Abstract
The pre-mRNA processing strategy of the B19 virus is unique among parvoviruses. B19 virus-generated pre-mRNAs are transcribed from a single promoter and are extensively processed by alternative splicing and alternative polyadenylation to generate 12 transcripts. Blockage of the production of full-length B19 virus transcripts at the internal polyadenylation site [(pA)p] was previously reported to be a limiting step in B19 virus permissiveness. We show here that in the absence of genome replication, internal polyadenylation of B19 virus RNAs at (pA)p is favored in cells which are both permissive and nonpermissive for B19 viral replication. Replication of the B19 virus genome, however, introduced either by viral infection or by transfection of an infectious clone into permissive cells or forced by heterologous replication systems in nonpermissive cells, enhanced readthrough of (pA)p and the polyadenylation of B19 virus transcripts at the distal site [(pA)d]. Therefore, replication of the genome facilitates the generation of sufficient full-length transcripts that encode the viral capsid proteins and the essential 11-kDa nonstructural protein. Furthermore, we show that polyadenylation of B19 viral RNA at (pA)p likely competes with splicing at the second intron. Thus, we conclude that replication of the B19 virus genome is the primary limiting step governing B19 virus tropism.
The human parvovirus B19 virus (B19V) is a member of the Erythrovirus genus in the subfamily Parvoviridae (8, 41). B19V causes a number of human diseases, such as fifth disease in children, arthropathy, particularly in women, transient aplastic crisis in individuals with a high red cell turnover rate, pure red cell aplasia in immunocompromised patients, and hydrops fetalis following infection of pregnant women (4).
The B19V virion is approximately 20 nm in diameter and contains a single-stranded DNA genome of either plus or minus polarity. The left-hand portion of the genome contains the NS-encoding region, which is essential for virus replication, transcription transactivation, and cytotoxicity. The right-hand portion harbors the sequences for the two structural proteins, VP1 and VP2. At least 12 transcripts are generated by alternative splicing and alternative polyadenylation from a single pre-mRNA that is transcribed from the single promoter at map unit 6 (P6) (Fig. 1) (24, 46). B19V transcripts polyadenylated at the proximal polyadenylation site [(pA)p] accumulate as either spliced or unspliced mRNAs. As unspliced mRNAs, they encode the nonstructural NS1 protein, which is essential for virus replication. Spliced RNAs polyadenylated internally at (pA)p encode a 7.5-kDa protein whose function is unknown. All B19V transcripts detected so far that are polyadenylated at the distal polyadenylation site [(pA)d] excise the first intron (D1 to A1-1 or D1 to A1-2) (Fig. 1). Those that are not additionally spliced encode the capsid protein VP1. RNAs in which the second intron (D2 to A2-1) is additionally spliced encode the capsid protein VP2, while those additionally spliced between D2 and A2-2 encode the nonstructural 11-kDa protein that is essential for virus export from the nuclei (38, 48). Two internal polyadenylation sites, (pA)p1 and (pA)p2, have been identified. (pA)p1, which accounts for approximately 90% of the internal polyadenylation events, has a noncanonical cleavage and polyadenylation specificity factor binding motif (AUUAAA) located at nucleotide (nt) 2819. (pA)p2, which accounts for approximately 10% of the internal polyadenylation events, has a canonical signal of AAUAAA located at nt 3115 (46).
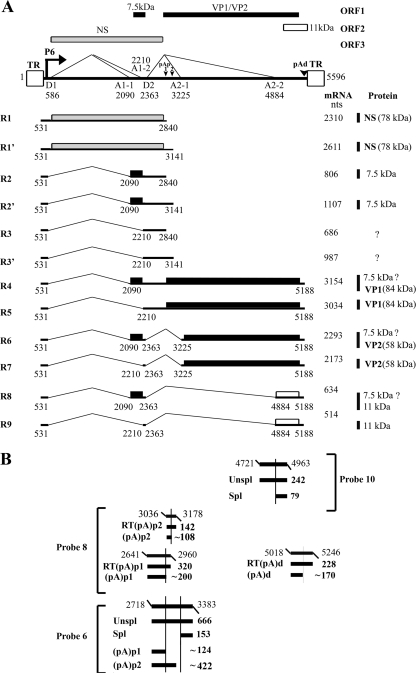
FIG. 1.
Transcription map of B19V and probes used. (A) The B19V genome of the J35 virus (GenBank accession no. AY386330) is depicted, including the locations of the P6 promoter, the intron donors (D1 and D2) and acceptors (A1-1, A1-2, A2-1, and A2-2), the internal polyadenylation sites [(pA)p1 and (pA)p2], the distal polyadenylation site [(pA)d], and the ITRs. The four open reading frames (ORFs) are diagramed at the top of the figure. The different open reading frames that were used are shown with different shading patterns. The major transcript species and the proteins they encode are shown below the diagramed B19V genome (46). (B) The probes used for RPA in this study, i.e., probe 6, probe 8, and probe 10, are diagramed at the bottom, and their protected bands are shown with relative sizes and identities. Probe 6 has been described previously (46).
B19V has been shown to undergo productive replication in erythroid progenitors of human hematopoietic stem cells (26, 37). Different disease manifestations are due to the direct cytotoxicity of the virus in susceptible cells (6). The tropism of B19V infection is controlled by a number of determinants. B19V infection is restricted primarily to erythroid progenitor cells by the distribution of the viral receptor, the P antigen (5), and reported coreceptors, namely, integrin α5β1 (42) and the KU80 protein (22). Productive B19V infection has also been shown to be restricted by limited capsid protein expression. In nonpermissive cells, capsid expression is impaired due to a blockage in full-length transcription of the viral genome (17). Because in nonpermissive cells all RNAs transcribed from the P6 promoter are polyadenylated at (pA)p, the majority of mRNAs present are NS1 encoding, with only limited production of the capsid-encoding mRNAs. NS1 causes cell death by its cytotoxic or apoptotic characteristics (19, 25). In contrast, in permissive cells, more B19V RNAs read through the (pA)p sites, which results in sufficient capsid-encoding mRNAs (17). Studies have also shown that B19V tropism is restricted intracellularly, but the mechanism of this restriction has not been clearly established (12, 26, 36). In some cases, conversion of single-stranded DNA to double-stranded DNA has been shown to be blocked, resulting in aborted infection. In other cases, conversion of the single-stranded viral DNA to double-stranded replicative forms (RF) has been detected, but only within a small population of cells.
The target cells for B19V replication in human bone marrow are the erythrocyte precursors CFU-E and BFU-E (26, 37). A few semipermissive myeloblastoid cell lines have been isolated (e.g., UT7/Epo-S1 [20] and KU812Ep6 [18]) and support B19V infection with a limited efficiency (only a 10-fold increase of B19V genomes) (3, 44). CD36+ erythroid progenitor cells (CD36+ EPCs) are highly permissive to B19V infection and support B19V replication at least 100-fold (44). They can be generated ex vivo on a large scale from primary hematopoietic stem cells with a combination of cytokines for erythroid lineage differentiation (11, 13).
To understand the limiting factors of B19V tropism, we studied the relationship of B19V replication and its RNA processing. In particular, we examined alternative polyadenylation during viral infection in permissive CD36+ EPCs, as well as viral infection and transfection of the B19V infectious clone and nonreplicating B19V constructs in semipermissive UT7/Epo-S1 cells. Our findings show that more B19V transcripts read through (pA)p in virus infections of CD36+ EPCs and UT7/Epo-S1 cells and in transfection of the infectious clone into UT7/Epo-S1 cells than in transfection of nonreplicating B19V plasmid constructs into UT7/Epo-S1 cells. Moreover, replication of the B19V genomes supported by artificial replication systems also strongly induced a significant increase of transcripts reading through (pA)p. Our findings indicate that replication of the B19V genome affects B19V pre-mRNA processing, which results in an efficient production of the capsid protein- and 11-kDa protein-encoding mRNAs. We concluded that blockage of the full-length transcripts is determined by the replication of the genome, which is the key determinant for B19V tropism.
MATERIALS AND METHODS
Cell lines.
COS-7 cells (CRL-1651; ATCC) and 293 cells (CRL-10852; ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) at 37°C in 5% CO2. UT7/Epo-S1 cells (20) were cultured in DMEM with 10% FCS and 2 units/ml of erythropoietin (Epo) (Epogen; Amgen, Thousand Oaks, CA) at 37°C in 5% CO2.
Generation of CD36+ EPCs.
CD133+ cells were isolated from granulocyte colony-stimulating factor-mobilized peripheral blood stem cells from healthy donors (a gift from Richard Childs, NHLBI, NIH). A total of 104 cells/ml were grown in an expansion medium (44). The composition of the expansion medium was medium BIT 9500 (StemCell Technologies Inc.) diluted with the alpha modification of Eagle's medium with glutamine (Mediatech, Inc.) and supplemented with 900 ng/ml of ferrous sulfate (Sigma), 90 ng/ml of ferric nitrate (Sigma), 10−6 M of hydrocortisone (Sigma), 100 ng/ml of recombinant human stem cell factor (Invitrogen), 5 ng/ml of human interleukin-3 (Invitrogen), and 3 units/ml of Epo (Amgen).
On day 4 of culture in expansion medium, the CD36+ EPCs were frozen at 2.5 × 105 cells/vial in 0.25 ml of 90% FCS plus 10% dimethyl sulfoxide in liquid nitrogen. The frozen cells were thawed and maintained for another 4 days in expansion medium at a concentration of <1 × 106 cells/ml. Large numbers of CD36+ EPCs, which were used for B19V infection, were obtained on day 8 or day 9.
Virus and infection.
B19V viremic plasmas were obtained from ViraCor Laboratories (Lee's Summit, MO) and were quantified for B19V genomic copies by a real-time PCR method. Briefly, virus DNA was extracted from 20 μl of the plasma by use of a QIAamp DNA blood mini kit (Qiagen) and finally eluted in 200 μl of deionized H2O. The pN8 plasmid, which contains the B19V sequence of nt 199 to 5410 (49), was used as a control (1 genomic copy = 5.4 × 10−12 μg) to establish a standard curve for absolute quantification by using TaqMan technology with an Applied Biosystems 7500 system (Foster City, CA). The amplicon primers and the TaqMan probe were designed by Primer Express 2.0.0 software (Applied Biosystems). Their sequences are as follows: forward primer, 5′CCTGGGCAAGTTAGCGTACAA3′ (nt 2975 to 2995); reverse primer, 5′ATGAATCCTTGCAGCACTGTCA3′ (nt 3061 to 3082); and TaqMan probe, 5′CCGGTACTAACTATGTTGGGCATGCAA3′ (nt 3040 to 3067). TaqMan universal PCR master mix (Takara Bio USA, Madison, WI) was used for amplification following a standard protocol. All of the nucleotide numbers for the B19V genome used in this study refer to the B19V sequence of the J35 isolate (GenBank accession no. AY386330).
Plasma samples that contained genomic copies at over 1011/ml were chosen for infection studies. Plasma 14 (P14) had the highest infectivity in UT7/Epo-S1 cells, detected by an RNase protection assay (RPA) using probe 8 (described below). The B19V genome in P14 contained the BamHI site at nt 4077 that was used for digestion to identify B19V replicated forms as previously reported (26, 49).
Twenty microliters of the P14 B19V was incubated with 2 × 106 cells in a volume of 500 μl DMEM with slow rotation at 4°C for 2 h. Infected cells were either washed with DMEM or not washed and then cultured at a concentration of 2 × 105 cells/ml at 37°C with 5% CO2. Infected cells were taken at various time points postinfection (p.i.) for preparation of total RNA and Hirt DNA samples and for immunofluorescence assay as previously described (29).
Construction of plasmids. (i) B19V constructs.
The B19V infectious clone pM20 and its NS1 knockout mutant, pM20NS(−), were described previously (48, 49). The pB19NSCap plasmid was made by inserting nt 383 to 5410 into an EcoRI-XhoI digest of pBluescript (Stratagene). The pSKB19NS(−)Cap plasmid was constructed by inserting nt 199 to 5410 of the B19V genome into an EcoRI-XhoI digest of pBluescript (Stratagene) in which nt 754 and 755 were deleted to abolish NS1 expression. p5TRB19NS(−)Cap was made by inserting the same B19V NS(−)Cap sequence into an adeno-associated virus 5 (AAV5) inverted terminal repeat (ITR)-containing plasmid, pAV5ITR; thus, the B19V NS(−)Cap sequence was flanked with AAV5 ITRs at two ends in p5TRB19NS(−)Cap. pAV5ITR was constructed by introducing EcoRV sites at the 3′ end of the left ITR and the 5′ end of the right ITR and inserting the resulting fragment into pAV5 (32).
pSVOB19NS(−)Cap was made by cloning the B19V NS(−)Cap sequence into a modified pSVOd plasmid, pSVOdΔtet, which was constructed by deleting the B19V genome and the tetracycline gene from pSVOdΔ170B19 (1). Deleting the B19V sequence between the D1 donor and the A1-1 acceptor (nt 586 to 2090) in both pSVOB19NS(−)Cap and pSKB19NS(−)Cap resulted in plasmids pSKB19ΔD1A1Cap and pSVOB19ΔD1A1Cap, respectively. Further deletion of the B19V sequence from nt 3426 to 4781 (Δ1300) between the A2-1 and A2-2 acceptors resulted in plasmids pSKB19ΔD1A1CapΔ1300 and pSVOB19ΔD1A1CapΔ1300, respectively. pSKB19P6Cap and pSVOB19P6Cap were made by deleting the B19V sequence from nt 657 to 2250 in pSKB19NS(−)Cap and pSVOB19NS(−)Cap, respectively.
(ii) Clones used to generate probes for RPA.
Probe 6 was described previously (46). Probe 8 was constructed by cloning nt 2641 to 2960, nt 3036 to 3178, and nt 5018 to 5246 of the B19V genome into EcoRI-KpnI-, BamHI-SalI-, and PstI-HindIII-digested pGEM3Z (Promega), respectively. Probe 10 was constructed by cloning nt 4721 to 4963 of the B19V genome into BamHI-HindIII-digested pGEM3Z (Promega).
Transfection.
COS-7 cells were transfected with 2 μg of DNA per 60-mm plate, using Lipofectamine and Plus reagent (Invitrogen) as previously described (33). For cotransfection, 293 cells were transfected with 1 μg of pSKB19NS(−)Cap or p5TRB19NS(−)Cap, 0.5 μg of the AAV5 Rep plasmid pHIVAV5Rep (32), and 2 μg of the pHelper plasmid (Stratagene).
UT7/Epo-S1 cells were transfected with 2 μg of DNA per 2 × 106 cells, using Nucleofector (Amaxa, MD) and a universal transfection reagent (DNAproject Inc., Seattle, WA), with program X005. After transfection, cells were maintained at 2 × 105 cells/ml in DMEM with 10% FCS and 2 units/ml of Epo.
RNA isolation and RPA.
Total RNAs were isolated from transfected cells 2 days after transfection and from infected cells at various time point p.i., using Trizol reagent (Invitrogen).
RPAs were performed as previously described (23, 35). Probes were generated from EcoRI-digested templates by in vitro transcription with SP6 polymerase, using a MAXIscript kit (Ambion) following the protocol provided. RNA hybridizations for RPAs were done with a substantial probe excess, and RPA signals were quantified with a Storm 856 phosphorimager and Image Quant TL software v2005 (GE Healthcare). Relative molar ratios of individual species of RNAs were determined after adjusting for the number of 32P-labeled uridines (U) in each protected fragment, as previously described (35).
To test the stability of the B19V RNAs, we infected CD36+ EPCs with B19V P14 as described above and, 72 h later, added actinomycin D (Sigma) to a final concentration of 5 μg/ml. Total RNAs were isolated 0, 2, 4, and 6 h after treatment and were subjected to RPA analysis.
DNA isolation and analysis.
DNA of low molecular weight (Hirt DNA) (14) was isolated to visualize RF by Southern blot analysis. After B19V infection or transfection, paired dishes of cells were collected for Hirt DNA extraction, except for one which was used for total RNA isolation as described above. Cells were washed twice with phosphate-buffered saline. Cells were then lysed in 2% sodium dodecyl sulfate and treated with proteinase K (0.5 mg/ml). DNA was collected by binding to a DNA miniprep spin column (Zymo, Orange, CA) and being eluted with Tris-EDTA buffer. Prepared Hirt DNA samples were digested with appropriate restriction enzymes where necessary, as indicated in the figure legends, and samples were run in a 1% agarose gel. Southern blot analysis was performed essentially as described previously (23, 28), using the B19V NSCap probe (nt 199 to 5410). Blots were exposed to a GE phosphorimaging screen, and the DNA forms were quantified by a Storm 856 phosphorimager and Image Quant TL software v2005 (GE Healthcare) where indicated.
RESULTS
The majority of B19V transcripts read through the (pA)p sites during viral infection of permissive CD36+ EPCs.
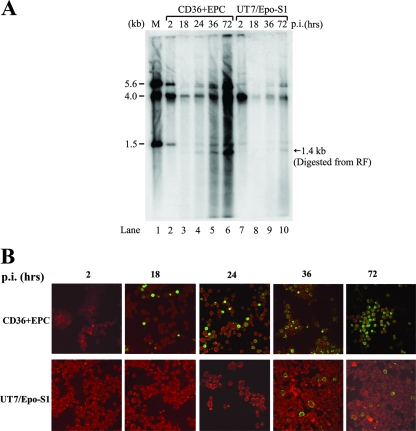
CD36+ EPCs were expanded and differentiated in large quantities in the presence of a combination of cytokines as previously reported (11, 13). Cells were infected with B19V at a multiplicity of infection of ∼103 genomic copies/cell on day 9 postdifferentiation, within the days that cells were previously shown to be most susceptible to B19V replication (44). Total RNA or Hirt DNA samples were prepared at 2, 18, 24, 36, and 72 h p.i. and were analyzed by RPA or Southern blotting. As shown by the 1.4-kb bands that were present following digestion of DNA samples by BamHI (Fig. 2A, lanes 2 to 6), replication of the B19V genome was detectable at 24 h p.i. (Fig. 2A, lane 4) and progressed to the highest level at 72 h p.i. (Fig. 2A, lane 6) in CD36+ EPCs. There was an approximately 20-fold increase in the RF (1.4-kb band) from 24 to 72 h p.i. Assembled capsids were also detected by immunofluorescence (Fig. 2B), confirming that the CD36+ EPCs were highly permissive for B19V replication and capsid production, as previously reported (44). The rate of appearance of B19V capsids in infected CD36+ EPCs (Fig. 2B, top panels) paralleled the appearance of the viral RF, as shown in Fig. 2A. At 18 h p.i., the 1.4-kb RF was barely detectable (Fig. 2A, lane 3). However, it seems that a low level of replication, or at least second-strand synthesis, must have occurred by this time, since capsid proteins were detected by immunofluorescence (Fig. 2B, top panels, compare 18 h p.i. to 2 h p.i.).
FIG. 2.
B19V infection of CD36+ EPCs and UT7/Epo-S1 cells. CD36+ EPCs or UT7/Epo-S1 cells were infected with B19V. (A) Southern blotting. Hirt DNAs were isolated at different time points p.i., as indicated at the top of the figure. BamHI-digested fragments were resolved in 1% agarose and were transferred to a nitrocellulose membrane. The blot was hybridized with the B19V NSCap probe spanning the B19V genome at nt 199 to 5410. Lane 1 shows the BamHI-digested bands of B19V infectious DNA (nt 1 to 5596) as a control and a size marker. (B) Immunofluorescence assay. B19V-infected cells were collected at different time points p.i., as indicated at the top, and were cytocentrifuged onto slides by use of a Shandon Cytospin 3 centrifuge (Thermo Scientific, MA). Fixed cells were incubated with a mouse monoclonal antibody that reacted only with assembled B19V capsids (clone 5215D; Millipore) and were stained with fluorescein isothiocyanate-conjugated secondary antibody. The background was counterstained with Evans blue. Images were acquired by an Eclipse C1 Plus confocal microscope (Nikon) at a magnification of ×40.
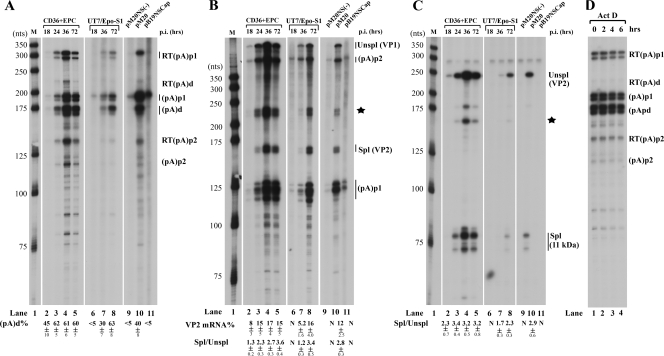
RPA results using probe 8, which was able to display B19V RNAs polyadenylated at (pA)p1, (pA)p2, and (pA)d, showed that at 24, 36, and 72 h p.i., approximately 60% of the B19V transcripts read through the (pA)p sites [(pA)p1 and (pA)p2] and were ultimately polyadenylated at (pA)d (Fig. 3A, lanes 3 to 5). However, at 18 h p.i., half of the B19V transcripts were blocked at (pA)p (Fig. 3A, lane 2). As the infection progressed, the total amount of B19V transcripts increased significantly and accumulated to a maximum level at 36 h p.i. (Fig. 3A, lanes 3 and 4). B19V transcripts decreased after 36 h p.i. (Fig. 3A, lane 5), although replication of the B19V genome continued and reached its highest level at 72 h p.i. (Fig. 2A, lane 6). Analysis of B19V RNAs by probe 6 revealed that there were approximately two- to threefold more B19V RNAs spliced at the A2-1 acceptor site (Fig. 3B, lanes 3 to 5) after 24 h p.i.; however, this increase was less at 18 h p.i., early in infection (Fig. 3B, lane 2). B19V RNAs spliced at the A2-1 acceptor encode the VP2 capsid protein. The steady-state level of the VP2 mRNA shown in Fig. 3B indicates that approximately 15 to 17% of B19V RNAs were VP2-encoding mRNAs at 24 h p.i., which is approximately twice more than what was present at 18 h p.i. (Fig. 3B, compare lanes 3 to 5 with lane 2). B19V RNAs were spliced at the A2-2 acceptor at a relatively consistent level throughout the course of infection. The ratio of RNA spliced at the A2-2 junction to unspliced B19V RNA across this region was approximately 2 to 3 (Fig. 3C, lanes 2 to 5), indicating that a high level of mRNA encoding the 11-kDa protein was generated during B19V infection of CD36+ EPCs.
FIG. 3.
Polyadenylation of B19V RNAs shows a shift from (pA)p early in infection to (pA)d late during infection of CD36+ EPCs or UT7/Epo-S1 cells. In addition, B19V RNAs generated following transfection of nonreplicating B19V genomes did not generate significant levels of B19V RNAs polyadenylated at (pA)d. CD36+ EPCs were infected with B19V, and total RNAs were isolated at 18, 24, 36, and 72 h p.i.; UT7/Epo-S1 cells were infected with B19V, and total RNAs were isolated at 18, 36, and 72 h p.i. Plasmids pM20, pM20NS(−), and pSKB19NSCap were transfected into UT7/Epo-S1 cells, and 72 h later, total RNA was isolated. All RNA samples were protected by probe 8, probe 6, or probe 10, and results for a representative experiment are shown. (A) RPA using probe 8. Bands representing RNAs that either read through or were polyadenylated at (pA)p1, (pA)p2, and (pA)d are designated RT(pA)p1, RT(pA)p2, and RT(pA)d or (pA)p1, (pA)p2, and (pA)d, respectively. Quantification of the ratio of B19V RNA at (pA)d to total polyadenylated B19V RNA is shown as a percentage [(pA)d%]. (B) RPA using probe 6. Bands representing RNAs that were either spliced or unspliced at the A2-1 acceptor are designated Unspl (for unspliced RNAs) and Spl (for spliced RNAs). Alternative representations for these bands are shown with their coding capabilities in parentheses. Quantification of the ratio of spliced RNA (VP2 mRNA) versus unspliced RNA (VP1 mRNA) is shown as the Spl/Unspl ratio, and quantification of the VP2 mRNA is shown as a percentage of VP2 mRNA (VP2 mRNA%) in the total RNA protected by probe 6. The identities of the bands indicated with a star are unknown, as previously described (46), and therefore they were not included in the quantification. (C) RPA using probe 10. Bands representing RNAs that were either spliced or unspliced at A2-2 are designated Unspl (for unspliced RNAs) and Spl (for spliced RNAs). Alternative representations for these bands are shown with their coding capabilities in parentheses. Quantification of the ratio of spliced RNA (11-kDa protein mRNA) versus unspliced RNA (VP2 mRNA) is shown as the Spl/Unspl ratio. The bands indicated with a star are likely nonspecific hybridized products. (D) B19V RNAs were stable. CD36+ EPCs were infected with B19V. After 72 h, cells were treated with actinomycin D, and total RNAs were isolated at the times indicated and were protected by probe 8. All of the quantifications represent average results for at least three individual experiments, with standard deviations underneath. N, not determined because there were nearly no spliced RNAs detected. Lanes 1 in panels A, B, and C, a 32P-labeled RNA ladder with the respective sizes indicated to the left, as previously described (32).
Similar to AAV2 and AAV5 RNAs (21, 32), B19V RNA transcripts generated in CD36+ EPCs that were polyadenylated either at (pA)p or at (pA)d were stable after 6 h of treatment with actinomycin D (Fig. 3D). B19V RNAs generated during infection of UT7/Epo-S1 cells and following transfections of 293 cells and COS-7 cells were also stable (data not shown).
Blockage of full-length B19V transcripts was observed early during B19V infection and in transfection of B19V nonreplicative plasmid constructs in semipermissive UT7/Epo-S1 cells.
We further investigated the expression of B19V RNAs generated during infection of the semipermissive cell line UT7/Epo-S1 (20). In contrast to the case for B19V infection of CD36+ EPCs, we could not detect B19V transcripts polyadenylated at (pA)d at 18 h p.i. (Fig. 3A, lane 6). Approximately 30% of the B19V transcripts were polyadenylated at (pA)d at 36 h p.i. (Fig. 3A, lane 7). Surprisingly, at 72 h p.i., a higher level (approximately 60%) of B19V RNAs were polyadenylated at (pA)d (Fig. 3A, lane 8), similar to levels generated by B19V at later time points during infection of CD36+ EPCs. These results suggested that as infection progressed in UT7/Epo-S1 cells, there was a switch to increased levels of B19V transcripts that read through (pA)p. There also was a gradual increase of the steady-state level of VP2 mRNA as infection progressed, which was accompanied by an increase in the ratio of spliced/unspliced RNA in the region of the A2-1 acceptor site (Fig. 3B, lanes 6 to 8) and the A2-2 site (Fig. 3C, lanes 6 to 8). Approximately 16% of the B19V RNAs generated at 72 h p.i. in UT7/Epo-S1 cells and protected by probe 6 were VP2-encoding mRNAs (Fig. 3B, lane 8), similar to the levels generated late during B19V infection of CD36+ EPCs.
The difference we observed between CD36+ EPCs and UT7/Epo-S1 cells at early time points p.i. could potentially have been due to a more rapid onset of viral replication in CD36+ EPCs. Replication of B19V in UT7/Epo-S1 cells is restricted to a small population (12). We found that replication of the B19V genome in UT7/Epo-S1 cells was slower than that in CD36+ EPCs. At 72 h p.i., only a limited amount of viral RF could be detected in UT7/Epo-S1 cells, comparable to the level in CD36+ EPCs at 24 h p.i. (Fig. 2A, compare lane 10 to lane 4). Between 36 and 72 h p.i., the number of infective cells shown by immunofluorescence was not significantly increased.
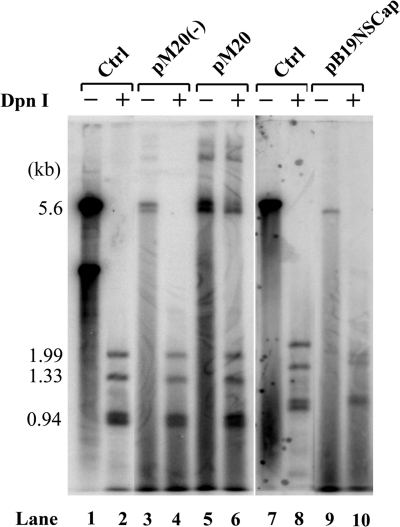
Potentially, efficient B19V replication in CD36+ EPCs could facilitate readthrough of (pA)p early during infection, and reduced replication of the B19V genome in UT7/Epo-S1 cells did result in an increase in polyadenylation at (pA)d over time. Our results with both CD36+ EPCs and UT7/Epo-S1 cells support the previous observation that in permissive cells B19V transcripts read through the internal (pA)p site, thereby generating capsid-encoding RNAs and supporting a productive infection, and that in nonpermissive cells polyadenylation at (pA)p is preferred, preventing extension into the capsid gene (17). Thus, we attempted to determine whether viral replication itself was able to facilitate readthrough of (pA)p or, alternatively, whether the permissive cell environment per se was able to support efficient polyadenylation at (pA)d. We transfected the B19V infectious clone pM20 (49) and two nonreplicating B19 plasmids, the infectious clone in which the NS1 gene was mutated [pM20NS(−)] and the pB19NSCap plasmid, into UT7/Epo-S1 cells. The infectious clone pM20 replicated with a low efficiency (Fig. 4, lane 6) in UT7/Epo-S1 cells; replication of pM20NS(−) and B19NSCap was not detected (Fig. 4, lanes 4 and 10). Approximately 40% of the B19V RNAs generated by pM20 following transfection of UT7/Epo-S1 cells were polyadenylated at (pA)d (Fig. 3A, lane 10). In contrast, more than 95% of the B19V RNAs generated by transfection of the two nonreplicative plasmids, pM20NS(−) and pB19NSCap, were polyadenylated at (pA)p1 (Fig. 3A, lanes 9 and 11), indicating a block to the generation of full-length transcripts in the semipermissive UT7/Epo-S1 cells. Since there were few readthrough transcripts following transfection of the nonreplicative plasmids, neither VP2-encoding mRNAs (Fig. 3B, lanes 9 and 11) nor 11-kDa protein-encoding mRNAs (Fig. 3C, lanes 9 and 11) could be detected. The efficiency of splicing at A2-1 for B19V RNAs generated by transfection of the infectious clone was similar to that seen in viral infection (Fig. 3B, compare lane 10 with lane 8). Similar levels of B19V RNAs spliced at A2-2 (the 11-kDa protein-encoding mRNAs) were also seen for both transfection of the infectious clone and viral infection (Fig. 3C, compare lane 10 to lane 8). These results indicate that in UT7/Epo-S1 cells, B19V RNAs were efficiently polyadenylated at the internal (pA)p site both early in B19V infection and in transfection of the nonreplicative B19V constructs. Therefore, replication of the viral genome, initiated either by viral infection or by transfection of the infectious clone, facilitates the readthrough of (pA)p, producing RNAs that can subsequently be spliced to generate the capsid- and 11-kDa protein-encoding mRNAs.
FIG. 4.
Southern blot analysis of UT7/Epo-S1 cells transfected with B19V infectious or noninfectious templates. Hirt DNAs were prepared at 72 h posttransfection and were digested with EcoRI (−) or DpnI (+) as indicated at the top of the blot. Digested DNA samples were resolved in 1% agarose gel and were subjected to Southern blot hybridization with the B19V NSCap probe. Lanes 1 and 7 and lanes 2 and 8 show the 5.6-kb band and the DpnI-digested bands of the B19V infectious DNA (nt 1 to 5596), respectively, which serve as controls for DpnI digestion and also as size markers.
Transfection of the pM20 infectious clone into CD36+ EPCs was attempted following a method reported previously (44). Similar to what was observed with UT7/Epo-S1 cells, approximately one-half of the B19V RNAs read through the (pA)p site and ultimately were polyadenylated at (pA)d (data not shown). However, we were unable to obtain sufficient RNAs to perform RPA from transfections of the nonreplicative plasmids, pM20NS(−) and pB19NSCap, into CD36+ EPCs, since a large portion of the cells after transfection were dead (data not shown).
Replication of the B19V genome using AAV5 ITRs resulted in efficient readthrough of B19V transcripts at (pA)p.
To study whether replication of the B19V genome itself can facilitate increased readthrough of (pA)p, we used a heterologous replication origin, the AAV5 ITRs, to support replication of the B19NS(−)Cap genome (nt 199 to 5410). In addition, we provided all the necessary genes for replication of this construct in 293 cells. In this construct, NS1 expression was abolished by a frameshift, and half of the B19V ITRs at both ends were deleted. The NS gene, which is not involved in B19V pre-mRNA processing (46; data not shown), was mutated in these constructs to avoid toxicity (19, 25). AAV5 is a member of the Dependovirus genus; it is a human parvovirus that is dependent on adenovirus for replication (41). Like AAV5, B19V has identical terminal repeat structures at both ends, so its replication should employ a similar model. Replication of the AAV genome involves hairpin-primed conversion of the single-stranded parental genome into a double-stranded replicative intermediate, followed by resolution of the terminal hairpins by the large nonstructural protein Rep78 and final separation of the single-stranded progeny viral genome (45).
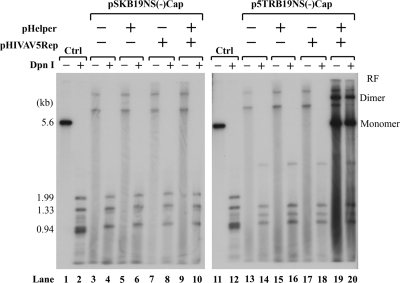
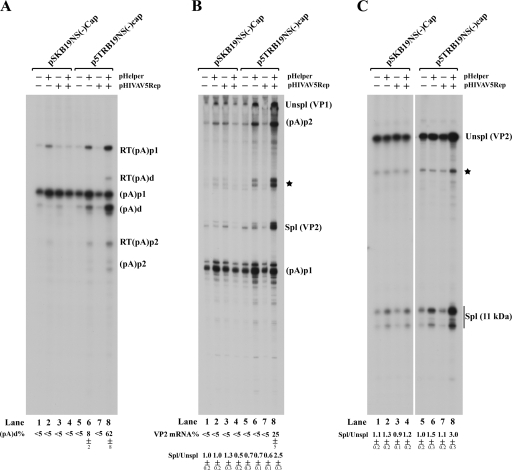
Transfection of the AAV5 ITR-B19NS(−)Cap chimera, p5TRB19NS(−)Cap, but not pSKB19NS(−)Cap, which did not contain the AAV5 ITRs, together with the AAV5 Rep gene and the pHelper plasmid, which contains the adenovirus E2, E4orf6, and VA RNA genes, resulted in highly efficient replication of the B19V genome in 293 cells (Fig. 5, compare lanes 19 and 20 to lanes 9 and 10). As expected, more than 90% of the B19V RNAs generated from the nonreplicating construct were polyadenylated at (pA)p (Fig. 6A, lanes 1 to 7), while approximately 60% of B19V RNAs generated from the replicating constructs read through (pA)p and were polyadenylated at (pA)d (Fig. 6A, lane 8). These results suggested that the replication process is the critical parameter that allows extension of B19V RNA past (pA)p. Interestingly, we also observed an increase in splicing at the A2-1 acceptor under replication conditions. The ratio of spliced to unspliced RNA in this region was approximately 1 under all nonreplicating conditions (Fig. 6B, lanes 1 to 7) and approximately 2.5 under the replication conditions described here (Fig. 6B, lane 8). Twenty-five percent of the steady-state spliced RNAs under these conditions were VP2-encoding mRNAs (Fig. 6B, lane 8), resulting in efficient capsid production (data not shown). Thus, increased genome replication resulted in the generation of greater levels of VP2-encoding mRNAs. Splicing at the A2-2 site (which generates mRNAs encoding the 11-kDa protein) was also slightly increased under replication conditions (the ratio of spliced to unspliced RNA in this region was approximately 3) (Fig. 6C, lane 8) compared with that under nonreplicating conditions (where the ratio was 1) (Fig. 6C, lanes 1 to 7). These results indicated that replication of the B19V genome supported by the AAV5 ITRs was sufficient to facilitate increased readthrough of (pA)p and the subsequent splicing of the second intron, similar to what was found in B19V infection of CD36+ EPCs and UT7/Epo-S1 cells (Fig. 3). Surprisingly, we did not observe a proportional increase of transcripts relative to the replicated levels of the 5TRB19NS(−)Cap template (Fig. 6A, B, and C, lanes 8). The reason for this discrepancy is currently unknown.
FIG. 5.
Replication of the B19V genome is supported by AAV5 replication elements. Plasmids pSKB19NS(−)Cap and p5TRB19NS(−)Cap were transfected into 293 cells with different combinations of the pHIVAV5Rep and pHelper plasmids, as indicated at the top of the figure. After 48 h, total RNAs and Hirt DNAs were isolated. Hirt DNAs were digested with EcoRI (−) or DpnI (+) and were resolved in 1% agarose gel. After transfer, the membrane was probed with the B19V NSCap probe. Lanes 1 and 2 and lanes 11 and 12 are controls for DpnI digestion and size markers, respectively. The RF of both monomers and dimers in lanes 19 and 20 are indicated.
FIG. 6.
Replication of the B19V genome initiated by AAV5 ITRs resulted in significant increases in B19V RNAs polyadenylated at (pA)d. Plasmids pSKB19NS(−)Cap and p5TRB19NS(−)Cap were transfected into 293 cells with different combinations of the pHIVAV5Rep and pHelper plasmids, as indicated at the top of the figure. After 48 h, total RNAs were isolated and were protected by probe 8 (A), probe 6 (B), or probe 10 (C). All of the protected bands are designated as described in the legend to Fig. 3, and all of the quantifications were also performed as described in the legend to Fig. 3.
Minimal replication in COS-7 cells of the B19V genome in constructs containing the SV40 replication origin increased readthrough of (pA)p.
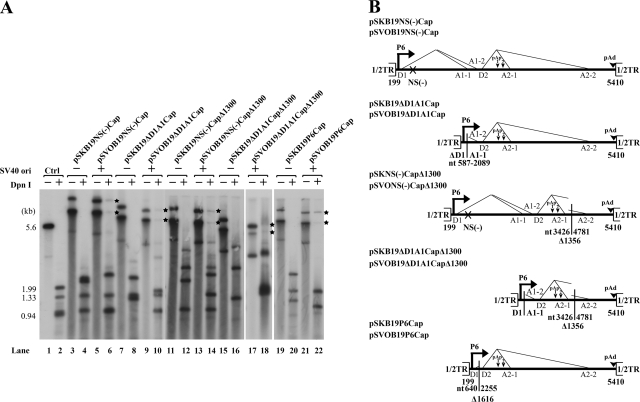
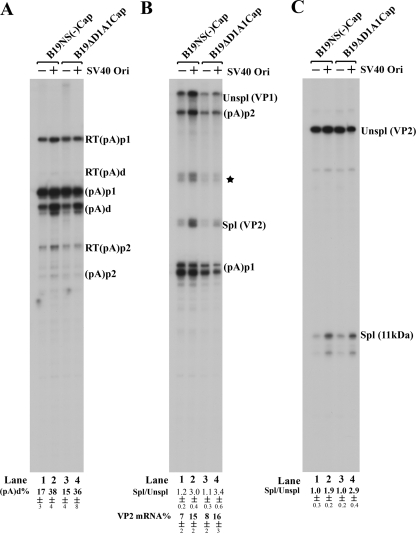
COS-7 cells are normally nonpermissive for B19V replication. It has been reported that the simian virus 40 (SV40) origin of replication supports replication of a plasmid containing the B19V genome in COS-7 cells that express the SV40 large T antigen (1). In our experiments, however, replication of the previously analyzed pSVOdB19Δ170 construct (1), as well as various other SV40 origin-B19V genome-containing constructs, was quite low (Fig. 7A). To determine whether this low level of replication still had effects on the relative usage of (pA)p, we compared two variant B19V genomes in both the poorly replicating pSVOd and nonreplicating pSK plasmid backgrounds. B19V RNAs generated by transfection of these constructs into COS-7 cells were analyzed with probe 8, probe 6, and probe 10 (Fig. 1B). Surprisingly, polyadenylation at (pA)d increased from 17% to 38% in the replicating background for B19V RNAs generated from a B19V genome with an NS mutation [pSKB19NS(−)Cap and pSVOB19NS(−)Cap] (Fig. 8A, compare lane 2 with lane 1) and from 15% to 36% for transcripts from a B19V genome in which the large intron from D1 to A1-1 had been deleted (pSKB19ΔD1A1Cap and pSVOB19ΔD1A1Cap) (Fig. 8A, compare lane 4 with lane 3). These results suggested that even weak replication of the B19V genome can still influence the polyadenylation of B19V RNAs at (pA)p and that upon replication, the level of readthrough of (pA)p in nonpermissive COS-7 cells approaches that for B19V infection of human bone marrow cells (1). Interestingly, this was the case even if replication was within a circular double-stranded DNA molecule.
FIG. 7.
COS-7 cells support only poor replication of SV40 ori-containing B19V plasmids. (A) Southern blot. COS-7 cells were transfected with the constructs that are diagramed in panel B. After 48 h, Hirt DNAs were prepared, digested with EcoRI (−) or DpnI (+), and resolved in 1% agarose gel. After transfer, the membrane was probed with the B19V NSCap probe. Lanes 1 and 2 are a control for DpnI digestion and size markers, respectively. The bands of newly synthesized B19V DNAs in COS-7 cells that were resistant to DpnI digestion are indicated with asterisks. (B) Diagrams of B19V constructs. The B19V genome from nt 199 to 5140, which contains only half of the terminal repeat (1/2TR), and the locations of the P6 promoter, the intron donors (D1 and D2) and acceptors (A1-1, A2-2, A2-1, and A2-2), and the polyadenylation sites [(pA)p and (pA)d] are shown. The regions of deletion in individual B19V minigenomes are indicated by the nucleotide numbers underneath. The B19V genomes or deletions were inserted into either an SV40 ori-containing plasmid backbone (pSVOd) or a regular backbone (pBluescript) and are termed the pSVOB19 or pSKB19 series of plasmids, as described in Materials and Methods.
FIG. 8.
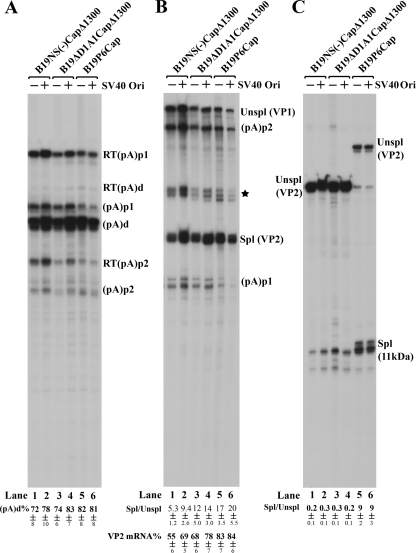
Despite poor replication of the B19V genome in COS-7 cells, it did moderately increase polyadenylation of B19V RNAs at (pA)d. COS-7 cells were transfected with pSVOd-based (pSVO) or pBluescript-based (pSK) constructs, as indicated by SV40 Ori (+) or SV40 Ori (−) at the top. The B19V genomes of B19NS(−)Cap and B19ΔD1A1Cap are diagramed in Fig. 7B. After 48 h, total RNAs were prepared and were protected by probe 8 (A), probe 6 (B), and probe 10 (C). All of the protected bands are designated as described in the legend to Fig. 3, and all of the quantifications were performed as described in the legend to Fig. 3.
As observed above, greater levels of VP2-encoding mRNAs (approximately 15%) were also seen under replication condition in COS-7 cells, which was consistent with increased splicing (approximately threefold more) at the A2-1 site (Fig. 8B, compare lanes 2 and 4 with lanes 1 and 3). Interestingly, increased splicing (approximately two- to threefold more) at the A2-2 site was also found under replication conditions (Fig. 8C, compare lanes 2 and 4 with lanes 1 and 3), similar to that seen in viral infection, indicating an increased 11-kDa protein-encoding mRNA level.
Splicing of the second intron at A2-1 and polyadenylation of B19V RNAs at (pA)p1 compete for the pool of B19V pre-mRNAs.
We previously demonstrated that splicing and polyadenylation at (pA)p compete for the pool of AAV5 pre-mRNAs (31, 34). Because for B19V both the (pA)p1 and (pA)p2 sites are located in the functional D2 and A2-1 intron, we investigated whether splicing and polyadenylation at (pA)p compete with each other for the processing of B19V mRNAs.
To test this possibility, we first made the construct pSKB19Cap, in which the distance between the P6 promoter and the D2 donor site was shortened, because we have previously shown that the size of the first exon is a critical factor for the splicing of the downstream intron of AAV5 (30). Following transfection of pSKB19P6Cap into COS-7 cells, splicing at A2-1 was significantly increased (Fig. 9B, lane 5), and as expected, polyadenylation of B19V RNAs at (pA)d was significantly decreased (Fig. 9A, lane 5). There was no apparent difference observed for both polyadenylation and splicing of the B19V mini-RNAs generated from the replicative construct of pSVOB19P6Cap in COS-7 cells (Fig. 9A, B, and C, lanes 6), suggesting that the increased splicing of B19V RNAs at A2-1 (spliced/unspliced ratio = 20) had driven the percentage of B19V RNAs polyadenylated at (pA)d to a maximum level (81%). Replication of pSVOB19P6Cap in COS-7 cells was not improved, although its size was reduced (Fig. 7A, lane 22).
FIG. 9.
Splicing and polyadenylation at the (pA)p site compete for the processing of B19V pre-mRNAs. COS-7 cells were transfected with pSVOd-based (pSVO) or pBluescript-based (pSK) constructs, as indicated by SV40 Ori (+) or SV40 Ori (−) at the top. The variant B19V minigenomes of B19NS(−)CapΔ1300, B19ΔD1A1CapΔ1300, and B19P6Cap are diagramed in Fig. 7B. After 48 h, total RNAs were prepared and were protected by probe 8 (A), probe 6 (B), and probe 10 (C). All of the protected bands are designated as described in the legend to Fig. 3, and all of the quantifications were performed as described in the legend to Fig. 3.
We next generated two constructs, pSKB19NS(−)CapΔ1300 and pSKB19ΔD1A1CapΔ1300, in which a region of approximate 1,300 nt between the A2-1 and A2-2 acceptors was deleted (Fig. 7B). This deletion resulted in an increase in the level of B19V RNAs spliced at A2-1 following transfection of these two constructs into COS-7 cells, similar to the levels seen with pSKB19P6Cap (Fig. 9B, compare lanes 1 and 3 to lane 5). Interestingly, pSKB19NS(−)CapΔ1300 and pSKB19ΔD1A1CapΔ1300, which were nonreplicative (Fig. 7A, lanes 12 and 16), already generated more than 70% of the B19V transcripts polyadenylated at (pA)d in COS-7 cells (Fig. 9A, lanes 1 and 3). Thus, the differences observed did not result from differences in replication of the two sets of constructs, as they generated similar levels of B19V mini-RNAs polyadenylated at (pA)d (Fig. 9A, lanes 2 and 4).
Taken together, these results indicate that increased splicing of the D2 to A2-1 intron, as a result of either introduction of a short first exon or a deletion in the last exon, was accompanied by a decrease in RNA polyadenylated at (pA)p and an increase in polyadenylation at (pA)d, which would be predicted to ultimately result in more production of VP2-encoding mRNA (Fig. 9B, lanes 1 to 6). Interestingly, the relative level of B19V RNAs spliced at the A2-2 acceptor site was decreased approximately fivefold following transfection of the two Δ1300 constructs (Fig. 9C, lanes 1 to 4), suggesting that splicing at A2-2 is unlikely involved in competition with polyadenylation at (pA)p.
DISCUSSION
In this study, we have shown that in the absence of replication, polyadenylation of B19V RNAs at (pA)p is predominant in cells both permissive and nonpermissive for viral replication, although the core poly(A) signal of (pA)p1 is noncanonical (AUUAAA). However, replication of the B19V genome, either during viral infection or following transfection of an infectious clone or transfection of the B19V genome replicated via heterologous origins of replication, resulted in an increased readthrough of (pA)p, increasing polyadenylation at (pA)d in both cell types. B19V mRNAs polyadenylated at (pA)d encode the capsid proteins, VP1 and VP2, and the essential nonstructural 11-kDa protein.
We found that polyadenylation at (pA)p1 and splicing of the second intron (D2 to A2-1) are in competition with each other for the pool of B19V pre-mRNAs. Increased readthrough of (pA)p1 was always accompanied with increased splicing of the second intron at A2-1 in virus infection, transfection of an infectious clone, and heterologous replication systems. On the other hand, “artificial” increased splicing of the second intron by shortening the first or the last exon during transfection of variant B19V plasmid mutants was able to induce B19V RNAs to pass (pA)p1. For AAV5, polyadenylation at (pA)p also competes with splicing; however, the mechanism underlying this effect must be different. In AAV5, the 3′ polypyrimidine tract and the (pA)p downstream element overlap. Thus, U2AF65, which binds to the 3′ polypyrimidine tract, and CstF65, which binds to the downstream element, likely compete for binding to the same RNA sequence. For AAV5, U1 snRNP binding to the small intron donor upstream of (pA)p has been shown to inhibit polyadenylation at the internal site (34). Our results with pSVOB19P6Cap, presented here, in which shortening of the 5′ exon increases readthrough of (pA)p, may indicate that B19V (pA)p shares this feature of regulation. The distance between the P6 promoter and the D2 donor was critical for efficient polyadenylation at (pA)p of B19V RNAs generated following transfection of the pSVOB19P6Cap construct. When this distance was shorter than 200 nt, which was the case for pSVOB19P6Cap, polyadenylation at (pA)p1 was inhibited. When this distance was large (∼400 nt), which was the case for pSVOB19ΔD1A1Cap, polyadenylation at (pA)p1 was not inhibited. Conceivably, as in AAV5 (30), a shorter distance between the 5′ end of the RNA and the first donor site stabilizes U1 snRNP binding to the D2 donor, which inhibits downstream polyadenylation at (pA)p1. Presumably, as previously reported for AAV5 (34), the distance between the D2 donor and (pA)p1, which is approximately 450 nt, was still able to allow for the upstream binding of U1 snRNP to moderately inhibit polyadenylation at (pA)p.
We have not ruled out the possibility that deletion of the 1,300-nt region between the A2-1 and A2-2 acceptors directly increases polyadenylation at (pA)d, thus reducing polyadenylation at (pA)p and increasing splicing at A2-1. However, polyadenylation of B19V RNAs at (pA)p and splicing at the D2 to A2-1 intron must compete within the same pool of B19V pre-mRNAs generated from the single P6 promoter. How splicing and polyadenylation compete in B19V RNA processing is under further investigation.
Our experiments do not allow us to determine whether replication of the B19V genome increases B19V RNA splicing, which then competes with and reduces polyadenylation at (pA)p, or alternatively, whether replication directly enhances polyadenylation at (pA)d. Replication of DNA viral genomes often takes place in replication centers, e.g., in adenovirus infection (27). It has been shown that replication of the AAV2 genome occurs within adenovirus replication centers, where RNA processing factors are concentrated (43). It is possible that if B19V replicates in such replication centers, then the replication center or replication factors that are recruited to the replication center help with B19V RNA splicing, which therefore competes with polyadenylation at (pA)p, or vice versa. It is further possible that when replication of the B19V genome is mediated via AAV5 ITRs in the presence of the AAV5 Rep and pHelper plasmids, it takes place in an adenovirus-like replication center formed by five adenovirus helper products (E1A, E1B, E2, E4orf6, and VA RNA). SV40 chromatin replicates in discrete subnuclear factories that reside near promyelocytic leukemia protein bodies (15, 16, 40). Thus, it is likely that replication of the B19V genome by the SV40 ori and the large T antigen also takes place in replication-like centers in COS-7 cells. Although the replication efficiency is low for these large SV40 ori-containing plasmids, it is possible that most of the SV40 ori-containing DNA templates were relocated to the replication center by the large T antigen. Whether replication of the B19V genome during natural infection occurs in a replication center, like AAV2 replication (43), or whether it reorganizes in a nuclear compartment like the case for minute virus of mice (47) needs to be investigated further. Apparently, a significantly higher percentage of polyadenylation at (pA)d was seen for these nonreplicative B19V templates in COS-7 cells (Fig. 8, lanes 1 and 3), and a slight increase of polyadenylation at (pA)d was seen for transfection of p5TRB19NS(−)Cap in the presence of pHelper expression (Fig. 6A, lane 6). These results imply that other factors or changes to the intranuclear architecture or location of the B19V transcription templates, in addition to the replication of the genome, may also be involved in regulation of polyadenylation, at least under these conditions.
mRNA processing occurs cotranscriptionally. Capping, splicing, and cleavage/polyadenylation of pre-mRNAs are interdependent events that are all stimulated in vivo by the carboxy-terminal domain of RNA polymerase II (2, 7, 10). It is possible that transcription of the B19V P6 promoter, which could be activated during replication of the B19V genome, confers efficient polyadenylation at (pA)p and thus increases the number of readthrough transcripts. Experiments in which the B19V P6 promoter is activated by heterologous activators via their cis elements are ongoing to investigate such a possibility.
Collectively, our study has demonstrated that replication of the B19V genome is the key determinant for the generation of sufficient B19V capsid-encoding mRNAs after B19V achieves proper entry into cells, an event which is mediated by the receptors and coreceptors. For parvoviruses, viral replication relies on cells cycling through the cellular S phase, for autonomous parvovirus (9), or occurs in conjunction with helper virus genes, for the dependoviruses (45). The remarkable tropism of B19V to erythroid progenitor cells supports the model that replication of the B19V genome requires cellular factors induced by differentiation directed toward the erythroid lineage. To date, the B19V-permissive cells identified all depend on the erythroid-specific cytokine Epo for proliferation and differentiation. The susceptibility of primary erythropoietic cells to B19V has been shown to increase with differentiation (39). Virus replication was detected only in Epo-containing cultures, such as UT7/Epo-S1 cells and CD36+ EPCs. However, we were recently able to show a significant increase of replication of the B19V infectious clone by adenovirus E2, E4orf6, and VA RNA in 293 cells (W. Guan and J. Qiu, unpublished data), suggesting that the tropism of B19V, currently thought to be restricted to the presence of cellular factors generated by erythroid differentiation, might be expanded by helper viruses to infect cells that express B19V receptors.
Acknowledgments
This work was supported by PHS grant RO1 AI070723 from NIAID and grant P20 RR016443 from the NCRR COBRE program to J.Q. and by PHS grants RO1 AI46458, RO1 AI56310, and RO1 AI21302 from NIAID to D.J.P.
We thank John Brunstein at University of British Columbia, Vancouver, for providing the pSVOdB19 plasmids.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Beard, C., J. St. Amand, and C. R. Astell. 1989. Transient expression of B19 parvovirus gene products in COS-7 cells transfected with B19-SV40 hybrid vectors. Virology 172659-664. [DOI] [PubMed] [Google Scholar]
- 2.Beyer, A. L., and Y. N. Osheim. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2754-765. [DOI] [PubMed] [Google Scholar]
- 3.Bonvicini, F., C. Filippone, S. Delbarba, E. Manaresi, M. Zerbini, M. Musiani, and G. Gallinella. 2006. Parvovirus B19 genome as a single, two-state replicative and transcriptional unit. Virology 347447-454. [DOI] [PubMed] [Google Scholar]
- 4.Brown, K. E. 2005. The genus Erythrovirus, p. 25-45. In J. Kerr, M. M. Fan, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 5.Brown, K. E., S. M. Anderson, and N. S. Young. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262114-117. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. E., and N. Young. 1997. Human parvovirus B19: pathogenesis of disease, p. 105-119. In L. J. Anderson and N. Young (ed.), Human parvovirus B19, vol. 20. Karger, Basel, Switzerland. [Google Scholar]
- 7.Calvo, O., and J. L. Manley. 2003. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 171321-1327. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1984. Characterization and molecular cloning of a human parvovirus genome. Science 2261161-1165. [DOI] [PubMed] [Google Scholar]
- 9.Cotmore, S. F., and P. Tattersall. 2006. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p. 171-181. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hoddler Arond, London, United Kingdom.
- 10.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 151783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freyssinier, J. M., C. Lecoq-Lafon, S. Amsellem, F. Picard, R. Ducrocq, P. Mayeux, C. Lacombe, and S. Fichelson. 1999. Purification, amplification and characterization of a population of human erythroid progenitors. Br. J. Haematol. 106912-922. [DOI] [PubMed] [Google Scholar]
- 12.Gallinella, G., E. Manaresi, E. Zuffi, S. Venturoli, L. Bonsi, G. P. Bagnara, M. Musiani, and M. Zerbini. 2000. Different patterns of restriction to B19 parvovirus replication in human blast cell lines. Virology 278361-367. [DOI] [PubMed] [Google Scholar]
- 13.Giarratana, M. C., L. Kobari, H. Lapillonne, D. Chalmers, L. Kiger, T. Cynober, M. C. Marden, H. Wajcman, and L. Douay. 2005. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat. Biotechnol. 2369-74. [DOI] [PubMed] [Google Scholar]
- 14.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26365-369. [DOI] [PubMed] [Google Scholar]
- 15.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jul-Larsen, A., T. Visted, B. O. Karlsen, C. H. Rinaldo, R. Bjerkvig, P. E. Lonning, and S. O. Boe. 2004. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp. Cell Res. 29858-73. [DOI] [PubMed] [Google Scholar]
- 17.Liu, J. M., S. W. Green, T. Shimada, and N. S. Young. 1992. A block in full-length transcript maturation in cells nonpermissive for B19 parvovirus. J. Virol. 664686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagawa, E., T. Yoshida, H. Takahashi, K. Yamaguchi, T. Nagano, Y. Kiriyama, K. Okochi, and H. Sato. 1999. Infection of the erythroid cell line, KU812Ep6 with human parvovirus B19 and its application to titration of B19 infectivity. J. Virol. Methods 8345-54. [DOI] [PubMed] [Google Scholar]
- 19.Moffatt, S., N. Yaegashi, K. Tada, N. Tanaka, and K. Sugamura. 1998. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J. Virol. 723018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita, E., K. Tada, H. Chisaka, H. Asao, H. Sato, N. Yaegashi, and K. Sugamura. 2001. Human parvovirus B19 induces cell cycle arrest at G(2) phase with accumulation of mitotic cyclins. J. Virol. 757555-7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouw, M. B., and D. J. Pintel. 2000. Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus. J. Virol. 749878-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munakata, Y., T. Saito-Ito, K. Kumura-Ishii, J. Huang, T. Kodera, T. Ishii, Y. Hirabayashi, Y. Koyanagi, and T. Sasaki. 2005. Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood 1063449-3456. [DOI] [PubMed] [Google Scholar]
- 23.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 61107-1119. [DOI] [PubMed] [Google Scholar]
- 24.Ozawa, K., J. Ayub, Y. S. Hao, G. Kurtzman, T. Shimada, and N. Young. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 612395-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozawa, K., J. Ayub, S. Kajigaya, T. Shimada, and N. Young. 1988. The gene encoding the nonstructural protein of B19 (human) parvovirus may be lethal in transfected cells. J. Virol. 622884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozawa, K., G. Kurtzman, and N. Young. 1986. Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233883-886. [DOI] [PubMed] [Google Scholar]
- 27.Pombo, A., J. Ferreira, E. Bridge, and M. Carmo-Fonseca. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 135075-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu, J., F. Cheng, L. R. Burger, and D. Pintel. 2006. The transcription profile of Aleutian mink disease virus in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J. Virol. 80654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, J., F. Cheng, and D. Pintel. 2006. Molecular characterization of caprine adeno-associated virus (AAV-Go. 1) reveals striking similarity to human AAV5. Virology 356208-216. [DOI] [PubMed] [Google Scholar]
- 30.Qiu, J., F. Cheng, and D. Pintel. 2007. Distance-dependent processing of adeno-associated virus type 5 RNA is controlled by 5′ exon definition. J. Virol. 817974-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu, J., R. Nayak, and D. J. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by both a downstream element within the intron 3′ splice acceptor and an element upstream of the P41 initiation site. J. Virol. 7883-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu, J., R. Nayak, G. E. Tullis, and D. J. Pintel. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 7612435-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, J., and D. J. Pintel. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 223639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu, J., and D. J. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by the distance between the promoter and the intron and is inhibited by U1 small nuclear RNP binding to the intervening donor. J. Biol. Chem. 27914889-14898. [DOI] [PubMed] [Google Scholar]
- 35.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 18122-34. [DOI] [PubMed] [Google Scholar]
- 36.Srivastava, A., E. Bruno, R. Briddell, R. Cooper, C. Srivastava, K. van Besien, and R. Hoffman. 1990. Parvovirus B19-induced perturbation of human megakaryocytopoiesis in vitro. Blood 761997-2004. [PubMed] [Google Scholar]
- 37.Srivastava, A., and L. Lu. 1988. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J. Virol. 623059-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Amand, J., and C. R. Astell. 1993. Identification and characterization of a family of 11-kDa proteins encoded by the human parvovirus B19. Virology 192121-131. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, T., K. Ozawa, K. Takahashi, S. Asano, and F. Takaku. 1990. Susceptibility of human erythropoietic cells to B19 parvovirus in vitro increases with differentiation. Blood 75603-610. [PubMed] [Google Scholar]
- 40.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 749694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tattersall, P. 2006. The evolution of parvovirus taxonomy, p. 5-14. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arond, London, United Kingdom.
- 42.Weigel-Kelley, K. A., M. C. Yoder, and A. Srivastava. 2003. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood 1023927-3933. [DOI] [PubMed] [Google Scholar]
- 43.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 701845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong, S., N. Zhi, C. Filippone, K. Keyvanfar, S. Kajigaya, K. E. Brown, and N. S. Young. 2008. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J. Virol. 822470-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Word, P. 2006. Replication of adeno-associated virus DNA, p. 189-212. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hoddler Arond, London, United Kingdom.
- 46.Yoto, Y., J. Qiu, and D. J. Pintel. 2006. Identification and characterization of two internal cleavage and polyadenylation sites of parvovirus B19 RNA. J. Virol. 801604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, P. J., K. T. Jensen, L. R. Burger, D. J. Pintel, and C. L. Lorson. 2002. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 763892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhi, N., I. P. Mills, J. Lu, S. Wong, C. Filippone, and K. E. Brown. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrate essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J. Virol. 805941-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhi, N., Z. Zadori, K. E. Brown, and P. Tijssen. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318142-152. [DOI] [PubMed] [Google Scholar]