Abstract
Retroviral Gag proteins are synthesized as soluble, myristoylated precursors that traffic to the plasma membrane and promote viral particle production. The intracellular transport of human immunodeficiency virus type 1 (HIV-1) Gag to the plasma membrane remains poorly understood, and cellular motor proteins responsible for Gag movement are not known. Here we show that disrupting the function of KIF4, a kinesin family member, slowed temporal progression of Gag through its trafficking intermediates and inhibited virus-like particle production. Knockdown of KIF4 also led to increased Gag degradation, resulting in reduced intracellular Gag protein levels; this phenotype was rescued by reintroduction of KIF4. When KIF4 function was blocked, Gag transiently accumulated in discrete, perinuclear, nonendocytic clusters that colocalized with endogenous KIF4, with Ubc9, an E2 SUMO-1 conjugating enzyme, and with SUMO. These studies identify a novel transit station through which Gag traffics en route to particle assembly and highlight the importance of KIF4 in regulating HIV-1 Gag trafficking and stability.
The Gag polyprotein is an essential structural component of all retroviruses and lentiviruses, playing a leading role throughout multiple stages of the viral life cycle (1, 28). For a viral particle to form, thousands of Gag proteins must multimerize, bind to membranes, and then assemble into a virion that is released from the cell. For human immunodeficiency virus type 1 (HIV-1) Gag, the components responsible for membrane binding have been mapped to the N-terminal matrix domain (MA) and include the N-terminal myristate and a cluster of basic residues (35, 55, 58). Regions within the capsid and nucleocapsid domains promote Gag-Gag multimerization, while the C-terminal p6 domain of Gag is needed for budding and release of the newly formed virion. Although Gag contains all the structural information needed to form a virus-like particle (VLP), assembly and release of HIV-1 require recruitment and interaction of Gag with multiple host cell proteins (4, 52).
A key unanswered question is how Gag proteins move from their site of synthesis on soluble, cytoplasmic polysomes to assembly points on cellular membranes. Recent studies point to the existence of intracellular trafficking intermediates for HIV-1 Gag (38-40). However, the molecular basis for regulation of Gag movement within the cell is not known. We hypothesized that cellular motor proteins might be important for Gag movement. This hypothesis is based on the finding that Gag proteins from HIV-1, simian immunodeficiency virus, Moloney murine leukemia virus, and Mason-Pfizer monkey virus (MPMV) bind to KIF4, a kinesin family motor protein (21). The interaction was mapped to the N-terminal MA domain of Gag and the C-terminal tail of KIF4 (48). To date, no studies have examined the functional importance of KIF4 interaction with HIV-1 Gag.
KIF4A (hereby denoted KIF4) is a plus-end-directed, microtubule (MT)-stimulated ATPase in the kinesin motor family (24, 34). KIF4 contains a characteristic N-terminal motor domain, which binds MTs and ATP and is responsible for force generation along MTs (10, 16, 45). It also contains a central stalk region involved in dimerization and a C-terminal tail believed to mediate binding of cargo. KIF4 localizes to both the cytoplasm and the nucleus (24) and has been implicated in organelle and vesicle transport, midzone formation, chromosome segregation during mitosis, and cytokinesis (22, 37, 59) as well as in regulation of programmed cell death in juvenile neurons (27). These studies establish KIF4 as a multifaceted motor protein that regulates the movement of multiple intracellular components.
Here we describe the first functional evidence linking KIF4 to early stages of HIV-1 Gag biosynthetic trafficking. We show that disruption of endogenous KIF4 slows progression of newly synthesized Gag through its trafficking intermediates and results in decreased particle production. In addition, we report a novel, unanticipated regulatory function for KIF4, i.e., control of Gag protein stability. When KIF4 is disrupted, degradation of intracellular Gag is dramatically increased. Finally, we identify the earliest defined trafficking intermediate as a perinuclear cluster of Gag that also contains KIF4, Ubc9, and SUMO. Failure to progress beyond this stage results in degradation of intracellular Gag protein. This knowledge could potentially be exploited as a means to develop therapeutic agents that block HIV-1 viral particle production.
MATERIALS AND METHODS
Antibodies and reagents.
Polyclonal rabbit anti-actin was purchased from Sigma, St. Louis, MO; monoclonal mouse anti-CD63, polyclonal rabbit anti-Fyn, monoclonal mouse anti-caveolin-1, and monoclonal mouse anti-EEA1 were purchased from BD Transduction Laboratories, San Diego, CA; monoclonal mouse anti-myc, polyclonal rabbit anti-extracellular-signal-regulated kinase 2 (anti-ERK2), polyclonal rabbit anti-green fluorescent protein (anti-GFP), monoclonal mouse anti-vimentin, monoclonal mouse anti-lamin A, and monoclonal mouse anti-gp120 were purchased from Santa Cruz Biotechnology, Santa Cruz, CA; monoclonal mouse anti-Src was purchased from Upstate Biotechnology/Millipore, Billerica, MA; monoclonal anti-CD4 (leu3A) was purchased from the MSKCC Core Antibody Facility; anti-mouse Alexa fluor 594 and anti-rabbit Alexa fluor 594 were purchased from Molecular Probes/Invitrogen, Carlsbad, CA; and HIV-1 p24 monoclonal antibody was obtained from the NIH AIDS Research & Reference Reagent Program, Germantown, MD. Polyclonal anti-p24CA was generated in rabbits immunized with p24CA. Antibody to KIF4 was a kind gift from Mariano Bisbal and Alfredo Caceres (Mercedes y Martin Ferreyra Institute for Medical Research, Cordoba, Argentina). Protein Plus A/G-agarose was purchased from Santa Cruz Biotechnology. Tran35S Cys/Met was purchased from MP Biomedicals (Solon, OH). Chloroquine and MG-132 were purchased from Sigma.
Plasmids, transfections, and metabolic labeling.
Plasmids encoding pCMV5 Gag, GagGFP, Gag(Δp6), pHXB2ΔBalD25S Gag, Fyn(10)Gag, c-Src, and Fyn have been described previously (15, 26, 50). Plasmids encoding KIF4-T and KIF3A-T were generated by reverse transcription of human A549 cell mRNA, PCR, and cloning into Gateway (Invitrogen) mammalian expression vectors. KIF3A-T cDNA construction has been described previously (17). KIF tail constructs were amplified by PCR from human A549 cells and cloned into mammalian Gateway (Invitrogen) expression vectors as described by the manufacturer. KIF4A-T (amino acids [aa] 741 to 1232) and KIF4A-TS (aa 875 to 1232) were amplified using forward primers 5′ GCTCGAGTGAAGAATTGGCTTGGA 3′ and 5′ TCCTCCAAAATACAGGTCAGCAAACTT 3′, respectively. The reverse primer was 5′ TCAGTGGGCCTCTTCTTCGATAGGGGA 3′. Lentiviral Gag was generated by cloning GagGFP into the ViraPower lentiviral expression system (Invitrogen). The c-SrcGFP plasmid contains chicken c-Src fused to GFP with a 7-aa linker between the C terminus of c-Src and the N terminus of GFP. pcDNA_vSrc(ΔMA)Gag-myc was a kind gift from Paul Spearman (Emory University School of Medicine, Atlanta, GA). The pNL4.3 HIV-1 cDNA was a kind gift from John P. Moore (Weill Graduate School of Medical Sciences of Cornell University, New York, NY). pEGFP-N2-CD4FL was a kind gift from John Wills (Pennsylvania State University College of Medicine, Hershey). pCMV.Myc-Ubc9 was a kind gift from Robert Weldon (University of Nebraska, Lincoln, NE). The GFP-250 plasmid was a kind gift from Elizabeth Sztul (University of Alabama at Birmingham, Birmingham). pGFP-SUMO1 was a kind gift from Pier Paolo Pandolfi (Memorial Sloan-Kettering Cancer Center, New York, NY). KIF3 and KIF4 small interfering RNAs (siRNAs) were obtained from the HTS/siRNA core facility at Memorial Sloan-Kettering Cancer Center. The KIF4 sequences used were as follows: KIF4_siRNA#1, 5′ GCUUCAAGAUUCUCUAGGA 3′; KIF4_siRNA#2, 5′ CAAUUGAUUACCCAGUUAU 3′; and KIF4_siRNA#3, 5′ CCACUGAAGUUGGAGUCAU 3′.
COS-1 cells were seeded to 90% confluence and transfected with cDNA, using Lipofectamine 2000 (Invitrogen). Metabolic labeling was performed after 24 h of incubation at 37°C as described previously (49), using 50 μCi/ml Tran35S label (MP Diagnostics). Briefly, cells were incubated in cysteine- and methionine-deficient Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (FBS) for 1 h. Cells were pulse labeled for 15 min at 37°C and either maintained in medium containing label or chased for various lengths of time in chase medium (Dulbecco's modified Eagle's medium plus 10% FBS plus 100 μM cysteine and methionine). VLPs were purified from the medium as described below. Cells were harvested by being washed twice in cold saline-Tris-EDTA buffer followed by lysis in 1× RIPA buffer (150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, 1% Triton X-100, and 10 mM Tris, pH 7.4) containing protease inhibitors (0.5 mM 4-(2-aminoethyl)benzenesulfonyl flouride, 0.5 μg/ml leupeptin, and 0.5 μg/ml aprotinin). The lysate was clarified at 55,000 × g at 4°C for 15 min and immunoprecipitated with rabbit anti-p24 antibody and protein A/G beads overnight at 4°C. Samples were washed three times with cold 1× RIPA buffer and then subjected to SDS-polyacrylamide gel electrophoresis. Analysis of radiolabeled Gag was performed by exposure of the dried gels to a phosphorimager screen, which was scanned using a Fuji FLA500 phosphorimager. Quantitation and preparation of visual images were performed with ImageQuant software (Molecular Dynamics).
VLP preparation.
VLPs were purified as previously described (26). Briefly, medium from transfected cells was clarified at 500 × g for 10 min at 4°C and filtered through a 0.2-μm-pore-size filter. The filtrate was layered on top of a 20% sucrose cushion and centrifuged for 2 h at 34,500 rpm in a Ti55 rotor. VLPs were solubilized in 1× RIPA buffer and immunoprecipitated using rabbit anti-p24 antibody (as described above).
Proteasomal and lysosomal inhibition.
COS-1 cells transfected with appropriate cDNAs and incubated for 24 h were then preincubated for 1 h with 10 μM MG-132 (proteasomal inhibition) or 20 mM NH4Cl plus 30 μM chloroquine (lysosomal inhibition). Cells were then pulsed with 50 μCi/ml Tran35S label, chased over a period of 7 h, and processed as described above.
Microinjection.
COS-1 cells were pressure microinjected intranuclearly with cDNAs in HKCl (10 mM HEPES, 140 mM KCl, pH 7.4), using a Narishige micromanipulator (Narishige, Greenvale, NY). cDNA concentrations were 20 μg ml−1 for GagGFP and 5 to 10 μg ml−1 for KIF tail constructs. After injection, cells were maintained at 37°C for 120 min to allow for expression of cDNAs. Cells were transferred to recording medium (phenol red-free Hanks balanced salt solution with 20 mM HEPES, 1% FBS, 4.5 g/liter glucose, and essential and nonessential amino acids). Cells were incubated in a thermally controlled chamber (Harvard Apparatus) at 37°C (an open chamber was used to permit oxygen exchange) on a Nikon TE-2000U microscope (Nikon Inc., Mellville, NY), and GagGFP trafficking was monitored by time-lapse fluorescence microscopy. Images were collected at the indicated intervals, using a ×40 objective and an ORCAII-ER microscope (Hamamatsu Photonics, Bridgewater, NJ). All devices were controlled by MetaMorph (Molecular Devices, Sunnyvale, CA).
KIF4 knockdown with siRNA.
COS-1 cells were transfected on two consecutive days with 20 nM siRNA in Lipofectamine 2000. On the second day of transfection, the siRNA was cotransfected with the indicated cDNA. KIF4 and Gag mRNA levels were quantified from reverse-transcribed mRNA by TaqMan real-time quantitative PCR with appropriately designed primers. Changes in KIF4 protein levels were quantified by comparing the integrated fluorescence intensities obtained with anti-KIF4 and anti-calnexin antibody staining in cells treated with KIF4_siRNA or LacZ_siRNA.
Immunofluorescence.
Immunofluorescence analysis was performed as previously described (38). Briefly, at 24 h posttransfection, COS-1 cells were washed with phosphate-buffered saline (PBS) and fixed with 4% formalin-PBS for 15 min. Cells were incubated with a 1:250 dilution of primary antibody in PBS plus 10% FBS for 1.5 h, washed in PBS, and then incubated with a 1:500 dilution of secondary antibody for 45 min. Cells were washed and mounted on microscope slides in Pro-Long mounting medium (Invitrogen). Laser scanning confocal microscopy was performed on a Zeiss LSM510 confocal microscope equipped with an Axiovert 100 M inverted microscope, using a ×63, 1.2-numerical-aperture water immersion lens for imaging. Single confocal images were processed in Adobe Photoshop.
RESULTS
Disruption of endogenous KIF4 decreases VLP production and alters intracellular Gag stability.
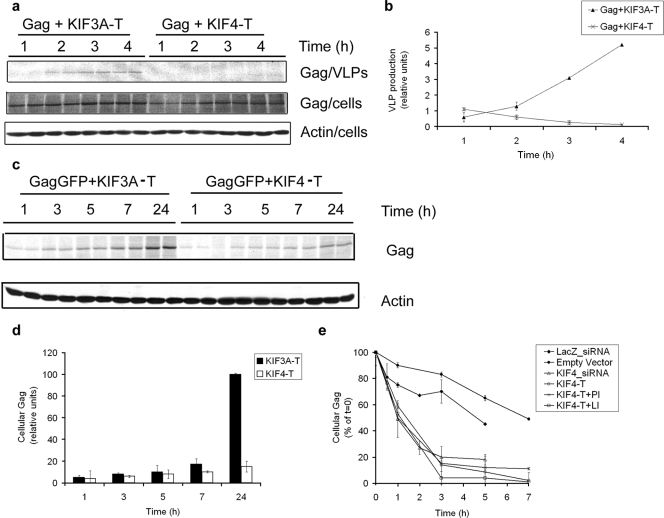
We first examined the effect of disrupting endogenous KIF4 on Gag function, specifically VLP production. Previous studies have established that constructs containing only the cargo-binding tail domain of various kinesins function as dominant-negative inhibitors of kinesin function (17, 46, 51, 54). We generated KIF4-T, a tail domain cDNA construct encoding aa 741 to 1232 of KIF4 fused to murine red fluorescent protein (mRFP) (17). As a control, we used a dominant-negative KIF3A tail construct encoding aa 601 to 702 of the KIF3A C-terminal tail (KIF3A-T) fused to mRFP. COS-1 cells were cotransfected with cDNAs encoding HIV-1 Gag and either myc-tagged KIF4-T or KIF3A-T. Cells were incubated with [35S]Cys/Met-containing medium 24 h after transfection, and incorporation of radiolabel into cell-associated Gag and Gag in VLPs was quantified over 4 h (Fig. 1a). The rates of radiolabel incorporation into Gag in cells expressing either KIF4-T or KIF3A-T were essentially equivalent during the 4-h labeling time course. However, VLP production was significantly reduced (∼10-fold) in the presence of the dominant-negative KIF4-T construct compared with that in cells expressing the control construct KIF3A-T (Fig. 1b).
FIG. 1.
Effects of KIF4 disruption on VLP production and intracellular Gag stability. (a and b) COS-1 cells were cotransfected with 2 μg Gag and 2 μg of either KIF3A-T or KIF4-T cDNA and radiolabeled with Trans35S label. Radiolabeled Gag from cell lysates and VLPs was immunoprecipitated with anti-p24 antibody, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and phosphorimager analysis. The amount of actin was determined by Western blotting. (c and d) COS-1 cells were cotransfected with Gag and KIF3A-T or KIF4-T, and levels of intracellular Gag were monitored over a 24-h period by Western blotting of cell lysates. (e) COS-1 cells were cotransfected with Gag and either empty vector, KIF4-T, LacZ_siRNA, or KIF4_siRNA. Transfected cells were then preincubated for 1 h in the presence or absence of either a proteasome inhibitor ([PI] MG-132) or lysosomal inhibitors ([LI] NH4Cl and chloroquine). The cells were pulse labeled for 15 min in medium containing Tran35S-labeled cysteine-methionine and then chased in nonradioactive medium with the respective inhibitor for the indicated times. The amount of labeled Gag remaining at each time point was quantified and plotted relative to the amount of Gag at time zero (100%). Each experiment was performed in duplicate a minimum of two times.
To better understand the effects of KIF4-T on Gag function, we monitored intracellular Gag levels over a 24-h period. COS-1 cells were cotransfected with HIV-1 Gag cDNA and either KIF4-T or control KIF3A-T cDNA, incubated for 24 h, and then radiolabeled in medium containing [35S]Cys/Met for up to 24 h. At early time points (up to 5 h), Gag levels were equivalent under the two conditions. However, at later time points, beyond 7 h, intracellular Gag levels were significantly reduced (∼5-fold at 24 h) in cells expressing KIF4-T compared with those in cells expressing KIF3A-T (Fig. 1c and d). To determine whether the reduction in intracellular Gag was due to decreased Gag stability, a pulse-chase experiment was performed to monitor rates of Gag degradation. Under control conditions, Gag degraded with a half-life of ∼6.5 h (Fig. 1e). In cells expressing KIF4-T, the half-life for Gag was considerably shorter, i.e., ∼1 h. The addition of either proteasomal (MG-132) or lysosomal (NH4Cl and chloroquine) inhibitors did not have any effect on Gag degradation rates (Fig. 1e). In contrast, these compounds did inhibit epidermal growth factor receptor degradation (see Fig. S1 in the supplemental material). Thus, expression of endogenous KIF4-T leads to an increase in the rate of Gag degradation, thereby reducing intracellular Gag protein stability.
Regulation of Gag stability by KIF4 is mediated through the N-terminal region of Gag.
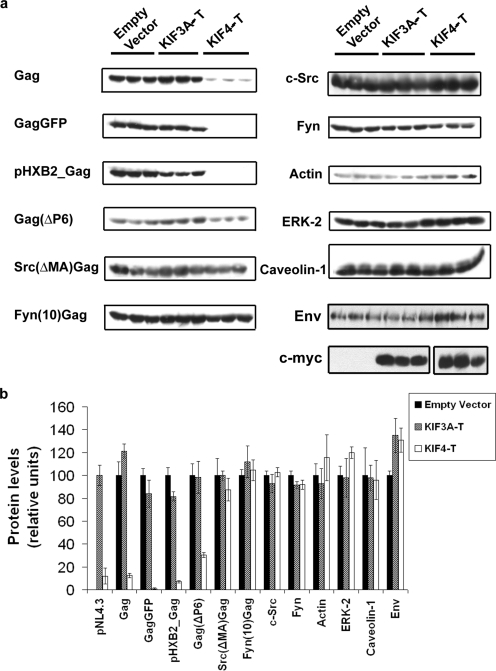
To determine whether KIF4 disruption altered host cell protein expression, we monitored the levels of several endogenous proteins in cells transfected with cDNAs encoding either KIF3-T or KIF4-T or with an empty vector. Western blotting with anti-myc antibody revealed equivalent expression levels of KIF3-T and KIF4-T. We detected no changes in levels of endogenous actin, ERK-2, or caveolin-1 (Fig. 2a and b). Next, we examined expression levels of two transfected proteins, c-Src and Fyn. Like Gag, these two proteins are myristoylated and targeted to the plasma membrane. Moreover, c-Src and Gag use a similar mechanism of membrane binding, a myristate-plus-basic motif in the N-terminal region (41, 58). Despite these similarities, the expression levels of c-Src and Fyn were not affected by the presence of KIF4-T (Fig. 2a and b).
FIG. 2.
Expression of dominant-negative KIF4 causes a reduction in intracellular Gag level. COS-1 cells were cotransfected with either empty vector, KIF3A-T, or KIF4-T along with constructs encoding the indicated Gag proteins, c-Src, or Fyn or with pHXB2ΔΒalD25S. The cells were incubated for 24 h. Levels of the indicated exogenous proteins, of endogenous actin, ERK-2, and caveolin-1 (in Gag-transfected cells), and of Env (anti-gp120) were monitored by Western blotting with the appropriate antibodies. (a) Representative panels (triplicates) showing expression of the indicated proteins. (b) Graphical representation of the combined experiments represented in panel a. Each experiment was performed a minimum of two times in triplicate.
The effects of KIF4-T on Gag derived from several different expression vectors were evaluated. We observed significant reductions in Gag expression for both the GagGFP fusion protein and untagged Gag. Both of these constructs encode codon-optimized Gag driven by a cytomegalovirus promoter. We also monitored Gag expressed from an HIV-1-derived lentiviral construct, pHXB2ΔBalD25S; in this plasmid, Gag expression is driven by the HIV-1 long terminal repeat. The presence of KIF4-T induced a dramatic decrease in the level of Gag expression (Fig. 2a and b). Finally, the effect of KIF4-T was tested in cells transfected with an infectious clone of pNL4.3 HIV-1 cDNA. Gag levels were strongly reduced by KIF4-T but not by KIF3-T (Fig. 2b). In contrast, no effect on another viral protein, Env, was detected (Fig. 2a and b).
The N-terminal matrix region of Gag has been shown to interact with KIF4 (48). We tested the ability of KIF4-T to alter expression levels of the following three Gag mutants: Gag(ΔP6), a construct with a C-terminal deletion of the p6 region of Gag; Src(ΔMA)Gag, where the matrix domain of Gag is replaced with the first 10 aa from Src; and Fyn(10)Gag, a chimera where the first 10 aa of Gag are replaced by those of Fyn (7, 26). In the presence of KIF4-T, Gag(ΔP6) expression was reduced significantly, although to a slightly lesser extent than that observed for full-length Gag. In contrast, KIF4-T had no effect on the level of Src(ΔMA)Gag or Fyn(10)Gag (Fig. 2a and b). These findings imply that the effects of disrupting endogenous KIF4 with dominant-negative mutants are specific to Gag and to the N-terminal region of Gag.
Intracellular Gag stability is reduced by KIF4_siRNA-mediated knockdown.
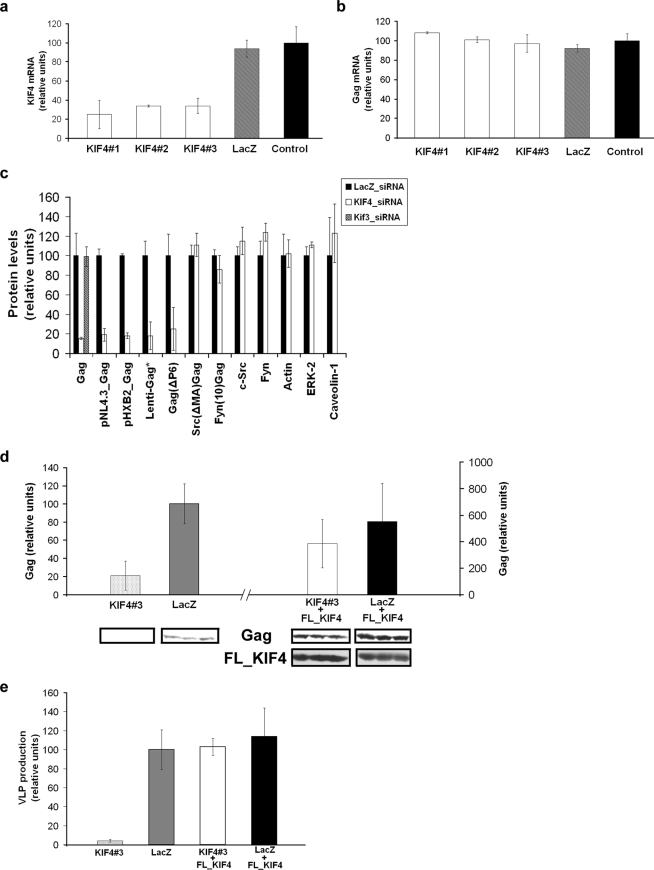
To confirm that the reduction in Gag protein levels induced by dominant-negative KIF4 was due to disruption of KIF4 function, we utilized siRNA technology to knock down endogenous KIF4. Three different siRNA sequences directed against human KIF4 were tested. The levels of endogenous KIF4 protein in COS-1 cells were undetectable by Western blotting. However, quantitative reverse transcriptase PCR (qPCR) revealed three- to fourfold decreases in endogenous KIF4 mRNA levels compared to those in LacZ and no-siRNA controls (Fig. 3a). No significant effect on Gag mRNA levels was detected in cells treated with any of the KIF4_siRNAs (Fig. 3b). In addition, we observed an ∼60% decrease in KIF4 immunofluorescence in cells treated with KIF4 siRNA compared with that in cells treated with a control LacZ siRNA.
FIG. 3.
siRNA-directed knockdown of KIF4 reduces intracellular Gag levels. (a and b) COS-1 cells were transfected with one of three different KIF4_siRNA constructs, with LacZ_siRNA, or with no siRNA. Levels of KIF4 mRNA (a) or Gag mRNA (b) were quantified by qPCR following reverse transcriptase PCR with KIF4- or Gag-specific TaqMan probes. Each experiment was performed in triplicate. (c) COS-1 cells were cotransfected with either control LacZ_siRNA or a KIF4_siRNA (20 nM) along with constructs encoding the indicated Gag proteins, c-Src, or Fyn. For Gag only, cells were also cotransfected with KIF3_siRNA (40 nM). The cells were incubated for 24 h. Levels of the indicated exogenous proteins or of endogenous actin, ERK-2, or caveolin-1 (in Gag-transfected cells) were monitored by Western blotting with the appropriate antibodies. Each experiment was performed a minimum of two times in triplicate. Lenti-Gag*, Jurkat T cells expressing Gag via a lentiviral delivery system. (d) Gag-transfected COS-1 cells were cotransfected with either LacZ_siRNA or KIF4_siRNA#3 and with either empty vector (left axis) or a vector expressing full-length rat KIF4 (right axis). The amounts of Gag and rat KIF4 in cell lysates were determined by Western blotting (representative blots are shown below the graphs). (e) The amount of Gag in VLPs isolated from cells treated as described for panel d was quantified.
The intracellular levels of several transfected and endogenous proteins were compared in COS-1 cells treated with KIF4_siRNA versus LacZ_siRNA controls. Treatment of cells with KIF4_siRNA resulted in decreased intracellular levels of Gag, pNL4.3 Gag, pHXB2ΔBalD25S Gag, and Gag(ΔP6) but had no effect on the intracellular levels of transfected Src(ΔMA)Gag, Fyn(10)Gag), c-Src, and Fyn or endogenous actin, ERK-2, and caveolin-1 (Fig. 3c). As an additional control, we monitored the effects of KIF3A_siRNA treatment of COS-1 cells. No change in levels of Gag were observed compared to those after LacZ_siRNA treatment (Fig. 3c). We also monitored the effects of KIF4_siRNA treatment in Jurkat T cells. In this case, Gag was expressed via a lentiviral infection delivery system (Lenti-Gag*). Again, KIF4_siRNA caused a strong reduction of Gag levels in Jurkat T cells, an effect equivalent to that observed in COS-1 cells (Fig. 3c).
The rat KIF4 sequence is 87% identical to that of human KIF4 and is predicted to be resistant to the effects of KIF4_siRNA#3 directed against human KIF4. Expression of full-length rat KIF4 rescued the Gag degradation phenotype and the defect in VLP production in KIF4_siRNA#3-treated cells (Fig. 3d and e). Moreover, intracellular Gag levels were increased in the presence of full-length rat KIF4 (Fig. 3d). Taken together, these data support the hypothesis that KIF4 regulates Gag protein stability. Knockdown of KIF4 induces Gag degradation, while increased KIF4 expression results in higher levels of intracellular Gag protein.
Gag intracellular trafficking is delayed when KIF4 function is disrupted.
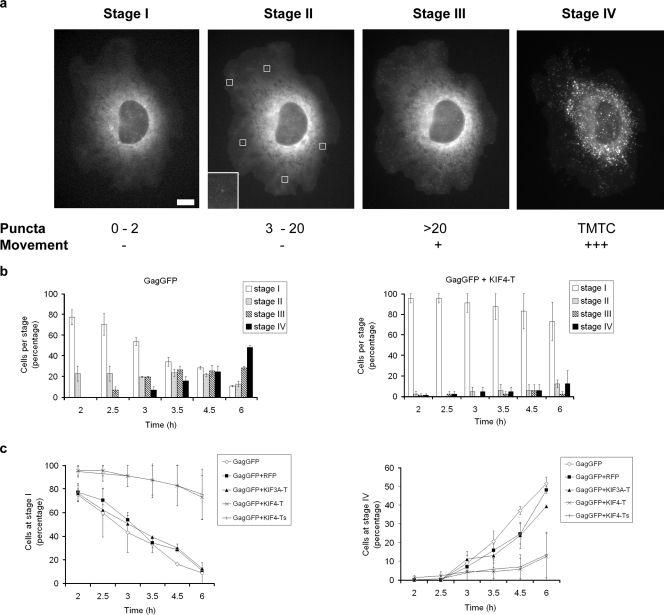
The GagGFP fusion protein has been exploited extensively as a stable, accurate reporter of Gag localization in transfected cells (15, 38). We microinjected cDNA encoding GagGFP into COS-1 cells and used time-lapse fluorescence microscopy to follow the intracellular trafficking of newly synthesized GagGFP. GagGFP fluorescence progressed through at least four phenotypically distinct stages, termed stages I to IV, over a period of 2 to 6 h postmicroinjection (Fig. 4a). The stages were characterized based on the number of GagGFP puncta and their mobility. GagGFP was initially dispersed diffusely throughout the cytoplasm (stage I). GagGFP puncta soon became apparent in the cytoplasm (stage II). There were typically 3 to 20 puncta per cell, and the puncta were relatively stationary. With time, the number of GagGFP puncta increased (>20 per cell), and they exhibited movement and began to disseminate throughout the cytoplasm (stage III). By 6 h, there were numerous, highly mobile GagGFP puncta (stage IV). The stage IV pattern was typical of the Gag and GagGFP distribution at steady state (31, 38). Quantification revealed that >50% of the cells had reached stage IV and only 10% remained at stage I by 6 h postinjection (Fig. 4b and c) (n = 56 cells).
FIG. 4.
Expression of dominant-negative KIF4 delays Gag trafficking. (a) COS-1 cells were microinjected with GagGFP cDNA and monitored for 6 h. Four distinct phenotypic stages were observed and were classified as stages I to IV, based on the number of GagGFP puncta and their movement within the cell. TMTC, too many to count. Puncta in stage II are indicated by white boxes; the inset represents an enlargement of one of the puncta. Bar = 10 μm. (b) At each time point, the percentage of cells with GagGFP at each of the four stages was quantified in the absence (left; n = 56) or presence (right; n = 59) of coinjected KIF4-T. (c) GagGFP was coinjected with one of three controls (empty vector, RFP, or KIF3A-T) or one of two dominant-negative KIF4 constructs (KIF4-T or KIF4-Ts). The percentage of cells with GagGFP at stage I (left) or stage IV (right) at the indicated time points was quantified.
The appearance of GagGFP fluorescence depends on folding and maturation of the GFP fluorophore. To determine whether additional Gag intermediates had formed but were not detected by GFP fluorescence, we analyzed GagGFP-microinjected cells that were fixed and stained with anti-GFP antibody. The staining patterns were identical to those observed by live imaging. Confocal imaging revealed that Gag and Gag puncta at early stages were localized within the interior of the cell and that by 6 h, many of the puncta were associated with the plasma membrane (see movies with three-dimensional image reconstruction in the supplemental material). Moreover, the stages observed in cells microinjected with GagGFP cDNA strongly resemble those reported recently for cells transfected with GagGFP (38). These results indicate that a series of intracellular Gag intermediates are visible before Gag appears at the plasma membrane.
We next examined the effect of interfering with KIF4 function on the kinetics of Gag trafficking. When GagGFP and mRFP-KIF4-T cDNAs were coinjected, Gag trafficking was significantly delayed. Nearly 80% of the cells remained at stage I and only 15% had progressed to stage IV by the 6-h time point (Fig. 4b and c). We obtained similar results when cells were coinjected with GagGFP and a shorter KIF4-T cDNA (KIF4-TS, encoding aa 875 to 1232) (Fig. 4c). In contrast, coexpression of mRFP or RFP-KIF3A-T cDNA did not affect GagGFP trafficking (Fig. 4c). These data suggest that KIF4 is involved in intracellular transport of HIV-1 Gag.
Disruption of endogenous KIF4 results in the accumulation of intracellular Gag in perinuclear clusters.
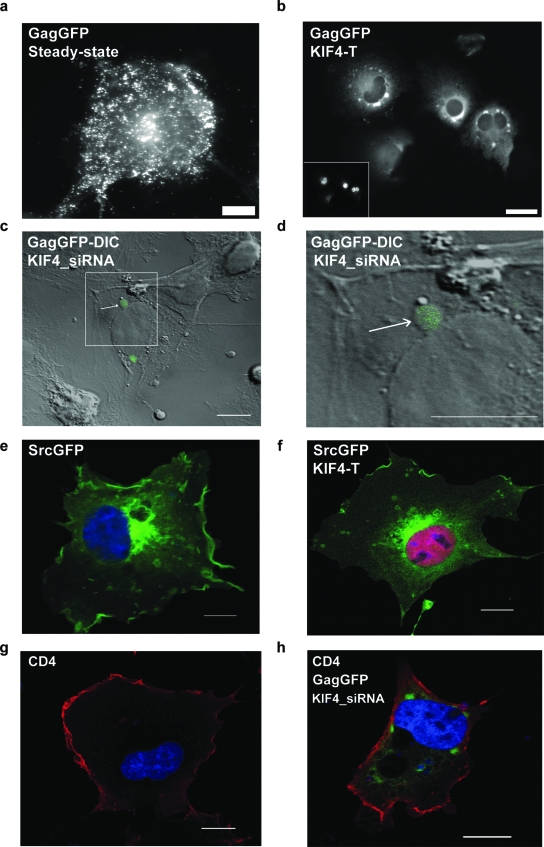
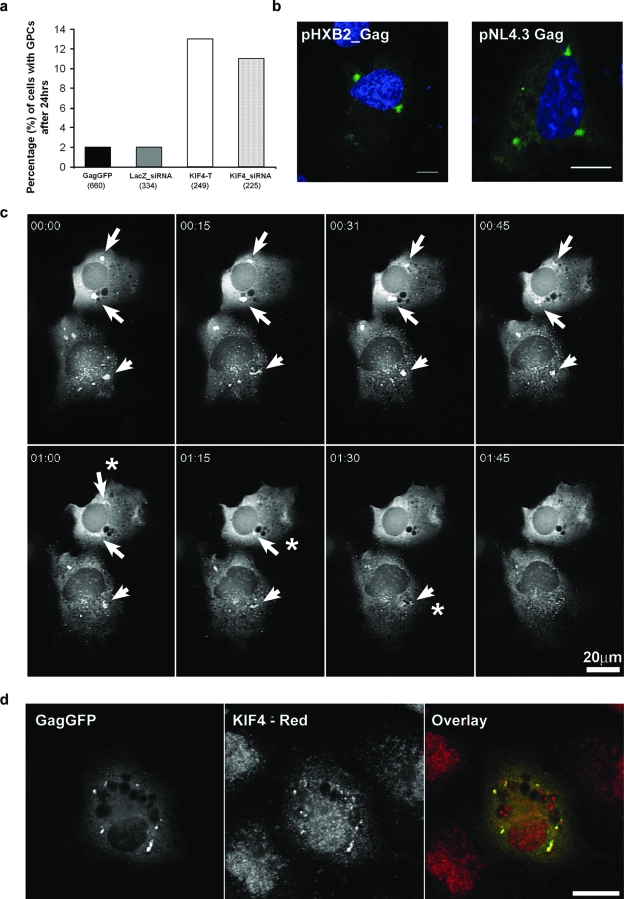
To further investigate the mechanism by which KIF4 disruption altered Gag levels, we performed a more detailed time course of GagGFP trafficking in COS-1 cells coinjected with GagGFP and mRFP-KIF4-T cDNAs. Time-lapse recordings revealed an accumulation of GagGFP into large clusters in the perinuclear region, which we termed Gag perinuclear clusters (GPCs) (Fig. 5b), compared to Gag at steady state (Fig. 5a). GPCs were also visible in GagGFP-transfected cells treated with KIF4_ siRNA (Fig. 5c and d) or cotransfected with KIF4-T (Fig. 6a). Quantitative analysis revealed a five- to sixfold increase in the number of cells with GPCs when Gag-GFP-expressing cells were treated with KIF4_siRNA or cotransfected with KIF4-T (Fig. 6a). Typically, GPCs appeared to be large structures of 0.5 to 1 μm in diameter and showed a clear outline by differential interference contrast imaging (Fig. 5d).
FIG. 5.
KIF4 disruption induces the formation of GPCs. (a and b) COS-1 cells were microinjected with GagGFP and either control vector (a) or vector encoding KIF4-T (b). The inset (b) shows the expression of RFP-tagged KIF4-T. (c and d) The differential interference contrast image of one cell transfected with GagGFP cDNA and KIF4_siRNA was magnified three times to visualize the size and outline of a single GPC (arrows). (e and f) COS-1 cells were cotransfected with cDNAs encoding c-SrcGFP and either KIF3A-T (e) or KIF4-T (f) constructs. (g and h) COS-1 cells were cotransfected with CD4 cDNA and either LacZ_siRNA (g) or KIF4_siRNA and GagGFP cDNA (h). Bar = 10 μm (a and c to h) or 15 μm (b).
FIG. 6.
GPCs form during the natural progression of intracellular Gag trafficking. (a) The percentage of GagGFP-positive cells (total number of cells counted is shown in parenthesis) that contained GPCs after 24 h was determined in COS-1 cells cotransfected with GagGFP and either KIF3A-T, LZ_siRNA, KIF4-T, or KIF4_siRNA. (b) COS-1 cells were transfected with either pHXB2ΔBalD25S or pNL4.3 (full-length infectious clone) and stained with anti-p24 antibody and Alexa 488 to detect GPCs (green). (c) Sequential images showing that GagGFP perinuclear clusters form (arrows) and disperse (*) over a period of 2 h in untreated cells. (d) GagGFP-expressing cells were stained with anti-KIF4 antibody to detect endogenous KIF4 (red). Bar = 20 μm.
To determine whether formation of GPCs was specific to Gag, the effects of KIF4 disruption on localization of two other proteins were evaluated. No obvious differences in the localization of c-Src were observed in the presence of KIF4-T compared to that in the KIF3A-T control (Fig. 5e and f). Moreover, localization of CD4, a protein that traffics to the plasma membrane via the secretory pathway, was unaffected by knockdown of KIF4 with KIF4_siRNA (Fig. 5g and h). Thus, the host cell trafficking pathways for c-Src and CD4 appear to remain intact when KIF4 function is disrupted.
We next examined whether GPCs formed during the natural progression of Gag intracellular trafficking. Cells were microinjected with GagGFP cDNA, and 4.5 h later, they were transferred to recording medium containing cycloheximide. Over the next 2 h, transient formation and dispersion of multiple GPCs were clearly evident (Fig. 6c). GPCs were also identified in cells transfected with pHXB2ΔBalD25S Gag and in cells transfected with a full-length infectious clone of pNL4.3 (Fig. 6b). Staining with anti-KIF4 antibody revealed significant colocalization between GPCs and endogenous KIF4 (Fig. 6d). These data imply that newly synthesized Gag transiently forms clusters that also contain KIF4.
Gag colocalizes with the E2-sumoylating enzyme Ubc9 in perinuclear clusters.
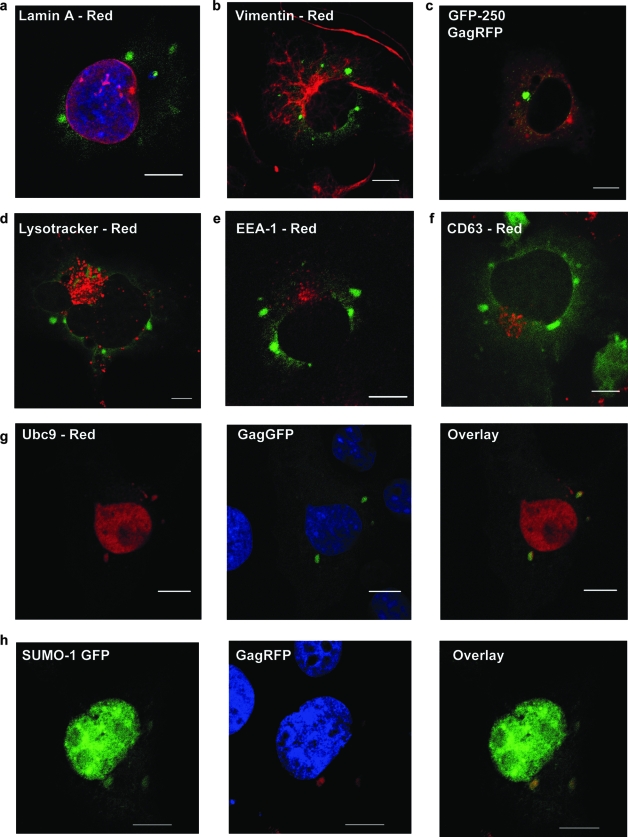
The next set of experiments was designed to identify the cellular structure or compartment(s) that contained the GPCs. Staining for lamin A revealed that GPCs, while often adjacent to the nucleus, were not enclosed by the nuclear membrane (Fig. 7a). Another possibility is that GPCs are aggresomes. These structures are sometimes formed when misfolded proteins accumulate in the cytoplasm and aggregate near the MT organizing center; proteins in the aggresome are then degraded by the proteasome. Since aggresomes are known to induce the reorganization of vimentin filaments around the MT organizing center (18), we monitored vimentin staining in the presence of KIF4_siRNA. No apparent restructuring of vimentin filaments around the GPCs was evident (Fig. 7b). Moreover, no colocalization between GagGFP GPCs and GFP-250, an aggresome marker (9), was noted in the presence of KIF4 siRNA (Fig. 7c).
FIG. 7.
GPCs colocalize with the E2-SUMO-conjugating enzyme Ubc9 but not with endocytic markers. (a, b, d, e, and f) COS-1 cells were cotransfected with GagGFP and KIF4_siRNA and stained for lamin A (red) (a), vimentin (red) (b), Lysotracker (red) (d), EEA-1 (red) (e), or CD63 (red) (f). (c) Imaging of COS-1 cells that were triply transfected with GagRFP, GFP-250, and Kif4_siRNA. (g) COS-1 cells were cotransfected with GagGFP, myc-hUbc9 (red), and KIF4-siRNA. Cells were stained with anti-myc and Alexa 594 antibodies. (h) COS-1 cells were cotransfected with GagRFP, SUMO-1 GFP, and KIF4-siRNA and imaged directly. Bar = 10 μm.
Gag has been shown to utilize compartments of the endocytic pathway for trafficking and assembly (11, 32, 38, 47). A subset of Gag proteins colocalize with CD63, a marker for the multivesicular body (MVB) (36, 38). Accordingly, we stained COS-1 cells cotransfected with GagGFP and KIF4_siRNA with Lysotracker, a marker of cellular acidic, endocytic compartments, or with antibodies to the early endosome marker EEA-1 and the late endosome/MVB marker CD63. No significant colocalization between any of these endocytic markers and GPCs was detected (Fig. 7d to f).
A previous study reported colocalization of MPMV Gag with the E2-sumoylating enzyme Ubc9 in structures described as Gag perinuclear foci (53). Ubc9 localizes to the nucleus and to the cytoplasm and functions as an E2-sumoylating enzyme for multiple sumoylated proteins (57). Given the similarity between these perinuclear foci and HIV-1 Gag-containing GPCs, we examined the localization of Ubc9 and HIV-1 Gag GPCs in cells cotransfected with GagGFP, myc-tagged Ubc9, and KIF4_siRNA. As depicted in Fig. 7g, GPCs colocalized with perinuclear cytoplasmic clusters containing Ubc9. In addition, in the presence of Ubc9, there was significant colocalization of GPCs with SUMO-GFP or myc-SUMO (Fig. 7h). In contrast, no colocalization between c-Src and myc-Ubc9 was detected in KIF4 siRNA-treated cells (see Fig. S2 in the supplemental material). Taken together, these data establish the GPC as a Ubc9- and SUMO-positive compartment that participates in the intracellular trafficking pathway for HIV-1 Gag.
DISCUSSION
KIF4 regulates Gag stability and trafficking.
In this study, we identify a novel role for the cellular motor protein KIF4 as a regulator of HIV-1 Gag protein stability and intracellular trafficking. Our results imply that Gag accumulates in GPCs as part of its intracellular trafficking pathway. When KIF4 is functional, GPC formation is transient. When KIF4 function is disrupted, a subpopulation of Gag accumulates in GPCs, and the remainder is degraded. Our data imply that GPCs, KIF4, and potentially other cellular components in GPCs play a role in controlling the stability of intracellular Gag protein.
Regulation of Gag protein stability.
We and others have shown previously that a subpopulation of newly synthesized HIV-1 Gag is degraded shortly after synthesis (44, 49). This population first appears in detergent-resistant, cytoplasmic complexes that fractionate in the middle of a 0 to 18% Optiprep gradient (49). Thus, degradation is part of the natural course of Gag biosynthesis. The studies in this report reveal a mechanism for increasing Gag degradation. Disruption of KIF4, either with KIF4-T or KIF4_siRNA, resulted in a loss of intracellular Gag protein due to an increased rate of Gag degradation (Fig. 1 to 3). The protease responsible for Gag degradation has yet to be identified, but the mechanism is neither proteasome nor lysosome dependent (Fig. 1e). Because KIF4 disruption did not alter levels of endogenous proteins (actin, ERK2, and caveolin-1), other exogenous, transfected proteins (c-Src and Fyn), or Gag chimeras with altered N termini, we concluded that the effect of KIF4 is specific to the N-terminal MA region of HIV-1 Gag. This is the region of Gag that has been shown to interact directly with KIF4 (48).
At least three other examples of increased Gag turnover have been documented. First, overexpression of a chimeric protein containing the cytoplasmic tail of the HIV-1 Env protein gp41 fused to β-galactosidase (6) was shown to induce HIV-1 Gag downregulation. Complexes of Gag and the β-galactosidase-Env fusion protein were detected in the perinuclear region of the transfected cells. Second, in HIV-1-expressing cells, siRNA-directed knockdown of heterogeneous nuclear ribonucleoprotein A1 resulted in reduced Gag expression (25). Third, siRNA-mediated depletion of SOCS1, or suppressor of cytokine signaling 1, decreased levels of intracellular Gag and caused Gag to accumulate in perinuclear aggregates (43). However, these Gag aggregates differ from the GPCs reported here, as they colocalized with lysosomal markers and were subject to lysosome-mediated degradation. Regardless of the mechanism of Gag degradation, strategies that reduce Gag protein levels are of great interest because they have the potential to serve as a novel form of antiviral therapy.
GPCs as an intracellular trafficking intermediate.
Formation of perinuclear clusters of intracellular Gag was previously documented by our laboratory, using live- and fixed-cell imaging of Gag- and GagGFP-transfected cells (38). Perinuclear clusters of Gag were generated early during Gag trafficking, within 1.5 h after cells were released from a cycloheximide block. These clusters then dispersed into multiple puncta, which spread throughout the cell and ultimately concentrated at the plasma membrane. The results reported here, obtained using microinjected GagGFP cDNA, confirm this finding and establish the GPC as the earliest detectable Gag intermediate. We do not know what percentage of the Gag in the GPC ultimately traffics to the plasma membrane. It is possible that a fraction of Gag traffics straight from the cytosol to the plasma membrane without passing through the GPC. It is also not known if Gag molecules multimerize within the GPC or, alternatively, are transported as monomers and multimerize at the plasma membrane.
What is the identity of the GPC-containing compartment? GPCs are unlikely to represent aggregated, misfolded Gag in an aggresome, since no colocalization between GPCs and GFP-250, an aggresome marker, was detected (Fig. 7b). Moreover, no colocalization of GPCs with markers of the endocytic pathway was noted (Fig. 7). GPCs were present, although transiently, even in cells where KIF4 function was unperturbed, arguing that their formation is part of the natural course of Gag intracellular trafficking. The presence of a discrete number of GPCs per cell (typically three or four) suggests that GPCs represent a distinct subcellular compartment that forms and is visible during Gag biosynthesis.
To assemble an infectious particle, HIV-1 Gag needs to recruit other viral proteins as well as viral RNA. In COS-1 cells expressing full-length, wild-type HIV-1 Gag, Gag colocalizes with viral genomic RNA in the perinuclear region (39). Thus, it is possible that GPCs comprise the region of Gag-RNA interaction. Recruitment of other viral components could occur as Gag continues along its trafficking pathway. Several studies have documented the presence of Gag in endocytic compartments, particularly the MVB (11, 32, 36, 38, 47). At least three different models for Gag trafficking have been proposed. One school of thought argues for the MVB as a Gag trafficking intermediate. Several lines of evidence support this model. HIV-1 Gag as well as infectious HIV-1 particles localize within the MVB (11, 32, 36, 38, 47). MVBs contain the ESCRT components that are required for virion egress (1). Moreover, Gag binds to the δ subunit of AP-3. When this interaction is disrupted, Gag no longer associates with MVBs, and viral particle formation is inhibited (7). A second model postulates that Gag accumulates and buds particles from tetraspanin-enriched microdomains. These regions of the plasma membrane are enriched in ESCRT components and tetraspanin proteins and are thought to be derived from MVBs as a result of fusion with the plasma membrane (19, 33). A variation on this theme is a third model proposing that newly synthesized Gag transits directly to the plasma membrane, where it assembles and buds particles (8, 14, 20). The presence of Gag in MVBs is attributed to endocytosis of virus particles, regulated by the viral accessory protein Vpu (14, 31). Differences in the data supporting these three models and their interpretations have not yet been reconciled. Regardless of the model(s) that ultimately proves to be most accurate, the GPC could serve as an early intermediate step of Gag trafficking in each scenario.
Ubc9 interacts with several retroviral Gags, including those of HIV-1, MPMV, and Moloney murine leukemia virus, and each of these Gag proteins is modified by sumoylation (12, 53, 56). Both MPMV Gag (53) and HIV-1 Gag (this study) colocalize with Ubc9 in perinuclear clusters. It is interesting to speculate that GPCs might provide a staging area for Ubc9 to interact with and sumoylate Gag. Sumoylation of Gag is not required for assembly, since mutation of the SUMO-1 attachment site on HIV-1 or murine leukemia virus Gag does not affect particle production (12, 56). However, the particles released from cells expressing these mutant Gag proteins exhibit reduced infectivity. It is possible that during intracellular trafficking Gag acquires posttranslational modifications that facilitate entry or postentry steps.
KIF4: motor or scaffold?
How does KIF4 function to regulate Gag stability and trafficking? An obvious explanation is that Gag utilizes the motor function of KIF4 to propel assembly intermediates along MTs. Since KIF4 is a plus-end-directed motor, movement would proceed centrifugally from the cell center toward the plasma membrane, the direction of viral egress. It is well established that MTs are also needed to transport MVBs and secretory lysosomes out to the plasma membrane (3).
Thus, Gag that assembled in these exocytic organelles would also be dependent on MTs for release from the cell. A role for MTs in assembly is supported by the finding that treatment of T cells with inhibitors of MT polymerization prevents formation of Gag-containing assembly platforms on the plasma membrane and reduces cell-to-cell transmission of HIV-1 across the virological synapse (19). On the other hand, no apparent interaction of Gag with tubulin was noted by fluorescence resonance energy transfer (39), and treatment of cells with the MT-depolymerizing agent nocodazole did not affect particle release (19, 20). However, it is important that cells contain a stable pool of MTs that are nocodazole resistant (13). In addition, transport events that normally occur along MTs can be bypassed in the absence of MTs (42). The mechanism underlying this bypass is thought to involve centrifugal reorganization of secretory and endocytic membrane compartments. The lack of an effect of nocodazole is therefore not sufficient to rule out a role for MTs in HIV-1 Gag trafficking. Moreover, it was recently shown that stable MTs are required for HIV-1 infection (30). Thus, Gag might preferentially utilize the stable pool of MTs for transport, and this selection may be enhanced by association with KIF4.
An alternate possibility is that KIF4 acts as a scaffold to coordinate Gag assembly and movement. Kinesins are often linked to their cargo via scaffolding or adaptor proteins (2). Interaction with such scaffolding proteins may promote assembly and/or packaging of protein complexes. Moreover, KIF4 interacts with multiple cellular components, including chromosomes, L1-containing vesicles, retroviral Gag proteins, and the cellular proteins PRC1 and PARP1 (22, 23, 27, 37, 48, 59). Thus, KIF4 could serve as a scaffold to recruit newly synthesized Gag and Gag-associated molecules.
It is important that disruption of KIF4 inhibits cytokinesis (59). Thus, the effects we observed on Gag stability and trafficking could somehow be related to a cytokinesis defect. At present, there are no reports directly linking HIV-1 assembly and cytokinesis. However, two proteins that participate in HIV-1 budding, Tsg101 and the ESCRT-associated protein ALIX, are recruited to the midbody during cell division and are required for the abscission step of cytokinesis (5, 29). One could postulate that the cytokinesis defect in KIF4-disrupted cells results in retention of ESCRT components at the midbody and a functional depletion of these components at sites of HIV-1 budding. We consider this possibility unlikely. Loss of ESCRT proteins causes tethered HIV-1 particles to accumulate at the plasma membrane. In contrast, in KIF4-depleted cells, Gag accumulates in the GPC in the perinuclear region. We cannot exclude other indirect effects of KIF4 depletion on Gag function.
Model for Gag trafficking and KIF4 function.
Based on previously published reports and the data presented here, we propose the following model. HIV-1 Gag is synthesized on soluble polysomes. Upon release into the cytosol, the newly synthesized protein accumulates in GPCs. Formation of GPCs is transient. Dispersion of GPCs is mediated, either directly or indirectly, by KIF4. When KIF4 function is blocked or when KIF4 levels are depleted, Gag begins to accumulate in GPCs. We propose that GPCs are discrete structures that have a limited capacity for Gag. When this capacity is reached, newly synthesized Gag proteins can no longer associate in GPCs and are instead targeted for degradation. This model explains the ability of KIF4-T and KIF4_siRNA to block Gag trafficking at early time points after synthesis and also for these treatments to promote downregulation of Gag. Induction of Gag degradation is a unique antiviral strategy that could be exploited as a therapeutic target for intervention during HIV infection.
Supplementary Material
Acknowledgments
We thank Agnes Viale for assistance with qPCR, John Moore for use of biosafety level 2 laboratory facilities, Alan Hall and Tatiana Omelchenko for assistance with microinjection, and Raisa Louft-Nisenbaum for technical assistance. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 monoclonal antibody (183-H12-5C), from Bruce Chesebro and Kathy Wehrly.
This research was supported by NIH grant CA72309 to M.D.R., NIH MSTP grant GM07739, NCI grant P30-CA08748 (which provides partial support for the Molecular Cytology Core), and American Cancer Society grant RSG-06-142-01-CSM to G.K.
Footnotes
Published ahead of print on 6 August 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adamson, C. S., and E. O. Freed. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 55347-387. [DOI] [PubMed] [Google Scholar]
- 2.Almenar-Queralt, A., and L. S. Goldstein. 2001. Linkers, packages and pathways: new concepts in axonal transport. Curr. Opin. Neurobiol. 11550-557. [DOI] [PubMed] [Google Scholar]
- 3.Blott, E. J., and G. M. Griffiths. 2002. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 3122-131. [DOI] [PubMed] [Google Scholar]
- 4.Camus, G., C. Segura-Morales, D. Molle, S. Lopez-Verges, C. Begon-Pescia, C. Cazevieille, P. Schu, E. Bertrand, C. Berlioz-Torrent, and E. Basyuk. 2007. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol. Biol. Cell 183193-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 3161908-1912. [DOI] [PubMed] [Google Scholar]
- 6.Chan, W. E., and S. S. Chen. 2006. Downregulation of human immunodeficiency virus type 1 Gag expression by a gp41 cytoplasmic domain fusion protein. Virology 348418-429. [DOI] [PubMed] [Google Scholar]
- 7.Dong, X., H. Li, A. Derdowski, L. Ding, A. Burnett, X. Chen, T. R. Peters, T. S. Dermody, E. Woodruff, J. J. Wang, and P. Spearman. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120663-674. [DOI] [PubMed] [Google Scholar]
- 8.Finzi, A., A. Orthwein, J. Mercier, and E. A. Cohen. 2007. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 817476-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Mata, R., Z. Bebok, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1461239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein, L. S., and Z. Yang. 2000. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu. Rev. Neurosci. 2339-71. [DOI] [PubMed] [Google Scholar]
- 11.Grigorov, B., F. Arcanger, P. Roingeard, J. L. Darlix, and D. Muriaux. 2006. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 359848-862. [DOI] [PubMed] [Google Scholar]
- 12.Gurer, C., L. Berthoux, and J. Luban. 2005. Covalent modification of human immunodeficiency virus type 1 p6 by SUMO-1. J. Virol. 79910-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurland, G., and G. G. Gundersen. 1993. Protein phosphatase inhibitors induce the selective breakdown of stable microtubules in fibroblasts and epithelial cells. Proc. Natl. Acad. Sci. USA 908827-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harila, K., I. Prior, M. Sjoberg, A. Salminen, J. Hinkula, and M. Suomalainen. 2006. Vpu and Tsg101 regulate intracellular targeting of the human immunodeficiency virus type 1 core protein precursor Pr55gag. J. Virol. 803765-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 748670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirokawa, N. 1998. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279519-526. [DOI] [PubMed] [Google Scholar]
- 17.Jaulin, F., X. Xue, E. Rodriguez-Boulan, and G. Kreitzer. 2007. Polarization-dependent selective transport to the apical membrane by KIF5B in MDCK cells. Dev. Cell 13511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1431883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolly, C., and Q. J. Sattentau. 2007. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J. Virol. 817873-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jouvenet, N., S. J. Neil, C. Bess, M. C. Johnson, C. A. Virgen, S. M. Simon, and P. D. Bieniasz. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, W., Y. Tang, Y. Okada, T. A. Torrey, S. K. Chattopadhyay, M. Pfleiderer, F. G. Falkner, F. Dorner, W. Choi, N. Hirokawa, and H. C. Morse III. 1998. Binding of murine leukemia virus Gag polyproteins to KIF4, a microtubule-based motor protein. J. Virol. 726898-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurasawa, Y., W. C. Earnshaw, Y. Mochizuki, N. Dohmae, and K. Todokoro. 2004. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 233237-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, Y. M., and W. Kim. 2004. Kinesin superfamily protein member 4 (KIF4) is localized to midzone and midbody in dividing cells. Exp. Mol. Med. 3693-97. [DOI] [PubMed] [Google Scholar]
- 24.Lee, Y. M., S. Lee, E. Lee, H. Shin, H. Hahn, W. Choi, and W. Kim. 2001. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem. J. 360549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levesque, K., M. Halvorsen, L. Abrahamyan, L. Chatel-Chaix, V. Poupon, H. Gordon, L. DesGroseillers, A. Gatignol, and A. J. Mouland. 2006. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic 71177-1193. [DOI] [PubMed] [Google Scholar]
- 26.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 757913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Midorikawa, R., Y. Takei, and N. Hirokawa. 2006. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell 125371-383. [DOI] [PubMed] [Google Scholar]
- 28.Miyaura, M., A. Yoshida, A. Sakurai, M. Fujita, A. H. Koyama, and A. Adachi. 2000. Mutational analysis of HIV-1 gag proteins. Int. J. Mol. Med. 6265-269. [DOI] [PubMed] [Google Scholar]
- 29.Morita, E., V. Sandrin, H. Y. Chung, S. G. Morham, S. P. Gygi, C. K. Rodesch, and W. I. Sundquist. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 264215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naghavi, M. H., S. Valente, T. Hatziioannou, K. de Los Santos, Y. Wen, C. Mott, G. G. Gundersen, and S. P. Goff. 2007. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. EMBO J. 2641-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neil, S. J., S. W. Eastman, N. Jouvenet, and P. D. Bieniasz. 2006. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic 4902-910. [DOI] [PubMed] [Google Scholar]
- 33.Nydegger, S., S. Khurana, D. N. Krementsov, M. Foti, and M. Thali. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh, S., H. Hahn, T. A. Torrey, H. Shin, W. Choi, Y. M. Lee, H. C. Morse, and W. Kim. 2000. Identification of the human homologue of mouse KIF4, a kinesin superfamily motor protein. Biochim. Biophys. Acta 1493219-224. [DOI] [PubMed] [Google Scholar]
- 35.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 734136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 781552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peretti, D., L. Peris, S. Rosso, S. Quiroga, and A. Caceres. 2000. Evidence for the involvement of KIF4 in the anterograde transport of L1-containing vesicles. J. Cell Biol. 149141-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perlman, M., and M. D. Resh. 2006. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic 7731-745. [DOI] [PubMed] [Google Scholar]
- 39.Poole, E., P. Strappe, H. P. Mok, R. Hicks, and A. M. Lever. 2005. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic 6741-755. [DOI] [PubMed] [Google Scholar]
- 40.Resh, M. D. 2005. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 784-91. [PubMed] [Google Scholar]
- 41.Resh, M. D. 2006. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2584-590. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Boulan, E., G. Kreitzer, and A. Musch. 2005. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 6233-247. [DOI] [PubMed] [Google Scholar]
- 43.Ryo, A., N. Tsurutani, K. Ohba, R. Kimura, J. Komano, M. Nishi, H. Soeda, S. Hattori, K. Perrem, M. Yamamoto, J. Chiba, J. Mimaya, K. Yoshimura, S. Matsushita, M. Honda, A. Yoshimura, T. Sawasaki, I. Aoki, Y. Morikawa, and N. Yamamoto. 2008. SOCS1 is an inducible host factor during HIV-1 infection and regulates the intracellular trafficking and stability of HIV-1 Gag. Proc. Natl. Acad. Sci. USA 105294-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert, U., L. C. Anton, J. Gibbs, C. C. Norbury, J. W. Yewdell, and J. R. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404770-774. [DOI] [PubMed] [Google Scholar]
- 45.Sekine, Y., Y. Okada, Y. Noda, S. Kondo, H. Aizawa, R. Takemura, and N. Hirokawa. 1994. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J. Cell Biol. 127187-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Setou, M., D. H. Seog, Y. Tanaka, Y. Kanai, Y. Takei, M. Kawagishi, and N. Hirokawa. 2002. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 41783-87. [DOI] [PubMed] [Google Scholar]
- 47.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4785-801. [DOI] [PubMed] [Google Scholar]
- 48.Tang, Y., U. Winkler, E. O. Freed, T. A. Torrey, W. Kim, H. Li, S. P. Goff, and H. C. Morse III. 1999. Cellular motor protein KIF-4 associates with retroviral Gag. J. Virol. 7310508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 745845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van't Hof, W., and M. D. Resh. 1997. Rapid plasma membrane anchoring of newly synthesized p59fyn: selective requirement for NH2-terminal myristoylation and palmitoylation at cysteine-3. J. Cell Biol. 1361023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhey, K. J., D. Meyer, R. Deehan, J. Blenis, B. J. Schnapp, T. A. Rapoport, and B. Margolis. 2001. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 987724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weldon, R. A., Jr., P. Sarkar, S. M. Brown, and S. K. Weldon. 2003. Mason-Pfizer monkey virus Gag proteins interact with the human sumo conjugating enzyme, hUbc9. Virology 31462-73. [DOI] [PubMed] [Google Scholar]
- 54.Wozniak, M. J., R. Milner, and V. Allan. 2004. N-terminal kinesins: many and various. Traffic 5400-410. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, X., X. Yu, T. H. Lee, and M. Essex. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 676387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yueh, A., J. Leung, S. Bhattacharyya, L. A. Perrone, K. de los Santos, S. Y. Pu, and S. P. Goff. 2006. Interaction of Moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J. Virol. 80342-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, H., H. Saitoh, and M. J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 226498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 682556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, C., and W. Jiang. 2005. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA 102343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.