantly different from that of RNA1 (Fig. 2). These results indicated that viral replication proteins were not necessary for the generation of SR1f. Thus, SR1f is not a transcript but a degradation intermediate of RNA1.
FIG. 2.
SR1f is not a transcript but a degradation intermediate of RNA1. Time course analysis of the accumulations of viral replicase proteins (p27) and viral RNAs in BYL incubated with RNA1 and R1-p27fs. RNA1 or R1-p27fs (2 pmol) was incubated in 110 μl of BYL translation reaction mixture at 17°C. At different times after incubation, 10-μl aliquots were used for Northern and Western blotting, respectively.
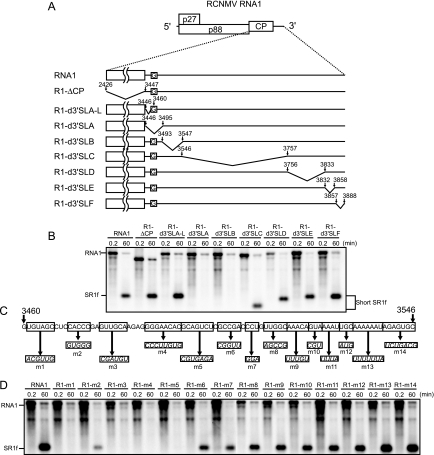
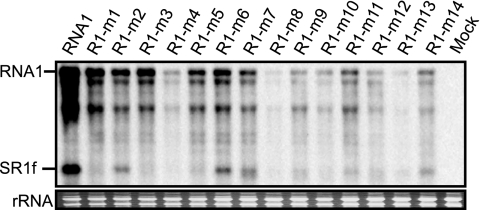
Fifty-eight nucleotides in the 3′ UTR of RNA1 are necessary and sufficient for the generation and accumulation of SR1f.
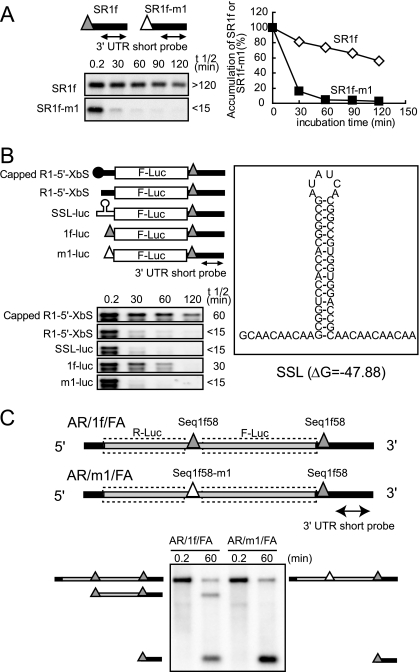
To define the nucleotide sequences required for the generation and accumulation of SR1f, RNA1 mutants with deletions or nucleotide substitutions (Fig. 3A and C) were incubated in BYL and the accumulation of SR1f was analyzed by Northern blotting (Fig. 3B and D). Nucleotide sequences from nt 3460 to 3503 in RNA1, except for five nucleotides from nt 3470 to 3474, were essential for the accumulation of SR1f (Fig. 3). We will refer below to the 58 nt from nt 3460 to 3517 of RNA1 as Seq1f58; this is a core nucleotide sequence required for SR1f generation (Fig. 4A).
FIG. 3.
Essential regions required for the generation of SR1f. (A) Schematic representation of deleted regions in the CP ORF and the 3′ UTR of RCNMV RNA1. The boldface lines indicate virus-derived sequences with the nucleotide numbers at the 5′ and 3′ ends. The bent lines indicate deleted regions. The Gs in the open boxes show the nucleotide residue in the 5′ end of SR1f. (B) Accumulation of small RNAs (SR1f) generated from RNA1 deletion mutants in BYL at 0.2 and 60 min after incubation. Genome RNAs and small RNAs (SR1f) were detected by RNA1/3′P. (C) Schematic representation of nucleotide substitution mutations in the 3′ UTR of RNA1. Italics in the open boxes show altered sequences. (D) Accumulation of small RNAs (SR1f) generated from RNA1 mutants in BYL at 0.2 and 60 min after incubation. Genome RNAs and small RNAs (SR1f) were detected by the 3′ UTR short probe.
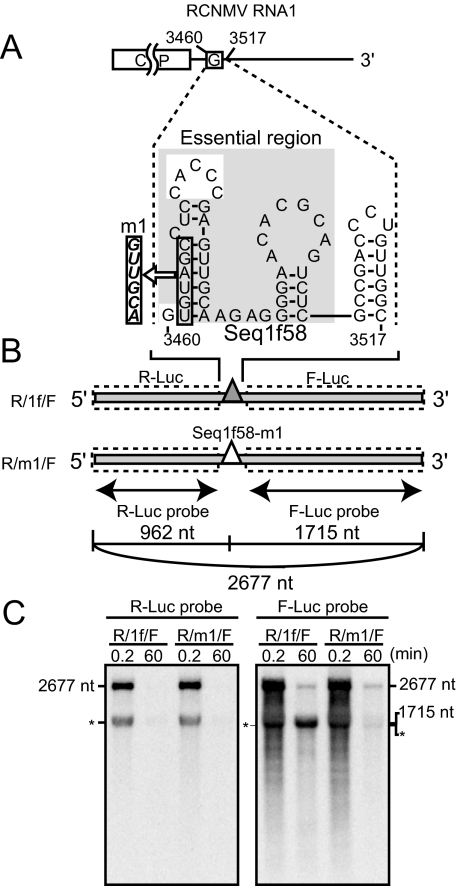
FIG. 4.
Seq1f58 is sufficient for the accumulation of its downstream nonviral RNA fragments. (A) Schematic representation of the essential nucleotide sequences required for the accumulation of SR1f (Seq1f58). The G's in the open boxes indicate the nucleotide residue at the 5′ end of SR1f. The nucleotide sequence in the gray shaded box indicates the essential nucleotide sequence required for the accumulation of SR1f. A secondary structure of Seq1f58 was predicted by using mfold (version 2.3) (50, 58) at 17°C. (B) Schematic representation of dicistronic mRNA. Seq1f58 was inserted between two different reporter ORFs, Renilla luciferase and firefly luciferase (R/1f/F). R/1f/F is 2,677 nt in length. The number of nucleotide residues from the 5′ end to just before the first guanine residue in the Seq1f58 is 962, and the number from the first guanine residue in the Seq1f58 to the 3′ end is 1,715. The filled and open triangles show Seq1f58 and Seq1f58 with the m1 mutation (Seq1f58-m1), respectively. The regions covered by RNA probes are shown as thin arrows. (C) Accumulation of degradation intermediates derived from R/1f/F or R/m1/F in BYL at 0.2 and 60 min after incubation. R/m1/F has the m1 mutation in Seq1f58. R/1f/F or R/m1/F (0.55 pmol) was incubated in 50 μl BYL at 17°C. Total RNA was extracted at 0.2 min and 60 min after incubation and used for Northern blotting with DIG-labeled RNA probes specific for the F-Luc ORF and R-Luc ORF. The sizes of the bands are indicated on the right or the left side. The asterisks indicate an unexpected band which was detected by both the R-Luc and F-Luc probes at 0.2 or 60 min after incubation.
In addition, we examined whether the 5′-proximal stem structure predicted in Seq1f58 (Fig. 4A) is important for the generation of SR1f by introducing compensatory mutations into R1-m1. This RNA1 mutant, R1-m1/m3, with the stem structure restored, failed to accumulate SR1f (data not shown). This suggested that nucleotide sequences in the stem rather than the predicted structure might be important for the generation of SR1f.
Next, to examine whether Seq1f58 would be sufficient for the generation and accumulation of its downstream nonviral RNA fragments containing Seq1f58, we designed a dicistronic RNA (R/1f/F), in which Seq1f58 was inserted between two different ORFs encoding Renilla luciferase (R-Luc) and firefly luciferase (F-Luc), respectively (Fig. 4B). If Seq1f58 works, an RNA fragment of 1,715 nt, which contains both Seq1f58 and the F-luc ORF, should be detected. As a control, we used R/m1/F, which has an m1 mutation in Seq1f58 (Fig. 3C and Fig. 4A). This mutation in RNA1 caused a loss of ability to accumulate SR1f (Fig. 3D and 4A). As expected, RNA fragments with a size of 1,715 nt were detected on incubation with R/1f/F but not after incubation with R/m1/F in BYL using a probe for the F-Luc ORF (Fig. 4C). On the other hand, degradation intermediates (962 nt) were not detectable using a probe for the R-Luc ORF on the inoculation of R/1f/F (Fig. 4C). These results indicated that Seq1f58 is sufficient for the accumulation of the 3′ portion of the transcript, suggesting that Seq1f58 protects its downstream nucleotide sequences from 5′→3′ decay.
Seq1f58 protects its downstream nucleotide sequences from 5′→3′ decay.
If Seq1f58 could protect its downstream nucleotide sequences from 5′→3′ decay, then SR1f must be more stable than SR1f derivatives with mutations in Seq1f58. To test this, we compared the stabilities of in vitro-transcribed SR1f and SR1f-m1 that had the m1 mutation in Seq1f58. The half-life of SR1f was longer than 240 min, whereas that of SR1f/m1 was shorter than 15 min (Fig. 5A). This result suggested that wild-type Seq1f58 stabilizes SR1f by protecting against 5′→3′ exoribonucleases.
FIG. 5.
Seq1f58 protects its downstream nucleotide sequences from 5′→3′ decay. (A) Stability of SR1f and mutant SR1f (SR1f-m1) in BYL. The filled and open triangles with bold lines indicate SR1f and SR1f-m1, respectively. SR1f or SR1f-m1 (4.3 pmol) was incubated in 100 μl of BYL at 17°C. Total RNA was extracted at the indicated times and used for Northern blotting with the 3′ UTR short probe. The signal intensities were quantified with the Image Gauge program (Fuji Photo Film, Japan). RNA half-life (t 1/2) was calculated with Microsoft Excel. (B) Stability of mRNAs with different RNA structures at the 5′ UTRs in BYL. mRNAs (0.53 pmol) were incubated in 30 μl of BYL at 17°C. Total RNA was extracted at the indicated times after incubation and used for Northern blotting with RNA1/3′P. For other conditions, refer to the legend of panel A. (C) Accumulations of degradation intermediates from AR/1f/FA and AR/m1/FA. RNAs were extracted at the indicated times and subjected to Northern blotting with the 3′ UTR short probe. Schematic representations of RNAs with Seq1f58 and its mutant (AR/1f/FA and AR/m1/FA) are shown above. AR/1f/FA is an R/1f/F derivative with the 5′ and 3′ UTRs of RCNMV RNA1. AR/m1/FA has an m1 mutation in Seq1f58. The filled and open triangles indicate Seq1f58 and Seq1f58-m1, respectively. The bold gray lines indicate nonviral sequences. The bold dark lines show the 5′ and 3′ UTRs of RCNMV RNA1. The rectangles with broken lines show the R-luc and F-luc ORFs.
To further confirm that Seq1f58 stabilizes its downstream RNA sequences, we compared the stabilities of RNAs with different RNA structures in the 5′ UTR but with the same 3′UTR (Fig. 5B). R1-5′-XbS was a reporter Luc mRNA with the 5′ and 3′ UTRs of RNA1 (27). SSL-luc had a stable stem-loop structure in the 5′ UTR. 1f-luc and m1-luc had Seq1f58 with or without the m1 mutation as the 5′ UTR, respectively. Uncapped transcripts of these four RNAs and capped R1-5′-XbS were incubated in BYL, and accumulations of full-length transcripts were monitored by Northern blotting using a probe specific to the 3′ UTR of RNA1. 1f-luc was the most stable (half-life, 30 min) among the uncapped transcripts tested, although the stability was less than that of capped R1-5′-XbS (half-life, 60 min) (Fig. 5B). These results indicated that Seq1f58 at the 5′ end confers stability to the 3′ portion of the transcript.
Next, to examine whether the 5′-proximal Seq1f58 would stall 5′→3′ exoribonucleases and consequently decrease the accumulation of 3′-proximal RNA fragments, we used an RNA with two Seq1f58 sequences (AR/1f/FA) (Fig. 5C). As expected, the accumulation of 3′-proximal RNA fragments was lower in AR/1f/FA than in AR/m1/FA, which had the m1 mutation in the 5′-proximal Seq1f58 (Fig. 5C).
In addition, the accumulation of SR1f was lower with the incubation of capped RNA1 than with uncapped RNA1 in BYL (data not shown). This suggested that the presence of either cap structure or Seq1f58 at the upstream region of the second Seq1f58 decreased the generation of SR1f from the downstream Seq1f58. Therefore, we concluded that Seq1f58 protects its downstream RNA sequences from degradation by 5′→3′ exoribonucleases. SR1f is likely a leftover of the activity of 5′→3′ exoribonucleases.
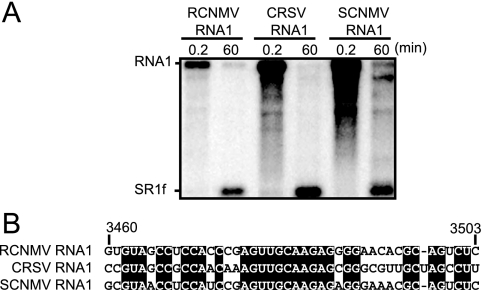
SR1f-like small RNAs are generated from the 3′ UTR of other dianthovirus RNA1.
To investigate whether SR1f-like RNA would be generated from RNA1s of other dianthoviruses, we incubated in vitro-transcribed RNA1s of CRSV and SCNMV in BYL. SR1f-like RNAs were detected using probes specific to their 3′ UTRs (Fig. 6A). These small RNAs were also detected in N. benthamiana and cowpea protoplasts infected with these viruses (data not shown). Comparing the essential sequences in Seq1f58 with the corresponding sequences of CRSV and SCNMV, 28 out of 43 nt (65%) were identical (Fig. 6B). These results suggest that the SR1f-like RNAs detected after the incubation of CRSV and SCNMV RNA1 are also degradation intermediates like SR1f.
FIG. 6.
SR1f-like RNAs are generated from the 3′ UTRs of other dianthoviruses. (A) Accumulations of SR1f and SR1f-like small RNAs generated from RCNMV, CRSV, and SCNMV RNA1 in BYL at 0.2 and 60 min after incubation. Genome RNAs and small RNAs were detected by mixed probes, which are specific to the 3′ UTRs of RNA1 of these viruses. (B) Comparison of the essential sequences for the accumulation of SR1f in RCNMV RNA1 with the corresponding sequences of two other dianthoviruses.
SR1f trans inhibits both cap-independent and cap-poly(A)-dependent translation in vitro and in vivo.
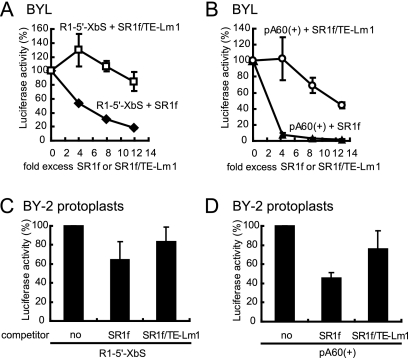
SR1f does not contain any protein-coding regions but contains RNA elements required for both cap-independent translation (14, 27) and negative-strand RNA synthesis (14). These RNA elements contained in SR1f may function as riboregulators of translation and RNA synthesis. First, we investigated the effects of SR1f on 3′TE-DR1-mediated cap-independent translation in vitro using R1-5′-XbS (see Fig. 5). The addition of in vitro-transcribed SR1f suppressed the translational activity of R1-5′-XbS in BYL (Fig. 7A). However, SR1f/TE-Lm1, which possesses a defective 3′TE-DR1, did not effectively inhibit the translational activity of R1-5′-XbS (Fig. 7A). This inhibition was dependent on the concentrations of SR1f added. A fivefold molar excess of SR1f inhibited the translational activity of R1-5′-XbS by 50% (Fig. 7A), whereas the same molar excess of SR1f/TE-Lm1 did not inhibit but rather upregulated the translational activity of R1-5′-XbS (Fig. 7A). We also tested whether SR1f affects cap-dependent translation using capped mRNA with poly(A) pA60(+) in BYL (Fig. 7B). A fourfold molar excess of SR1f inhibited the translational activity of pA60(+) by 95%, while the same molar excess of SR1f/TE-Lm1 did not effectively inhibit the translational activity of pA60(+) (Fig. 7B).
FIG. 7.
Effects of SR1f on cap-dependent and cap-independent translation in BYL and BY-2 protoplasts. (A and B) SR1f trans inhibited cap-independent and cap-dependent translation in BYL. R1-5′-XbS (2.7 pmol) and pA60(+) (2.5 pmol) were incubated with the indicated amount of SR1f or SR1f/TE-Lm1 in 25 μl of BYL at 17°C for 120 min. The luciferase activity of R1-5′-XbS or pA60(+) with no competitor was defined as 100%. The error bars indicate standard deviations. (C and D) BY-2 protoplasts were inoculated with R1-5′-XbS (8 pmol) and pA60(+) (8 pmol) with an eightfold molar excess of SR1f or SR1f/TE-Lm1. The luciferase activities of R1-5′-XbS and pA60(+) without competitor were defined as 100%, respectively. The error bars indicate standard deviations.
Next, to investigate whether SR1f would trans inhibit translation in vivo, BY-2 protoplasts were inoculated with R1-5′-XbS or pA60(+) together with eightfold molar excesses of SR1f or SR1f/TE-Lm1. SR1f inhibited the translational activity of R1-5′-XbS and pA60(+) by 40% and 60%, respectively, whereas SR1f/TE-Lm1 did not effectively inhibit the translational activity of either R1-5′-XbS or pA60(+) (Fig. 7C and D).
Together, these results indicated that SR1f, which has a functional 3′TE-DR1, suppressed both 3′TE-DR1-mediated cap-independent translation and cap/poly(A)-dependent canonical translation in vitro and in vivo. Interestingly, the results also suggest that SR1f suppresses cap-poly(A)-dependent canonical translation more efficiently than that of 3′TE-DR1-mediated cap-independent translation (Fig. 7).
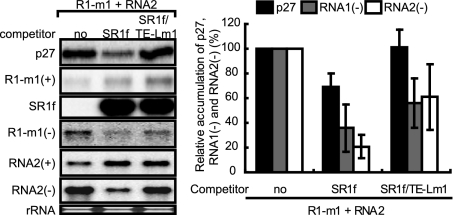
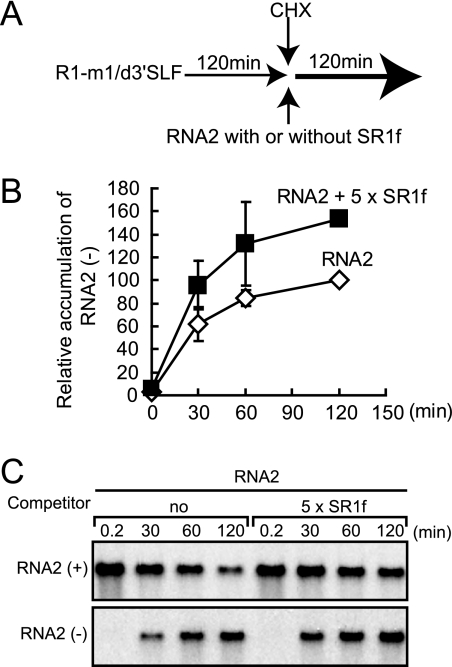
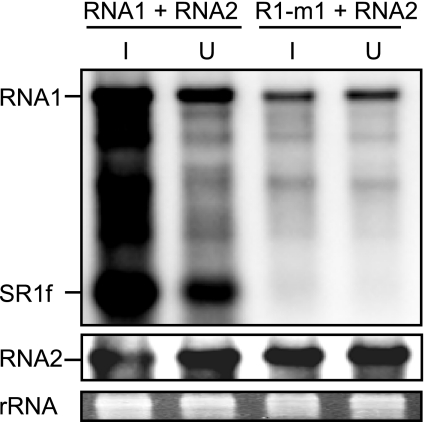
SR1f trans inhibits negative-strand RNA synthesis via the repression of replicase protein production but not via the competition of replicase proteins for cis elements in vitro.
Next, we tested the ability of SR1f to inhibit negative-strand RNA synthesis of RCNMV genomic RNAs, RNA1 and RNA2, in BYL. To distinguish the effects of SR1f supplied in trans from those of SR1f generated de novo from RNA1 during incubation, we used R1-m1 that generates no SR1f (Fig. 3C and 4A) in place of wild-type RNA1. Note that R1-m1 translated p27 efficiently and accumulated similar levels of negative-strand RNAs as wild-type RNA1 in BYL (data not shown). R1-m1 and RNA2 were incubated with a fivefold molar excess of SR1f or SR1f/TE-Lm1 in BYL. SR1f decreased the accumulation of p27 by approximately 30% and inhibited the negative-strand RNA synthesis of RNA1 and RNA2 by 60% and 80%, respectively (Fig. 8). SR1f/TE-Lm1 did not inhibit p27 accumulation, whereas it decreased the accumulation of negative-strand RNA of both RNA1 and RNA2 by 40% (Fig. 8). Therefore, these results also suggested that SR1f might compete with RNA1 and RNA2 for negative-strand RNA synthesis, because SR1f contains promoter sequences in the 3′ end conserved between RNA1 and RNA2, which are essential for negative-strand RNA synthesis (14, 48; M. An, H.-O. Iwakawa, and T. Okuno, unpublished data). These elements could recruit viral or host proteins required for negative-strand RNA synthesis, although p88 is required in cis for the replication of RNA1 (14). To examine whether SR1f would trans inhibit negative-strand RNA synthesis of RCNMV genomic RNAs by the competition of replicase proteins via these RNA elements, we separated the replication phase from the translation phase in BYL and analyzed the effects of SR1f on negative-strand RNA synthesis. First, an RNA1 mutant (R1-m1/d3′SLF) that lacks RNA elements required for SR1f generation and negative-strand RNA synthesis was incubated in BYL for 120 min for the production of viral replicase proteins (p27 and p88). Then, cycloheximide (CHX) and RNA2 with or without a fivefold molar excess of SR1f were added and incubated for an additional 120 min. The use of RNA2 allowed us to minimize the possible effects of the interaction between translation factors and RNA2, because RNA2 does not function as an effective mRNA in BYL and in protoplasts unlike RNA1; the cap-independent translation of RNA2 is strongly linked to RNA2 replication (26). The addition of SR1f did not decrease the accumulation of negative-strand RNAs of RNA2 (Fig. 9), suggesting that SR1f alone does not interact with replication factors and that the decrease in negative-strand RNA synthesis by either SR1f or SR1f/TE-Lm1 (Fig. 8) was not caused by the competition of replicase proteins (Fig. 8). The increase observed in the accumulation of negative-strand RNA2 in the presence of SR1f (Fig. 8 and 9C) could probably be attributed to the stabilization of input RNA2 by SR1f. These results suggest that SR1f trans inhibits negative-strand RNA synthesis of RCNMV via the repression of replicase protein production but not via the competition of replicase proteins by its promoter sequence.
FIG. 8.
SR1f trans inhibits negative-strand RNA synthesis of RNA1 and RNA2 in BYL. Accumulations of p27, positive-strand R1-m1, SR1f or SR1f/TE-Lm1, negative-strand R1-m1, positive-strand RNA2, and negative-strand RNA2 in BYL (25 μl) incubated with R1-m1, RNA2 (0.75 pmol), and a fivefold molar excess of SR1f or SR1f/TE-Lm1 at 17°C for 240 min (left panel). Total RNAs and proteins were extracted at 240 min after incubation and used for Northern and Western blotting, respectively. Quantification of the accumulations of p27 and negative-strand RNAs of R1-m1 and RNA2 was performed with the Image Gauge program (Fuji Photo Film, Tokyo, Japan) (right panel). Accumulations of p27, the negative-strand RNAs of R1-m1, and RNA2 in BYL incubated with R1-m1 and RNA2 were defined as 100%. The error bars indicate standard deviations.
FIG. 9.
SR1f does not decrease the accumulation of negative-strand RNA2 in BYL when the replication phase is separated from the translation phase. (A) First, an RNA1 mutant (R1/m1/d3′SLF) that lacks RNA elements required for negative-strand RNA synthesis and SR1f generation was incubated in BYL at 17°C for 120 min for the production of replicase proteins (thin horizontal arrow). Then, cycloheximide (CHX) was added to shut off the translation of R1/m1/d3′SLF, and RNA2 was incubated with or without a fivefold molar excess of SR1f in BYL at 17°C for an additional 120 min (bold horizontal arrow). (B and C) Total RNA was extracted at the indicated times and used for Northern blotting to detect positive- and negative-strand RNA2. The time when CHX was added was defined as 0 min. The Image Gauge program (Fuji Photo Film, Tokyo, Japan) was used for the quantification of negative-strand RNA2. The error bars indicate standard deviations.
In addition, SR1f/TE-Lm1 or both SR1f and SR1f/TE-Lm1 could also affect frameshift efficiency for p88 production, because accumulations of negative-strand RNAs in both RNA1 and RNA2 were lower than the control despite the same level of p27 accumulation (Fig. 8).
SR1f is not essential for replication and cell-to-cell or long-distance movement.
To investigate the biological functions of SR1f in RCNMV infection, first we tested RNA1 mutants with nucleotide substitutions between nt 3460 and 3495 in Seq1f58 (see Fig. 3) for the ability to replicate in single cells. To test this, BY-2 protoplasts were inoculated with RNA1s, and viral RNA accumulation was analyzed at 18 h after inoculation. All RNA1 mutants tested underwent replication but less efficiently than wild-type RNA1 (Fig. 10). This indicated that SR1f was not essential for the replication of RNA1. In addition, we failed to observe any associations between the accumulated levels of viral genomic RNAs and the ability to generate viral small RNAs. This result confirmed our previous results that the 5′-proximal region including Seq1f58 in the 3′ UTR of RNA1 contains cis-acting RNA elements required for the efficient replication of RNA1 (14).
FIG. 10.
Accumulations of RNA1 mutants with nucleotide substitutions in the 3′ UTR of RNA1 (Fig. 3C) in BY-2 protoplasts. Inoculated protoplasts were incubated at 17°C for 18 h. Total RNA was extracted from protoplasts and used for Northern blotting with the 3′ UTR short probe. For other conditions, refer to the legend for Fig. 1.
Finally, to investigate the functions of SR1f in the infection of plants, we inoculated N. benthamiana plants with several SR1f-deficient RNA1 mutants including R1-m1 together with RNA2 and analyzed viral RNAs in inoculated and uninoculated upper leaves by Northern blotting 10 days after inoculation. Systemic mosaic symptoms were induced, and viral RNA1 and RNA2 accumulated in both the inoculated and uninoculated upper leaves in the plants inoculated with SR1f-deficient RNA1 mutants, although the accumulated levels of viral RNA were lower than those seen following inoculation with wild-type RNAs (Fig. 11; data not shown). These results indicate that SR1f is not essential for virus cell-to-cell and long-distance movement in N. benthamiana.
FIG. 11.
Accumulations of RNA1, RNA2, and SR1f in inoculated (I) and uninoculated (U) upper leaves of N. benthamiana plants. The plants were inoculated with in vitro RNA transcripts and maintained at 17°C for 10 days. For other conditions, refer to the legend for Fig. 1.
DISCUSSION
Possible mechanisms for the generation of SR1f in the host cell.
Previously, we have predicted that SR1f would be an sgRNA, because SR1f accumulates abundantly in host cells following the accumulation of genomic RNAs during RCNMV infection, although it is packaged into virions unlike CP sgRNA. The present results show that SR1f is not an sgRNA but a degradation intermediate of RNA1 (Fig. 2). As far as we are aware, this is the first report of a virus-derived degradation intermediate that accumulates abundantly in the host cells, suppresses both cap-independent and cap-dependent translation, and is packaged into virions. Our results indicated that an RNA fragment of 58-nt sequences (Seq1f58) in the 3′ UTR of RNA1 plays an essential role in the generation and accumulation of SR1f by conferring stability to its downstream nucleotide sequences (Fig. 4 and 5). The lack of generation of SR1f from RNA1 in the absence of plant extracts (BYL) or in boiled BYL suggests the requirement of cellular proteins for the generation of SR1f (H.-O. Iwakawa and T. Okuno, unpublished results). Seq1f58 may protect its downstream nucleotide sequences from 5′→3′ exoribonucleases, resulting in the accumulation of SR1f (Fig. 5), although we cannot rule out the possible involvement of endoribonucleases in the early step of SR1f generation, followed by digestion of the 5′ portion of the RNA with 3′→5′ or 5′→3′ exoribonucleases or both. However, we think that endoribonucleases are not mainly involved in the initial step of SR1f generation, because the insertion of Seq1f58 upstream of SR1f decreased the accumulation level of SR1f (Fig. 5C).
An important question is how Seq1f58 protects its downstream nucleotide sequences from degradation. In yeast, poly(G) tracts or a highly stable RNA structure protects its downstream nucleotide sequences from a 5′→3′ exoribonuclease (XRN1p), resulting in the accumulation of the 3′ portion of the RNA (31, 49). However, poly(G) tracts do not work effectively as a 5′→3′ exoribonuclease stopper in higher eukaryotes, including plants, except for the interior of chloroplasts (7, 8, 15). Plant ribonucleases including XRNs may be more robust than those of yeast, or additional proteins such as RNA helicases might help those plant enzymes to function by resolving the structures formed by poly(G) sequences so that they can progress through the structure (15).
Unlike Seq1f58, a stable stem-loop structure (ΔG = −47.88 kcal/mol at 17°C) did not stabilize RNA in BYL (Fig. 5B), suggesting that strong secondary structures alone are insufficient to protect RNA from 5′→3′ exoribonucleases. The predicted secondary structure of Seq1f58 (Fig. 4A) was not as strong (ΔG = −28.83 kcal/mol at 17°C). Furthermore, compensatory mutations introduced into R1-m1 to restore the predicted stem structure failed to generate SR1f from the RNA1 mutant (R1-m1/m3). These results suggest that the strength of the RNA secondary structures at the 5′ end is not a main factor required for the Seq1f58-mediated stabilization of its downstream nucleotide sequences but rather that host factors, which might bind to Seq1f58, are involved in the generation and stabilization of its downstream nucleotide sequences. Thus, our results suggest that Seq1f58 is a novel RNA cis element that can stabilize its downstream nucleotide sequences in plant cells similar to the poly(G) tract in yeast.
Possible functions of SR1f in RCNMV infection.
SR1f started to accumulate later than the genomic and CP sgRNAs in RCNMV-infected BY-2 protoplasts. This suggests a possible role for SR1f in the later stage of virus infection. SR1f contains no ORF initiated with an AUG start codon, suggesting that SR1f does not function as mRNA. At present, we do not know the real roles of SR1f in RCNMV infection. However, the present results obtained with protoplasts and BYL in vitro systems present several possible functions of SR1f as a noncoding RNA which may be involved in the regulation of viral gene expression, because SR1f trans inhibited 3′TE-DR1-mediated cap-independent translation both in vivo and in vitro (Fig. 7).
Translational control of virus gene expression by viral small RNAs has been reported for BYDV. A small RNA transcript referred to as sgRNA2, which contains a BYDV-like cap-independent translation element (BTE) and a function-unknown small ORF, is generated in BYDV-infected cells (39). sgRNA2 is thought to be a riboregulator that functions as a switch from the early to late stages of gene expression, because sgRNA2 inhibits the translation of genomic RNA that encodes replicase proteins much more strongly than the translation of sgRNA1 that encodes a structural protein (40). BTE interacts specifically with the cap-binding initiation factor complexes eIF4F and eIFiso4F in wheat germ extract (47). Interestingly, RCNMV 3′TE-DR1 contains the same core 17-nt sequence as BTE (27). This suggests that the inhibition of 3′TE-DR1-mediated cap-independent translation by SR1f could likely result from the sequestration of translation initiation factors such as eIF4F and eIFiso4F. The inhibition of translation by sgRNA2 does not require the translation of a small ORF. Therefore, sgRNA2 and SR1f may be functionally similar in that both inhibit viral protein synthesis, although their mechanisms of generation differ.
Interestingly, SR1f trans inhibited cap/poly(A)-dependent translation more effectively than 3′TE-DR1-mediated cap-independent translation both in vivo and in vitro (Fig. 7). The suppression of host protein synthesis may provide a favorable environment for RCNMV in viral protein synthesis and RNA replication in host cells as reported for many animal viruses (3).
Viral RNA fragments are reported to accumulate in flavivirus (West Nile virus and Japanese encephalitis virus)-infected cells (20, 22, 37, 45). These RNAs are categorized into two types: one type contains the 3′-end sequence of genomic RNA (20, 37), and the other contains negative-sense RNA complementary to the 5′- and the 3′-end sequences of genomic RNA. Although the generation mechanism of the former type of positive-sense small RNA remains unclear, it is possible that the small RNA is generated by cis-element-mediated protection against RNases as with RCNMV SR1f, because a stem-loop structure predicted at the 5′ end of the small RNA may protect RNA from 5′→3′ exoribonucleases in animal cells (20, 37). Because another stem-loop structure at the 3′ end of flavivirus genomic RNA interacts with both viral and cellular proteins, and is required for RNA replication (4-6, 43), the small RNA of flavivirus may function as a negative regulator of RNA replication.
Small RNAs similar to SR1f have been observed for other dianthoviruses, including CRSV, SCNMV, and the Canadian strain of RCNMV (Fig. 6; data not shown). Furthermore, the cap-independent translation enhancer elements with the core 17 nt of RCNMV are conserved among these dianthoviruses (27, 34, 53; H. Nagano and T. Okuno, unpublished results). These imply that the small RNAs are important for the survival of dianthoviruses in nature. To survive and to evolve, viruses must have long-term fitness in both current hosts and new hosts. Moreover, they need the ability to be transmitted to their hosts, the ability to compete or cooperate with other viruses in mixed infections in the hosts, the ability to minimize damage to the hosts, and the ability to replicate efficiently and infect the host systemically (33). Therefore, these small RNAs of dianthoviruses might play important roles in any of these situations.
In addition to translational control, the presence of the small RNAs in virions may imply some roles for the RNA in the packaging of viral RNAs to form virions. Moreover, packaging the small RNA may alter the physical properties of the virions, which will affect virion stability and transmission efficiency through either biological or nonbiological means, including vectors for their survival in nature, although little information is available on the transmission of dianthoviruses by vectors (13). A correlation between the ability of satellite RNA to stimulate the encapsidation of groundnut rosette umbravirus (GRV) by groundnut rosette assistor luteovirus CP and its capacity to promote aphid transmission of GRV has been reported (32).
Novel aspects of our results are as follows: (i) the RNA fragment (SR1f) containing most of the 3′ UTR of RCNMV RNA1 accumulates stably and is packaged into virions in RCNMV-infected cells; (ii) SR1f is not an sgRNA but a degradation intermediate generated by cis-RNA element-mediated protection against 5′→3′ decay, and a 58-nt sequence (Seq1f58) is necessary and sufficient for the protection against 5′→3′ decay; (iii) SR1f trans inhibits both cap/poly(A)-dependent and 3′TE-DR1-mediated cap-independent translation in vivo and in vitro; and (iv) SR1f trans inhibits negative-strand RNA synthesis of RCNMV in vitro by repressing replicase protein production but not by competing with RCNMV RNAs for replicase proteins through cis-acting replication elements.
SR1f seems to play important roles in RCNMV infection and survival, although the precise roles of SR1f in RCNMV infection in nature remains to be solved.
Acknowledgments
We thank S. A. Lommel for the RNA1 and RNA2 cDNA clones of RCNMV Australian strain and the RNA1 cDNA clone of CRSV.
This work was supported in part by grants-in-aid (13306005 and 18208004) from the Japan Society for the Promotion of Science; by a grant-in-aid for scientific research on priority area “Spatiotemporal Network of RNA Information Flow” from the Ministry of Education, Culture, Sports, Science and Technology, Japan; and in part by a grant-in-aid for JSPS fellows.
Footnotes
Published ahead of print on 13 August 2008.
REFERENCES
- 1.Allison, R. F., M. Janda, and P. Ahlquist. 1988. Infectious in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with Brome mosaic virus. J. Virol. 623581-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, M. G., P. C. J. Haasnoot, N. Xu, S. Berenjian, B. Berkhout, and G. Akusjarvi. 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 799556-9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. B. W. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 4.Blackwell, J. L., and M. A. Brinton. 1995. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 695650-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 716433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. J., M. D. Kuo, L. J. Chien, S. L. Hsu, Y. M. Wang, and J. H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 713466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drager, R. G., J. Girard-Bascou, Y. Choquet, K. L. Kindle, and D. B. Stern. 1998. In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 1385-96. [DOI] [PubMed] [Google Scholar]
- 8.Drager, R. G., D. C. Higgs, K. L. Kindle, and D. B. Stern. 1999. 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J. 19521-531. [DOI] [PubMed] [Google Scholar]
- 9.Eckerle, L. D., and L. A. Ball. 2002. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 296165-176. [DOI] [PubMed] [Google Scholar]
- 10.Ge, Z. M., and C. Hiruki. 1993. The infectious transcripts of Sweet clover necrotic mosaic virus bipartite genome constructed by the polymerase chain reaction. Proc. Jpn. Acad. 69113-118. [Google Scholar]
- 11.Gould, A. R., R. I. B. Francki, T. Hatta, and M. Hollings. 1981. The bipartite genome of Red clover necrotic mosaic virus. Virology 108499-506. [DOI] [PubMed] [Google Scholar]
- 12.Hiruki, C. 1987. The dianthoviruses: a distinct group of isometric plant viruses with bipartite genome. Adv. Virus Res. 33257-300. [DOI] [PubMed] [Google Scholar]
- 13.Hiruki, C., Y. Furuya, G. Figueiredo, and A. L. N. Rao. 1983. Serological relationship among some members of dianthovirus group as determined by means of monoclonal and polyclonal antibodies. Phytopathology 73959-960. [Google Scholar]
- 14.Iwakawa, H. O., M. Kaido, K. Mise, and T. Okuno. 2007. cis-Acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3′ untranslated region of Red clover necrotic mosaic virus RNA1. Virology 369168-181. [DOI] [PubMed] [Google Scholar]
- 15.Kastenmayer, J. P., and P. J. Green. 2000. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. USA 9713985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, K. H., and S. A. Lommel. 1994. Identification and analysis of the site of −1 ribosomal frameshifting in Red clover necrotic mosaic virus. Virology 200574-582. [DOI] [PubMed] [Google Scholar]
- 17.Kim, K. H., and S. A. Lommel. 1998. Sequence element required for efficient −1 ribosomal frameshifting in Red clover necrotic mosaic dianthovirus. Virology 25050-59. [DOI] [PubMed] [Google Scholar]
- 18.Komoda, K., S. Naito, and M. Ishikawa. 2004. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. USA 1011863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin, E. V., G. H. Choi, D. L. Nuss, R. Shapira, and J. C. Carrington. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. USA 8810647-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, K. C., H. L. Chang, and R. Y. Chang. 2004. Accumulation of a 3′ terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. J. Virol. 785133-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lommel, S. A., M. Westonfina, Z. Xiong, and G. P. Lomonossoff. 1988. The nucleotide sequence and gene organization of Red clover necrotic mosaic virus RNA-2. Nucleic Acids Res. 168587-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, A., J. Maeda, H. Takagi, and I. Kurane. 2008. Detection of small RNAs containing the 5′- and the 3′-end sequences of viral genome during West Nile virus replication. Virology 371130-138. [DOI] [PubMed] [Google Scholar]
- 23.Mathews, M. B., and T. Shenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 655657-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of Brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature 31368-70. [DOI] [PubMed] [Google Scholar]
- 25.Mizumoto, H., Y. Hikichi, and T. Okuno. 2002. The 3′-untranslated region of RNA1 as a primary determinant of temperature sensitivity of Red clover necrotic mosaic virus Canadian strain. Virology 293320-327. [DOI] [PubMed] [Google Scholar]
- 26.Mizumoto, H., H.-O. Iwakawa, M. Kaido, K. Mise, and T. Okuno. 2006. Cap-independent translation mechanism of Red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J. Virol. 803781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizumoto, H., M. Tatsuta, M. Kaido, K. Mise, and T. Okuno. 2003. Cap-independent translational enhancement by the 3′ untranslated region of Red clover necrotic mosaic virus RNA1. J. Virol. 7712113-12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair, V., and M. Zavolan. 2006. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 14169-175. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, K., H. Nagano, H. Iwakawa, H. Mizumoto, A. Takeda, M. Kaido, K. Mise, and T. Okuno. 2008. cis-Preferential requirement of a −1 frameshift product p88 for the replication of Red clover necrotic mosaic virus RNA1. Virology 375205-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuno, T., C. Hiruki, D. V. Rao, and G. C. Figueiredo. 1983. Genetic determinants distributed in two genomic RNAs of sweet clover necrotic mosaic, red clover necrotic mosaic and clover primary leaf necrosis viruses. J. Gen. Virol. 641907-1914. [Google Scholar]
- 31.Poole, T. L., and A. Stevens. 1997. Structural modifications of RNA influence the 5′ exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 235799-805. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, D. J., E. V. Ryabov, S. K. Raj, I. M. Roberts, and M. E. Taliansky. 1999. Satellite RNA is essential for encapsidation of groundnut rosette umbravirus RNA by groundnut rosette assistor luteovirus coat protein. Virology 254105-114. [DOI] [PubMed] [Google Scholar]
- 33.Roossinck, M. J. 2005. Symbiosis versus competition in plant virus evolution. Nat. Rev. Microbiol. 3917-924. [DOI] [PubMed] [Google Scholar]
- 34.Ryabov, E. V., E. V. Generozov, T. L. Kendall, S. A. Lommel, and S. K. Zavriev. 1994. Nucleotide sequence of Carnation ringspot dianthovirus RNA-1. J. Gen. Virol. 75243-247. [DOI] [PubMed] [Google Scholar]
- 35.Samanta, M., D. Iwakiri, T. Kanda, T. Imaizumi, and K. Takada. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 254207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Scherbik, S. V., J. M. Paranjape, B. M. Stockman, R. H. Silverman, and M. A. Brinton. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 802987-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp, T. V., M. Schwemmle, I. Jeffrey, K. Laing, H. Mellor, C. G. Proud, K. Hilse, and M. J. Clemens. 1993. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VA(I)RNA. Nucleic Acids Res. 214483-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, R. Z., and W. A. Miller. 2004. Subgenomic RNA as a riboregulator: negative regulation of RNA replication by Barley yellow dwarf virus subgenomic RNA 2. Virology 327196-205. [DOI] [PubMed] [Google Scholar]
- 40.Shen, R., A. M. Rakotondrafara, and W. A. Miller. 2006. trans regulation of cap-independent translation by a viral subgenomic RNA. J. Virol. 8010045-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sit, T. L., P. R. Haikal, A. S. Callaway, and S. A. Lommel. 2001. A single amino acid mutation in the Carnation ringspot virus capsid protein allows virion formation but prevents systemic infection. J. Virol. 759538-9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281829-832. [DOI] [PubMed] [Google Scholar]
- 43.Ta, M., and S. Vrati. 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 745108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda, A., M. Tsukuda, H. Mizumoto, K. Okamoto, M. Kaido, K. Mise, and T. Okuno. 2005. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 243147-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takegami, T., and S. Hotta. 1990. Synthesis and localization of Japanese encephalitis virus RNAs in the infected cells. Microbiol. Immunol. 34849-857. [DOI] [PubMed] [Google Scholar]
- 46.Tatsuta, M., H. Mizumoto, M. Kaido, K. Mise, and T. Okuno. 2005. The Red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J. Virol. 79978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Treder, K., E. L. P. Kneller, E. M. Allen, Z. H. Wang, K. S. Browning, and W. A. Miller. 2008. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14134-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner, R. L., and K. W. Buck. 1999. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology 253115-124. [DOI] [PubMed] [Google Scholar]
- 49.Vreken, P., and H. A. Raue. 1992. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′-terminal part of the coding region. Mol. Cell. Biol. 122986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter, A. E., D. H. Turner, J. Kim, M. H. Lyttle, P. Muller, D. H. Mathews, and M. Zuker. 1994. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. USA 919218-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong, Z., K. H. Kim, D. Giesmancookmeyer, and S. A. Lommel. 1993. The roles of the Red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology 19227-32. [DOI] [PubMed] [Google Scholar]
- 52.Xiong, Z., K. H. Kim, T. L. Kendall, and S. A. Lommel. 1993. Synthesis of the putative Red clover necrotic mosaic virus RNA polymerase by ribosomal frameshifting in vitro. Virology 193213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong, Z., and S. A. Lommel. 1989. The complete nucleotide sequence and genome organization of Red clover necrotic mosaic virus RNA-1. Virology 171543-554. [DOI] [PubMed] [Google Scholar]
- 54.Xiong, Z. G., and S. A. Lommel. 1991. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology 182388-392. [DOI] [PubMed] [Google Scholar]
- 55.Xu, N., B. Segerman, X. Zhou, and G. Akusjarvi. 2007. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the RNA-induced silencing complex and associate with polyribosomes. J. Virol. 8110540-10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4757-768. [DOI] [PubMed] [Google Scholar]
- 57.Zavriev, S. K., C. M. Hickey, and S. A. Lommel. 1996. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology 216407-410. [DOI] [PubMed] [Google Scholar]
- 58.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]