Abstract
MicroRNAs (miRNAs) are potent RNA regulators of gene expression. Some viruses encode miRNAs, most of unknown function. The majority of viral miRNAs are not conserved, and whether any have conserved functions remains unclear. Here, we report that two human polyomaviruses associated with serious disease in immunocompromised individuals, JC virus and BK virus, encode miRNAs with the same function as that of the monkey polyomavirus simian virus 40 miRNAs. These miRNAs are expressed late during infection to autoregulate early gene expression. We show that the miRNAs generated from both arms of the pre-miRNA hairpin are active at directing the cleavage of the early mRNAs. This finding suggests that despite multiple differences in the miRNA seed regions, the primary target (the early mRNAs) and function (the downregulation of early gene expression) are evolutionarily conserved among the primate polyomavirus-encoded miRNAs. Furthermore, we show that these miRNAs are expressed in individuals diagnosed with polyomavirus-associated disease, suggesting their potential as targets for therapeutic intervention.
MicroRNAs (miRNAs) are small, ∼22-nucleotide RNA molecules that regulate gene expression (1). miRNAs bind to an mRNA and can repress translation or direct the cleavage of the target mRNA as part of the multiprotein RNA-induced silencing complex (RISC). The so-called seed region (nucleotides 2 to 8 of the 5′ ends of miRNAs) plays an important role in target selection by RISC-bound miRNAs (4). Host-encoded miRNAs have been shown previously to play a role in processes relevant to viral infection, such as apoptosis and the adaptive and innate immune responses (12). Additionally, members of several virus families have been reported to encode miRNAs (3, 5, 15, 17, 18, 20). Activities have been ascribed to a few such miRNAs; however, the functions of the majority of virus-encoded miRNAs remain poorly understood.
We have shown previously that the monkey polyomavirus simian virus 40 (SV40) encodes a pre-miRNA late during infection that is processed into two miRNAs (23). Both SV40-encoded miRNAs function to downregulate the expression of the viral early genes by directing their RISC-mediated cleavage. The two human polyomaviruses JC virus (JCV) and BK virus (BKV) cause significant morbidity and mortality in immunosuppressed patients (2, 16). JCV is the causative agent of a fatal central nervous system demyelinating disease, progressive multifocal leukoencephalopathy (PML). BKV is the causative agent of polyomavirus-associated nephropathy in renal transplant patients. Currently, there are no drugs that are effective against polyomaviral infection. In this report, we now show that the human polyomaviruses BKV and JCV also encode miRNAs. Interestingly, results from fine-mapping studies show that the JCV and BKV miRNAs contain multiple differences in their seed sequences compared to the seed sequences of the SV40 miRNAs. Despite this finding, these miRNAs show similarities to the SV40 miRNAs in processing and share a conserved autoregulatory function with the SV40 miRNAs. The evolutionary implications of this finding are discussed below.
MATERIALS AND METHODS
Computational prediction of viral miRNA precursors and miRNA cloning.
We used a new version of vMir (6, 21) with a vMir cutoff score of 175 to obtain candidate pre-miRNAs from the genomes of BKV and JCV (the genome accession numbers are as follows: BKV, NC_001538, and JCV, NC_001699). For the purpose of JCV miRNA cloning, we made an miRNA library from total RNA harvested from SVG cells infected with JCV. The library was generated utilizing a modified protocol of Pfeffer et al. (14). Briefly, total RNA was run on a denaturing 15% polyacrylamide gel, and small RNAs (approximately 10 to 40 nucleotides in length) were isolated. Small RNAs were eluted overnight from the gel slurries and ethanol precipitated using the Megaclear kit (Ambion, Austin, TX) according to the manufacturer's suggested protocol for harvesting small RNAs. Small RNAs were ligated to a preadenylated 3′ linker (5′-CTGTAGGCACCATCAATddCddCddC-3′, where “dd” is dideoxy ribose, which blocks the 3′ end) with T4 RNA ligase (Ambion, Austin, TX). Ligation products were gel purified, eluted, and ligated to the 5′ oligonucleotide (nonpreadenylated linker) (5′-CAGTTGATCAGAGCCCArGrGrG-3′, where “r” is ribose). Next, cDNAs were synthesized using reverse transcriptase (Ambion, Austin, TX), and the resulting products were cloned using a TA cloning kit (Invitrogen, Carlsbad, CA). Different primers specific to either the 5′ or 3′ portions of the predicted JCV miRNAs, combined with linker-specific primers, were utilized for PCR amplification. To amplify and map the JCV miRNAs, we performed nested PCRs. Resulting products were sequenced to define the 5′ and 3′ ends of the JCV 5′-arm (5p) and 3′-arm (3p) miRNAs. The PCR primers used were as follows: 5′ linker forward primer, GTTGATCAGAGCCCAGGG; 3′ linker backward primer, ATTGATGGTGCCTACAG; JCV 5p forward primer, TCTGAGA CCTGGG AAA; JCV 5p backward primer, AATCACAATGCTTTTCC; JCV 3p outer forward primer, AGAGCCCAGGGTGCTTG; JCV 3p inner forward primer, TGCTTGATCCATGTCCAGA; JCV 3p outer backward primer, GATGGTGCCTACAGGACTCT; and JCV 3p inner backward primer, GACTCTGGACATGGATCAAG.
Cell culture, virus infection, and PML brain tissues.
The human glial cell line U87 and the green monkey kidney cell line Vero were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and grown in Eagle's minimal essential medium supplemented with 10% fetal bovine serum. Cells were routinely tested to ensure that they were free of mycoplasma contamination. The cells were infected with 512 hemagglutination units of JCV (Mad-1/SVE delta strain) or BKV (Dunlop strain) for 1 h at 37°C in reduced (2%) serum. Posthumous clinical brain tissue samples were obtained from the National Neurological AIDS Bank (NNAB; Los Angeles, CA). All the samples came from pathologically normal brain tissues or the brains of patients diagnosed microscopically with PML. The diagnosis was made in all cases based on the presence of typical histological features of PML, along with the clinical history. Samples were also confirmed to be JCV positive via PCR analysis utilizing primers that amplified a region of the genome corresponding to a portion of JCV VP1. The PCR primers used were as follows: VP1 gene forward primer, CCCAGCAGTGGAGAGGACT, and VP1 gene backward primer, CAGCATTTTTGTCTGCAACTG.
RNA isolation and miRNA Northern blotting.
Total RNA was harvested from cultured cells infected with BKV or JCV or from solid brain tissue samples by using RNAbee according to the manufacturer's (TelTest Labs) directions. RNA was run on a Tris-borate-EDTA-urea-15% polyacrylamide gel. The gel was transferred onto a Hybond H+ membrane and probed for candidate miRNAs as described previously (6, 19). The blot was probed with radiolabeled oligonucleotides generated with P32-labeled radioactive gamma phosphate and T4 polynucleotide kinase (U.S. Biologicals, Cleveland, OH). The blot was first probed with the 5p probe, stripped with boiling hot stripping buffer (0.1% sodium dodecyl sulfate in double-distilled water), and then probed with the 3p probe, the internal-loop probe, or the control probe (hsa-let-7a). The probe sequences used were as follows: JCV 5p probe, CAATCACAATGCTTTTCCCAGGTCTCAGAAGCCTCT; JCV 3p probe, CAGAAGACTCTGGACATGGATCAAGCACTGAATCA; JCV internal-loop probe, AGCACTGAATCACAATCACAAT; BKV 5p probe, CAATCACAATGCTCTTCCCAAGTCTCAGATACTTCA; BKV 3p probe, ACTGAAGACTCTGGACATGGATCAAGCACTGAATCC; and hsa-let-7a probe, TGAGGTAGTAGGTTGTATAGTT.
Modified 5′ RACE for the detection of JCV and BKV early gene fragments.
mRNA cleavage fragments from JCV- and BKV-infected cells were identified utilizing a modified rapid amplification of cDNA ends (RACE) protocol (11) with a FirstChoice RNA-ligase-mediated-RACE kit (Ambion, Austin, TX). A 5′ RACE adaptor was ligated to total RNA extracted from infected cells. Reverse transcription followed by nested PCRs was performed using the 5′ RACE adapter primers and JCV- or BKV-specific primers. PCR products were cloned using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA). Twelve clones each from JCV and BKV were sequenced. The primer sequences used for the PCRs were as follows: 5′ adaptor, GCGAGCACAGAATTAATACGACTCACTATAGG(T)12VN; 5′ RACE outer primer, GCGAGCACAGAATTAATACGACT; 5′ RACE inner primer, CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG; JCV outer primer, GTAATA TGCAGTACATTTTAAT; JCV inner primer, TACATTTTAATAAAGTATAAC; BKV outer primer, GTACATATACATTTAATAAATGC; and BKV inner primer, ATTTAATAAATGCTGCTTTTG.
DNA constructs, transfection, and luciferase assay.
All DNA constructs were confirmed by sequence analysis. The plasmid pCDNA3.1JCV miRNA, which expresses JCV miRNA, was generated by cloning the PCR-amplified portion of the JCV genome into the BamHI/XhoI sites of the pCDNA3.1puro expression vector (19). A plasmid containing the entire JCV genome (kindly provided by Mike Imperiale, University of Michigan) served as the template to amplify the region of the JCV genome corresponding to the JCV pre-miRNA, as well as ∼1 kb of flanking regions. The primers used were as follows: JCV miRNA construction forward primer, CCTTGGATCCCTTCTTATAAGAGGAGGAGTAG, and JCV miRNA construction backward primer, GAGACTCGAGTTGGAAACCAAGTGTGAGGATG.
pCDNA3.1dsRluc and pCDNA3.1dsFFluc, expressing destabilized Renilla luciferase and firefly luciferase cDNAs, were constructed by PCR amplification of a luciferase gene insert generated from the plasmid templates pGL4.84hRlucCP/Puro (Promega; catalog no. E7521) and pMSCV-dsLuc2cp (a kind gift from Andrei Goga, University of California, San Francisco). PCR products were cloned into the KpnI/XhoI sites of the pCDNA3.1puro expression vector. The primers used were as follows: Renilla luciferase gene forward primer, ATTGGTACCATGGCTTCCAAGGTGTACGACCC; firefly luciferase gene forward primer, GCTGGTACCATGGAAGATGCCAAAAACATTAAG; and shared firefly and Renilla luciferase gene backward primer, AATCTCGAGTTAGACGTTGATCCTGGCGCTGGC.
A concatemerized region of the JCV genome corresponding to a portion of the large T antigen (TAg) was subcloned into the 3′ untranslated region of pCDNA3.1dsRluc. This 150-bp region of pCDNA3.1dsRluc contains sequences complementary to those of the JCV 5p and 3p miRNAs, as well as additional flanking regions. PCR utilizing primers that contain the nonpalindromic BanI restriction enzyme site was used to generate concatemers. The primers used were as follows: JCV TAg gene forward primer, GAAGGCACCAGACCCATTCTTGACTTTCCT, and JCV TAg gene backward primer, GCACCACGGACAGATGTGAAAGTGCAGTTT.
PCR products were digested with BanI, purified, and ligated with T4 DNA ligase (New England Biolabs, Inc., Beverley, MA). Taq polymerase (New England Biolabs, Inc., Beverley, MA) was used to fill in the overhanging nucleotides, and the resulting products were cloned using the TA cloning kit. Concatemers of the four copies of the JCV TAg construct (pCR2.1JCVTAg) were selected by restriction digest analysis and subcloned into the XhoI/XbaI sites of pCDNA3.1dsRluc vectors. PCR was used to generate inserts to subclone from the TA vector pCR2.1JCVTAg into pcDNA3.1. The primers used were as follows: JCV TAg concatemer forward primer, ATGCTCGAGCGGCCGCCAGTGTGATGGATA, and JCV TAg concatemer backward primer, GCATCTAGAGTAACGGCCGCCAGTGTGCTG.
293 cells were obtained from the ATCC and cultured in Dulbecco's modification of Eagle's medium supplemented with 10% fetal bovine serum. 293 cells were plated into 12-well dishes and transfected using the Lipofectamine 2000 reagent. Cells were transfected with the pCDNA3.1dsRlucJCVTAg plasmid, as well as pcDNA3.1dsFFluc as a transfection control, along with the pCDNA3.1 JCV miRNA expression vector. Transfections were conducted in the presence of a cocktail of 50 nM 2′-O-methylated inhibitors specific for the 5p and 3p miRNAs (see below) or an irrelevant control oligonucleotide. Cells were harvested 48 h after transfection and analyzed via a dual luciferase assay system (Dual-Glo luciferase assay system; catalog no. E2980) according to the recommendations of the manufacturer (Promega). The luciferase assay results were read on a Luminoskan Ascent luminometer (Thermo Electronic Corporation, Milford, MA). Results are presented with the Renilla luciferase levels normalized by the firefly luciferase levels.
Antisense oligonucleotides.
The modified antisense oligonucleotides were Dharmacon products synthesized by Thermo Fisher Scientific (Lafayette, CO). The antisense oligonucleotides were designed by a method similar to that described by Vermeulen et al. and contained 2′-O-methylated nucleotides (24). The primers used were as follows: irrelevant control antisense primer, AGAAGAGAGAAAUCUCUUCUUGGCCACUCGGGGGGACAACACUAAUCGCCAACAGACAUCUUCUCUUUCGAGAGAAGA; JCV 3p miRNA antisense primer, AGAAGAGAGAAAUCUCUUCUCAGAAGACUCUGGACAUGGAUCAAGCACUGAAUCACAAUCUUCUCUUUCGAGAGAAGA; and JCV 5p miRNA antisense primer, AGAAGAGAGAAAUCUCUUCUCUGAAUCACAAUCACAAUGCUUUUCCCAGGUCUCAUCUUCUCUUUCGAGAGAAGA.
Transfection with antisense oligonucleotides and Western blot analysis of JCV-infected cells.
Control and infected cells were transfected every 24 h, throughout the duration of infection, with either 50 nM irrelevant 2′-O-methylated oligonucleotide (control) or 50 nM (each) 2′-O-methylated antisense oligonucleotides directed at the JCV 3p and 5p miRNA sequences. Transfections were conducted using Lipofectamine 2000 according to the protocol of the manufacturer (Invitrogen, Carlsbad, CA). All samples were collected by scraping at 6 days postinfection and lysed for 3 h in 50 μl of 1× radioimmunoprecipitation assay buffer on ice. Protein concentrations were quantified by a Bradford assay, and results were read on an Eppendorf biophotometer. Fifty micrograms of protein from each sample was analyzed by electrophoresis on a 4 to 15% Tris-HCl gel for 1.5 h at 30 mA, and the proteins were transferred overnight onto nitrocellulose membranes at 4°C and 20 V. The membranes were then probed with an anti-SV40 TAg monoclonal antibody, diluted 1:1,000, that is known to cross-react with JCV TAg. Bound primary antibody was detected with goat anti-mouse horseradish peroxidase-conjugated secondary antibody diluted 1:20,000. The blots were then developed using the Rodeo sensitive Western blotting kit (U.S. Biologicals, Cleveland, OH). Bands were visualized by chemiluminescence on a ChemiDoc XRS system (Bio-Rad, Hercules, CA). Levels of TAg expressed in each sample were quantified using Bio-Rad Quantity One gel analysis software, and results were calculated relative to values for a loading control as percentages of the known amount of protein loaded in the standardized controls according to the manufacturer's directions. The results are displayed as percentages of TAg relative to multiple protein standards (Kaleidoscope; Bio-Rad).
RESULTS
JCV and BKV encode miRNAs homologous to SV40 miRNAs.
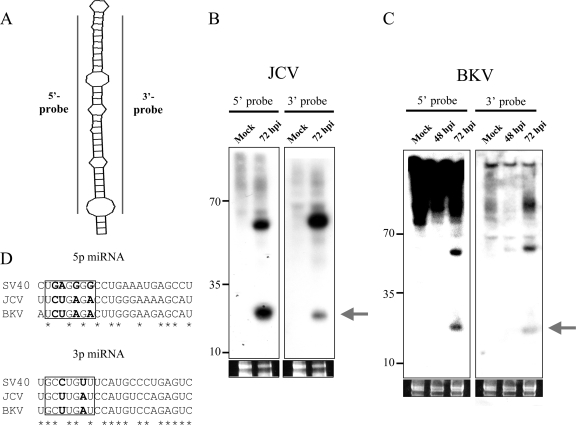
Previously, using the viral miRNA prediction software vMir, we predicted that JCV and BKV would encode pre-miRNAs homologous to SV40 pre-miRNA (23). However, given the high false-positive rate of vMir and the fact that the predicted candidates were only ∼65% identical, it was unclear if these predictions were valid (6). We utilized a newer version of vMir (6, 21) to examine the JCV and BKV genomes for candidate pre-miRNAs. This analysis identified four and two candidates for the BKV and JCV genomes, respectively (data not shown). Next, we looked for the possible derivatives from each predicted pre-miRNA via Northern blot analysis. Individual 5p and 3p probes were generated for each candidate, and Northern blot analysis was conducted (Fig. 1A). Only those candidates that showed sequence identity to SV40 miRNAs scored positive, and similar to SV40 miRNAs, these candidates were easily detected only at late times postinfection (Fig. 1B and C; also data not shown). Notably, for all three viruses, both arms of the pre-miRNA are processed into detectable miRNAs. Interestingly, like SV40, JCV and BKV each have a readily detectable band corresponding to the pre-miRNA. This results in a relatively higher ratio of the ∼60-nucleotide pre-miRNA band to the processed ∼22-nucleotide miRNA band than is detected for most cellular miRNAs (Fig. 1B and C) (23). Taken together, these results suggest that both JCV and BKV encode pre-miRNAs with notable similarities to SV40 pre-miRNA.
FIG. 1.
JCV and BKV encode miRNAs homologous to the SV40 miRNAs. (A to C) Northern blot analyses of the JCV and BKV miRNAs. (A) Diagram of probes used. (B) JCV miRNAs. (C) BKV miRNAs. The arrows indicate the bands corresponding to the miRNAs. Ethidium bromide staining of the low-molecular-weight RNA is shown as a loading control. Mock, uninfected. hpi, hours postinfection. (D) Alignment of the BKV and JCV 5p and 3p miRNAs with those of SV40. The 5p and 3p miRNAs are aligned with asterisks marking the conserved nucleotides. Note that the seed region (boxed; nucleotides 2 to 8), which is essential for target recognition (9, 10), is not conserved. Nucleotides that differ among the SEED sequences are in bold.
To map the derivative miRNAs with more precision, we generated a small RNA library from cells infected with either virus. To do this, we utilized a modified protocol of Pfeffer et al. (14). PCR primers specific to each arm of the vMir-predicted pre-miRNAs were used to individually map the 5′ and 3′ ends of the 5p and 3p miRNAs encoded by JCV and BKV. The results from these mapping studies showed some but not perfect nucleotide identity between the SV40 and BKV and JCV miRNAs. Importantly, both the 5p and 3p miRNAs from JCV and BKV have several nucleotide differences in their seed sequences compared to the SV40 miRNAs (Fig. 1D), suggesting different binding sites in their respective targets.
JCV and BKV miRNAs direct the cleavage of the early viral transcripts.
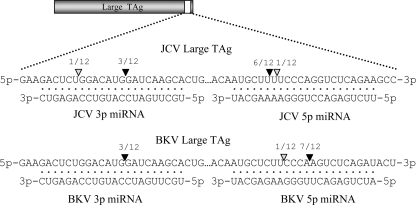
Since the 5p and 3p miRNAs are perfectly complementary to early mRNAs, we next examined whether they direct the cleavage of the early RNAs in a small interfering RNA-like fashion. We used a modified 5′ RACE analysis (11, 13, 25) to look for miRNA-mediated cleavage fragments. This analysis showed that, indeed, all four miRNAs direct the cleavage of their respective complementary early RNAs. The 5′ ends of the majority of the amplicons we sequenced (22 of 24) were complementary to the middle of an miRNA we identified (Fig. 2). Notably, for both JCV and BKV, more cleavage fragments mapped to the middle of the 5p miRNA than to the 3p miRNA. This result is consistent with our observation that the 5p miRNAs of both JCV and BKV are more abundant than the 3p miRNAs (Fig. 1B and C). To our knowledge, the polyomaviruses represent the only example in which different miRNAs, derived from both arms of a single pre-miRNA precursor, are active on the same molecular targets (the early mRNAs). These results show that multiple polyomaviruses encode miRNAs to autoregulate mRNA levels.
FIG. 2.
5′ RACE analysis demonstrates that each miRNA is active at directing the cleavage of the early mRNAs. A modified RACE protocol was utilized to screen for miRNA-mediated cleavage fragments. Portions of the early RNAs corresponding to the coding regions for the large TAgs are shown 5′ to 3′. The positions of the predominant 5′ ends of the cleavage fragments are denoted with black arrowheads, and the numbers of corresponding amplicons are shown. Gray arrowheads indicate the positions of the 5′ ends of less-common amplicons. miRNAs from the complementary strand are shown 3′ to 5′.
JCV miRNAs downregulate large-TAg expression late during infection.
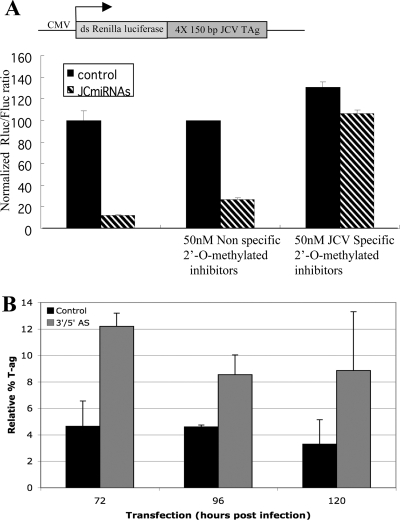
We predicted that the cleavage of the early mRNAs late during infection should result in a decrease in early protein levels. To test this hypothesis, we designed two different 2′-O-methylated antisense oligonucleotides complementary to either the 5p or 3p JCV miRNA by using a strategy similar to that of Vermeulen et al. (24). To quantitatively measure their inhibitory activities, we developed a luciferase-based reporter assay. The reporter construct contained four iterative copies of a region of the genome encoding the JCV large TAg that is complementary to the JCV miRNAs (Fig. 3A). As expected, the cotransfection of cells with the reporter and a plasmid expressing the JCV pre-miRNA resulted in a dramatic decrease in luciferase levels, while no decrease was detected with a control reporter (lacking the JCV complementary region) (Fig. 3A and data not shown). However, when cells were cotransfected with both the 5p and 3p inhibitors and the reporter and JCV miRNA constructs, almost no decrease in luciferase activity was observed. Transfection with an irrelevant control oligonucleotide had little effect in this assay. These results demonstrate that the JCV miRNAs can negatively regulate the expression of reporters containing 150 nucleotides of complementary early gene transcripts and that our antisense inhibitors blocked JCV miRNA-mediated mRNA cleavage. When we infected cells with JCV and then transfected them with the pooled 5p and 3p antisense inhibitors, a highly reproducible, albeit modest, increase in large-TAg protein levels was observable at late times postinfection (Fig. 3B), when the miRNAs are made. These results demonstrate that the JCV miRNAs downregulate early protein levels and that the antisense inhibitors we designed were effective at blocking, at least partially, the functions of both JCV miRNAs.
FIG. 3.
The JCV miRNAs expressed late during infection downregulate early protein levels. (A) A diagram of a Renilla luciferase reporter containing the region of the early genome complementary to the JCV miRNAs is shown (top). The graph shows luciferase levels from cells transfected with the reporter plus a plasmid expressing the JCV miRNAs (JCmiRNAs) or a negative control plasmid. The Renilla luciferase (Rluc) values were normalized to the values for firefly luciferase (Fluc; cotransfection control). CMV, cytomegalovirus; ds Renilla luciferase, double-stranded Renilla luciferase cDNA; 4X 150 bp JCV TAg, four copies of the JCV TAg construct. (B) A quantitative immunoblot analysis of the large TAg proteins shows that transfection with specific inhibitors (3′/5′ AS) of the JCV miRNAs increased early protein levels during infection. Results from transfection with an irrelevant control oligonucleotide are also shown. The data presented are the averages of results from three independent experiments, plotted as percentages of the protein standards loaded onto each gel.
JCV miRNAs are readily detectable in brain tissues of PML patients.
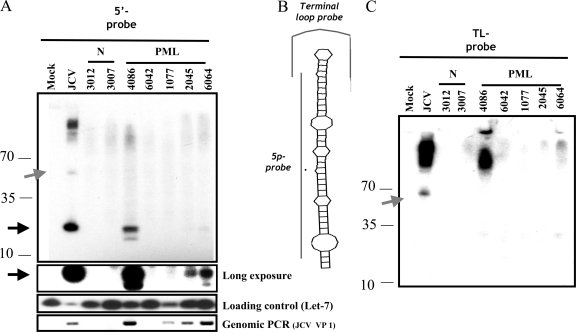
Most viral miRNAs have been identified by the infection of cultured cells in vitro. To examine whether the JCV miRNAs are made during infection in vivo, postmortem tissue samples from the brains of patients diagnosed with PML were obtained. We harvested total RNA and probed for the more abundant 5p JCV miRNA. Four of the five PML samples were positive for viral DNA, as assayed by PCR. Notably, the three samples with the highest levels of viral DNA (samples 4086, 2045, and 6064) scored positive for the JCV miRNA. One sample (4086) scored almost as high as the in vitro-infected cells (Fig. 4A and C, compare lanes 4086 to lanes JCV). The miRNA-specific band detected was unlikely to represent a degradation fragment, since a control probe for the loop region of the pre-miRNA (which corresponds to a sequence flanking the miRNAs and therefore should detect the pre-miRNA band but not the processed, stable ∼22-nucleotide miRNA band) (Fig. 4B) detected the pre-miRNA band without detecting any signal around ∼22 nucleotides (Fig. 4C). This result strongly suggests that there was no general enrichment of our samples with nonspecific cleavage fragments in the ∼22-nucleotide size range that may account for the miRNA-specific signal we detected. Due to the challenges of isolating PML lesions away from normal brain tissue, we believe that most of our PML lesion samples were contaminated with various amounts of normal, uninfected tissue. Thus, these results likely underrepresent the actual abundance of this miRNA in PML lesions. These data suggest that a majority of PML lesions in which JCV is actively replicating contain high enough levels of the JCV miRNA to be detected by Northern blot analysis. To our knowledge, this is the first report of a viral miRNA being detected in vivo in noncultured human tissue samples.
FIG. 4.
JCV miRNAs are detected in the brain tissue of PML patients. Total RNA was harvested from sections of brain tissue from patients diagnosed with PML. Northern blot analyses with probes specific for the JCV 5p and the loading-control let-7 miRNAs are shown. Northern blotting with a probe specific for the loop region of pre-miRNA showed that the JCV miRNA bands detected in brain tissue were not degradation fragments. (A) Full-length blot showing the detection of the JCV miRNA and pre-miRNA with the 5p probe. (B) Diagram of probes used. (C) Full-length blot from panel A, stripped and reprobed with the terminal loop (TL)-probe. Note that the control loop probe and the 5p probe both detect the pre-miRNA but that only the 5p probe detects bands at ∼22 nucleotides. This pattern rules out the possibility that degradation may account for the miRNA-specific bands detected. The miRNAs detected with the 5p probe are indicated with black arrows. The band corresponding to the pre-miRNA is indicated with a gray arrow. Numbers to the left of the blots indicate sizes in nucleotides. N, normal tissue samples.
DISCUSSION
Human polyomaviruses cause serious disease in immunocompromised patients, and effective therapies are lacking. We set out to explore whether the human polyomaviruses JCV and BKV encode miRNAs, a class of small RNA regulatory molecules that bind to and (typically) inactivate target mRNAs. Here, we have conclusively shown that homologous pre-miRNAs expressed by JCV and BKV show striking similarities to a pre-miRNA encoded by the related monkey polyomavirus SV40. JCV, BKV, and SV40 encode homologous pre-miRNA hairpins, both arms of which are processed into mature miRNAs. Despite being only ∼65% identical (5p miRNAs are ∼55% identical, and 3p miRNAs are ∼75% identical), all three pre-miRNAs share several atypical properties in terms of processing and abundance. Significantly, all produce two different miRNAs, both of which regulate early gene expression at late times of infection. To our knowledge, the pre-miRNAs encoded by the polyomaviruses are the only pre-miRNAs known to produce miRNAs from both strands of the precursor hairpin that are active on the same target (the early RNAs). Few viral miRNAs identified so far are evolutionarily conserved, and it is unclear if any have conserved functions. Notably, the seed region (the 5′ nucleotides of an miRNA, from positions 2 to 8) (Fig. 1D), which plays a crucial role in determining miRNA target specificity (9, 10), is not conserved between SV40 miRNAs and the human polyomavirus miRNAs. This suggests that a major function of the polyomaviral miRNAs is their conserved abilities to regulate their corresponding early RNAs, which necessarily have perfect complementarity to the miRNAs (including the seed region) generated from the opposite strand.
At least one of these miRNAs (from JCV) is expressed at robust levels during pathological infection in humans. Currently, there are no effective therapies against polyomaviral infection. As the numbers of immunosuppressed transplant and AIDS patients grow, so does the need for antipolyomaviral drugs. We have previously shown in in vitro assays that miRNA-mediated autoregulation of early gene expression can reduce cytotoxic T-cell-mediated lysis of SV40-infected cells. These data suggest a model in which the primate polyomaviral miRNAs may function to evade the immune response (22, 23). The implications of this model, combined with the recent successes in delivering anti-miRNA inhibitors directly into the brains of mice (7, 8), suggest that a similar approach directed against the JCV miRNAs may provide a treatment strategy for PML.
In summary, we have demonstrated that multiple members of the polyomavirus family encode a pre-miRNA with a function that has been conserved throughout millions of years of evolution. This finding suggests the likelihood that some miRNAs in other virus families may also have conserved functions.
Acknowledgments
We thank the NNAB for graciously providing the brain samples utilized in this study, Adam Grundhoff for sharing the latest version of vMir, Mike Imperiale for JCU and BKV plasmids, and members of the Sullivan lab for helpful comments regarding the manuscript.
This work was supported by University of Texas start-up funds and an ICMB fellowship to C.S.S., by NIH grants R01NS43097 and R01CA71878 to W.J.A., and by NIH grant NS 38841 to the NNAB.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 2.Berger, J. R. 2007. Progressive multifocal leukoencephalopathy. Curr. Neurol. Neurosci. Rep. 7461-469. [DOI] [PubMed] [Google Scholar]
- 3.Cullen, B. R. 2006. Viruses and microRNAs: is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Genet. 38(Suppl.)S25-S30. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz, W., S. N. Bhattacharyya, and N. Sonenberg. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9102-114. [DOI] [PubMed] [Google Scholar]
- 5.Grey, F., L. Hook, and J. Nelson. 2008. The functions of herpesvirus-encoded microRNAs. Med. Microbiol. Immunol. 197261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krutzfeldt, J., S. Kuwajima, R. Braich, K. G. Rajeev, J. Pena, T. Tuschl, M. Manoharan, and M. Stoffel. 2007. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 352885-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krutzfeldt, J., N. Rajewsky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438685-689. [DOI] [PubMed] [Google Scholar]
- 9.Lai, E. C. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30363-364. [DOI] [PubMed] [Google Scholar]
- 10.Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, and J. M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433769-773. [DOI] [PubMed] [Google Scholar]
- 11.Llave, C., Z. Xie, K. D. Kasschau, and J. C. Carrington. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2972053-2056. [DOI] [PubMed] [Google Scholar]
- 12.Lodish, H. F., B. Zhou, G. Liu, and C. Z. Chen. 2008. Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 8120-130. [DOI] [PubMed] [Google Scholar]
- 13.Mansfield, J. H., B. D. Harfe, R. Nissen, J. Obenauer, J. Srineel, A. Chaudhuri, R. Farzan-Kashani, M. Zuker, A. E. Pasquinelli, G. Ruvkun, P. A. Sharp, C. J. Tabin, and M. T. McManus. 2004. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 361079-1083. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer, S., M. Lagos-Quintana, and T. Tuschl. 2003. Cloning of small RNA molecules. Curr. Prot. Mol. Biol. 200326.4.1-26.4.18. [DOI] [PubMed] [Google Scholar]
- 15.Pfeffer, S., and O. Voinnet. 2006. Viruses, microRNAs and cancer. Oncogene 256211-6219. [DOI] [PubMed] [Google Scholar]
- 16.Randhawa, P., A. Vats, and R. Shapiro. 2006. The pathobiology of polyomavirus infection in man. Adv. Exp. Med. Biol. 577148-159. [DOI] [PubMed] [Google Scholar]
- 17.Samols, M. A., and R. Renne. 2006. Virus-encoded microRNAs: a new chapter in virus-host cell interactions. Future Virol. 1233-242. [Google Scholar]
- 18.Sarnow, P., C. L. Jopling, K. L. Norman, S. Schutz, and K. A. Wehner. 2006. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat. Rev. Microbiol. 4651-659. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan, C. S., and D. Ganem. 2005. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 797371-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan, C. S., and D. Ganem. 2005. MicroRNAs and viral infection. Mol. Cell 203-7. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan, C. S., and A. Grundhoff. 2007. Identification of viral microRNAs. Methods Enzymol. 4273-23. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan, C. S., A. Grundhoff, S. Tevethia, R. Treisman, J. M. Pipas, and D. Ganem. 2006. Expression and function of microRNAs in viruses great and small. Cold Spring Harbor Symp. Quant. Biol. 71351-356. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435682-686. [DOI] [PubMed] [Google Scholar]
- 24.Vermeulen, A., B. Robertson, A. B. Dalby, W. S. Marshall, J. Karpilow, D. Leake, A. Khvorova, and S. Baskerville. 2007. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA 13723-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yekta, S., I. H. Shih, and D. P. Bartel. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304594-596. [DOI] [PubMed] [Google Scholar]