Abstract
Sheep scrapie is the prototypical transmissible spongiform encephalopathy (prion disease), which has a fundamental pathogenesis involving conversion of normal cellular prion protein (PrPC [C superscript stands for cellular]) to disease-associated prion protein (PrPSc [Sc superscript stands for sheep scrapie]). Sheep microglial cell cultures, derived from a prnp 136VV/171QQ near-term fetal brain, were developed to study sheep scrapie in the natural host and to investigate potential cofactors in the prion conversion process. Two culture systems, a primary cell culture and a cell line transformed with the large T antigen of simian virus 40, were developed, and both were identified as microglial in origin as indicated by expression of several microglial phenotype markers. Following exposure to PrPSc, sheep microglial cells demonstrated relatively low levels (transformed cell line) to high levels (primary cell line) of PrPSc accumulation over time. The accumulated PrPSc demonstrated protease resistance, an inferred beta-sheet conformation (as determined by a commercial enzyme-linked immunosorbent assay), specific inhibition by anti-PrP antibodies, and was transmissible in a dose-dependent manner. Primary microglia coinfected with a small-ruminant lentivirus (caprine arthritis encephalitis virus-Cork strain) and PrPSc demonstrated an approximately twofold increase in PrPSc accumulation compared to that of primary microglia infected with PrPSc alone. The results demonstrate the in vitro utility of PrPSc-permissive sheep microglial cells in investigating the biology of natural prion diseases and show that small-ruminant lentiviruses enhance prion conversion in cultured sheep microglia.
Prion diseases (transmissible spongiform encephalopathies [TSEs]) are a group of invariably fatal, transmissible, neurodegenerative diseases, which include scrapie in sheep and goats, bovine spongiform encephalopathy in cattle, chronic wasting disease in deer and elk, and Creutzfeld-Jakob disease and kuru in humans (38). The similarities between scrapie and Creutzfeld-Jakob disease have long been recognized (36), and the use of scrapie as an experimental model allows for the investigation of a natural prion disease in a natural host. The central feature of prion pathogenesis is the conversion of the normal cellular form of the host-encoded prion protein (PrPC [C superscript stands for cellular]) to an abnormal isoform, designated PrPSc (Sc superscript stands for sheep scrapie) (6, 10, 13). The conversion occurs posttranslationally and involves a conformational change resulting in the generation of a detergent-insoluble, partially protease-resistant molecule that aggregates in affected tissues and serves as the marker for prion diseases. The principal component of the transmissible agent is thought to be the abnormal prion protein and provides the basis for the protein-only hypothesis of prion diseases (50).
There are at least 21 cell lines that have been used to study prion diseases in vitro (59). However, only 4 of these are susceptible to PrPSc derived from a natural TSE host, while the remaining 17 cell lines are susceptible only to rodent-adapted strains of PrPSc. Further, only one of the cell lines is derived from a natural TSE host, mule deer (Odocoileus hemionus) (51). While rodent-derived cells have many benefits, including the availability of reagents and highly inbred genetics, much of the work accomplished in these cells has to be verified in a natural host-TSE system. The lack of a sheep-derived, scrapie-permissive cell line also prevents full investigation into species-specific phenomena, such as allelic usage variation, allele-predicted susceptibility, and species-specific cofactors. Therefore, the development of a sheep cell culture system would provide an excellent model for such studies.
In addition to creating cell lines that accumulate PrPSc, it is also desirable to use cells that contribute to the pathophysiology of the clinical disease. Microglia (resident brain macrophages) are such cells, which not only accumulate infectivity in vivo (5) but are also thought to play a role in the neuropathology by their activation and release of immune mediators, such as interleukin-6 (8, 52, 63). Additionally, peripheral macrophages have demonstrated both accumulation (9, 23, 28, 41, 49) and proteolysis of PrPSc (9, 28). Only one of the current cell culture systems demonstrates microglial or macrophage characteristics, and this cell line is derived from mice and overexpresses the murine prion gene (33). While overexpression of PrPC often increases the permissiveness of cells to PrPSc accumulation, it can also introduce spontaneous cell pathology (62) and is a confounding factor when trying to study the effects of PrPSc accumulation at the cellular level.
Another area that would benefit from a natural host-TSE cell culture system is the investigation into possible cofactors for the prion conversion process (55). Identification of these accessory molecules is still unresolved; however, several studies suggest that nucleic acids are a possible family of cofactors (2, 14, 15, 18, 19, 21, 64). Interestingly, recombinant prion protein has demonstrated the abilities to bind and to chaperone retroviral RNA, which are similar to retroviral nucleocapsid's function (18, 19, 42). Other interactions between prion protein and retroviruses have been identified; these interactions include an increase in murine leukemia virus (MLV) titers and replication and a shortened scrapie incubation period in the brains of coinfected mice (12, 40). In vitro coinfection studies have also demonstrated increased scrapie infectivity release into the cell culture supernatant in murine cell cultures coinfected with MLV (39).
Sheep, the natural host of scrapie, are also clinically affected by retroviruses, most notably the small-ruminant lentiviruses (SRLVs), visna-maedi virus (VISNA), and caprine arthritis-encephalitis virus (CAEV) (47). These viruses have a worldwide distribution and are the target of eradication programs (47). However, there has been little work published regarding any possible correlation between scrapie and infection with SRLV, with only one report to the authors' knowledge demonstrating a correlation between VISNA-induced lymphofollicular mastitis and resulting PrPSc accumulation within macrophages and follicular dendritic cells (41). Other studies have demonstrated that chronic inflammation of various organs results in PrPSc accumulation within those organs that normally lack PrPSc in prion-affected animals (26, 37), suggesting that the effect of VISNA on PrPSc in mastitis was indirect. To directly determine whether coinfection with an SRLV increases accumulation of PrPSc, a sheep microglial cell culture system was developed and utilized in coinfection studies with PrPSc and SRLV.
MATERIALS AND METHODS
Primary ovine brain cell cultures.
Primary mixed glial cell cultures were obtained from an ovine fetal brain using a mechanical dissociation technique for small ruminants previously described in our laboratory (7). The ovine fetus was obtained from a near-term pregnant Suffolk cross ewe that was housed and cared for in accordance with the guidelines of the Institutional Animal Care and Use Committee at Washington State University, Pullman. At approximately day 102 of gestation, the ewe was euthanized by administering an intravenous overdose of barbiturate, and the fetal brain was removed in toto. Approximately 250 mg of brain tissue was removed from the cerebral cortex and used for genotyping of the fetal prion gene as previously described (3). Periventricular white matter tissue from the remainder of the cerebral cortices and midbrain was collected, cut into approximately 5-mm cubes, and disassociated by mechanical triturating in a 25-ml pipette. The resulting brain tissue explants were plated into 75-cm2 tissue culture flasks with Dulbecco's modified Eagle's medium (Cellgro) supplemented with 20% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, 10 IU/ml of penicillin, 10 mg/ml streptomycin, and 2.5 μg/ml amphotericin B and left undisturbed for approximately 1 week. Once the cells reached confluence, aliquots of cells were frozen in 90% heat-inactivated FBS and 10% dimethyl sulfoxide and stored in liquid nitrogen. Aliquots of cells were thawed as needed and serially passaged, using standard techniques, in OPTI-MEM I reduced serum medium (Invitrogen) supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 10 IU/ml of penicillin, and 10 mg/ml of streptomycin (OMEM).
Transformation of brain cells.
Cells were transfected according to the manufacturer's instructions using the LF2000 reagent (Invitrogen), with a plasmid containing the simian virus 40 (SV40) large T antigen, as described previously (25, 27). Primary cells were split in triplicate into 24-well plates and allowed to grow to approximately 60% confluence. Three wells of cells were incubated with plasmid DNA in LF2000 reagent (Invitrogen) and OPTI-MEM for 8 hours at 37°C in 5% CO2, while the three remaining wells were sham transfected by incubating in LF2000 reagent and OPTI-MEM without plasmid DNA. Following 8 hours, the cell culture medium was changed to OMEM. Cells were fed every 4 days and allowed to grow for 12 days. Cells were then lifted from the plate and passed without dilution into 25-cm2 plastic tissue culture flasks. Cells were fed every 3 to 5 days as needed and serially passaged 1/10 after reaching confluence. Transfected cells demonstrated an increase in mitotic activity and a loss of contact inhibition. Immunocytochemistry, with the SV40 large-T-antigen-specific monoclonal antibody (MAb) DP02A (Oncogene Research Products, Cambridge, MA), was used to confirm transfection as previously described (27).
Characterization of microglial cell cultures.
To characterize brain cell cultures, the brain cell cultures were phenotyped using various cell markers and functional activity assays. The microglial and endothelial marker biotinylated Ricinus communis agglutinin-1 (RCA-1) (Dako Cytomation) was used in immunocytochemistry and flow cytometry as previously described (7). Nonspecific esterase activity (Sigma) was performed per the manufacturer's directions (7). Two additional markers, CD14 (catalog no. MM61A; VMRD, Inc.) (immunoglobulin G1 [IgG1]) and CD68 (EBM11; Dako) (IgG1), that are predominately found on cells of the monocytic lineage were tested by immunocytochemistry and flow cytometry. In cells of the monocyte lineage, CD14 is a membrane-bound receptor for lipopolysaccharide (34). CD68 is a lysosome-associated glycoprotein used to identify macrophages (30), and while the MAb EBM11 was raised against human CD68, it has previously demonstrated immunoreactivity against bovine macrophages (1), thus suggesting its utility in this study of sheep cells. For flow cytometric detection of CD14 expression, cells were trypsinized and incubated with the primary antibody. Following three washes, cells were incubated with a secondary fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin antibody. Cells were then washed twice and fixed in 2% formaldehyde. CD68 is a predominately intracellular antigen; therefore, cells were fixed for 2 days in 10% neutral buffered formalin, permeabilized in 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 5 min, washed, and then labeled as described above. Five thousand events were analyzed on a FACSort flow cytometer (Becton Dickinson), and counts were determined with Macintosh CellQuest software (BD Biosciences). Results were graphically analyzed using FCS Express (De Novo Software). The Kolmogorov Smirnov test (CellQuest), with a cutoff P value of 0.05, was used to determine significance. Negative controls included the use of isotype-matched antibodies raised against an irrelevant antigen, omission of the primary antibody, and omission of both the primary and secondary antibodies.
For immunocytochemistry, cells were grown in chambered glass slides (Nunc) and allowed to grow to approximately 70% confluence. Cells were rinsed in PBS and fixed in 100% ethanol for 10 minutes. Following quenching of endogenous peroxidase with hydrogen peroxide for 10 min, cells were assayed for expression of the antigens, using the Signet kit (Covance) per the manufacturer's instructions, and the antibodies listed above. The immunolabeling was visualized with 3-amino-9-ethylcarbazole (Dako) with nuclear counterstaining by Mayer's hematoxylin. Negative controls included the use of isotype-matched antibodies raised against an irrelevant antigen, omission of the primary antibody, and omission of both the primary and secondary antibodies.
Inoculation of primary microglia with PrPSc.
Rov9 cells, which are rabbit renal epithelial cells that are stably transfected with the sheep VRQ (Val-136, Arg-154, and Gln-171) allele of the prion gene under the control of a tetracycline-inducible promoter and are susceptible to sheep PrPSc (60), were used as the inoculum. Rov9 cells with detectable amounts of PrPSc (Rov9Sc) and Rov9 cells that were never exposed to PrPSc (Rov9C) were obtained (B. Caughey with permission from D. Vilette) and maintained in OMEM supplemented with 1 μg/ml doxycycline) as previously described (60). PrPSc within Rov9Sc cells was verified by PrPSc-specific immunoblotting and enzyme-linked immunosorbent assay (ELISA) (see below). For use as an inoculum, mechanical lysates of the Rov9Sc and Rov9C cells were prepared as previously described (60). Briefly, the Rov9 cells were grown to confluence in two 75-cm2 plastic tissue culture flasks. Rov9Sc and Rov9C cells were rinsed three times with sterile 1× Dulbecco's phosphate-buffered saline (D-PBS) and scraped into 10 ml of PBS. The cell pellets were collected by centrifugation at 170 × g at room temperature for 10 min and resuspended in 0.5 ml of filter-sterilized 5% glucose. The cell suspensions were frozen and thawed four times and then subjected to 1 to 2 min of sonication in a cup horn sonicator. The inoculum was stored at −20°C. For inoculation, primary microglia were passed into six-well plates and allowed to grow to approximately 60% confluence. Microglia were rinsed once with PBS and then overlaid with 200 μl of a 1/20 dilution of either the Rov9Sc lysate (microgliaSc) or the Rov9C lysate (microgliaC) in OPTI-MEM. MicrogliaSc and microgliaC were incubated for 6 hours, and then 200 μl of OMEM was added to each well. Following an additional 2 days of incubation, an additional 0.5 ml of OMEM was added to each well, and microglia were incubated for 4 days at which time they were expanded into 25-cm2 tissue culture flasks. Microglia were fed every 3 or 4 days with OMEM as necessary and serially passaged 1/5 after reaching confluence.
Detection of PrPSc by ELISA.
At selected passages following trypsinization, four-fifths of the microglial cell suspension from a 25-cm2 tissue culture flask was rinsed in D-PBS and then lysed in 120 μl of lysis buffer (0.5% Triton X-100, 0.5% sodium deoxycholate, 50 mM Tris-HCl [pH 8.0], 5 mM EDTA, and 150 mM NaCl) for 3 min at room temperature, followed by centrifugation at approximately 2,300 × g at room temperature for 5 min. One hundred microliters of the supernatant was then used for PrPSc detection by the HerdChek scrapie antigen test kit ELISA (IDEXX) following the manufacturer's instructions. The proprietary ELISA positive and negative controls were used per the manufacturer's directions. Α standard curve prepared from diluted Rov9Sc inoculum (1/20, 1/100, and 1/400) was used to normalize corrected optical density results. MicrogliaSc results for cells two passages after inoculation with PrPSc were set at 1, and all other results were normalized to this value. The PrPSc signal from cells five passages after inoculation with PrPSc was compared to cells two passages after inoculation using an independent t test with a cutoff P value of 0.05 (SigmaPlot). Cells were considered positive for PrPSc accumulation if the corrected optical density was greater than 0.18 plus the negative-control value (per the manufacturer's instructions). To assay for accumulation of PrPSc over time and to compare the amount of PrPSc within microgliaSc to the amount in the original inoculum, the PrPSc signal within late-passage microgliaSc lysates was compared to early passage microgliaSc lysates and to dilutions of the inoculum, respectively, using an independent t test with a cutoff P value of 0.05 (SigmaPlot). MicrogliaC lysates served as the experimental negative controls.
Demonstration of PrPSc infectivity from microgliaSc-derived lysates.
Inocula were derived from mechanical lysates of microgliaSc and microgliaC as described above for the preparation of inocula from Rov9 cells. Rov9C cells (the target cells) were then inoculated as described above with three dilutions (1/20, 1/200, and 1/2,000) of the microgliaSc and microgliaC inocula. The Rov9 cells were then expanded once and split 1/5 three times. Lysates from the Rov9 cells were analyzed for PrPSc accumulation in a single HerdChek scrapie antigen test kit ELISA (IDEXX) plate and reported as the corrected optical density. Cells were considered positive for PrPSc accumulation if the corrected optical density was greater than 0.18 plus the negative-control value (per the manufacturer's instructions). The PrPSc signal of each of the microgliaSc-exposed Rov9 cells was compared to the signal of the respective microgliaC-exposed Rov9 cells using an independent t test with a cutoff P value of 0.05 (SigmaPlot).
Inoculation of immortalized microglia with PrPSc and antibody-based inhibition.
Immortalized microglial cultures were cloned by limiting dilutions in 96-well, plastic, flat-bottom tissue culture plates. Twenty wells contained small colonies that were visualized and split 1/2 into replicate 96-well formats. One replicate was inoculated with 1 μl of the Rov9Sc lysate, and the other replicate was inoculated with 1 μl of the Rov9C lysate (negative control) in 100 μl of OMEM. After the cells reached confluence, the cells and medium (with residual inoculum) were expanded in toto into 24-well plates and cultured in a total volume of approximately 1 ml of OMEM. One week later, an additional 5 μl of Rov9Sc lysate (Rov9C lysate for negative-control cells) was added to the appropriate wells, and 4 days later, 0.5 ml of OMEM was added to each well. When the cells reached confluence, the cells and medium were first expanded into six-well tissue culture plates (approximately 3 ml total of OMEM per well) and then finally into 75-cm2 tissue culture flasks (25 ml total of OMEM), each time without dilution. Cells were then serially passaged 1/5 at confluence in OMEM using standard techniques. At select passage points, an aliquot of cells was collected, lysed, and evaluated for PrPSc accumulation by ELISA as described above. Cells were considered positive for PrPSc accumulation if the corrected optical density was greater than 0.18 plus the negative-control value (per the manufacturer's instructions) after five 1/5 splits or later. PrPSc was verified by immunoblotting (see below).
PrPSc accumulation within the PrPSc-positive SV40 large-T-antigen-transformed cell line (B6) was then inhibited by a prion-specific antibody. Similar to the previously described method, B6 cells were treated for 13 days with the recombinant anti-prion protein Fab D18 (InPro Biotechnology), which has demonstrated the ability to bind membrane-bound PrPC and inhibit PrPSc accumulation in murine neuroblastoma cells (ScN2a cells) (46). Treated B6 cells were then split 1/5, without antibody, every 4 days for 4 weeks and then collected for immunoblot analysis as described below. The recombinant anti-prion protein Fab R72 (InPro Biotechnology), which does not bind to cell surface PrPC and thus does not inhibit PrPSc accumulation (46), was used as the negative control. All antibody inhibition experiments were performed in triplicate.
Immunoblot detection of PrPSc in cell cultures.
Since the ELISA does not utilize the standard method of protease digestion to discriminate between PrPSc and PrPC, cells were also collected for immunoblot confirmation of protease resistance. Primary cells were immunoblotted by a method similar to the method previously described (32). Approximately 2 to 3 million microgliaSc and microgliaC were trypsinized into solution and washed with 1× D-PBS. The resulting pellet was lysed for 1 to 2 h in 1 ml of lysis buffer containing 0.5% (vol/vol) Nonidet P-40 (Roche) and 0.5% (wt/vol) sodium deoxycholate (Sigma) in 10 mM Tris buffer (pH 7.4). Aliquots (100 to 250 μl) of lysate were brought up to a total volume of 500 μl by using additional lysis buffer. An equal volume of 4% (wt/vol) N-lauroylsarcosine sodium salt (Sigma) solution in PBS was added to the lysate and incubated for 15 min at 37°C, followed by DNase I (100 μg/ml) (Roche) treatment for 30 min at 37°C. Samples were centrifuged 1,100 × g for 5 min at room temperature, and 1 ml of the supernatant was treated with proteinase K (50 μg/ml) (Roche) for 1 h at 37°C. Replicate samples were incubated for 1 h at 37°C without proteinase K. Eighty microliters of 4% (wt/vol) phosphotungstic acid in 170 mM MgCl2 was added, the samples were incubated for 1 hour at 37°C, and following centrifugation at 16,500 × g at room temperature, the pellet was resuspended in 16 μl of water. Each sample (16 μl) was mixed with 7 μl of 4× NuPAGE sample buffer and 2.5 μl of 10× reducing agent (Invitrogen) and then boiled for 10 min. The samples were electrophoresed for 50 min at 200 V using the NuPAGE precast 12% Bis-Tris-buffered sodium dodecyl sulfate-polyacrylamide gel system (Invitrogen) with morpholinepropanesulfonic acid (MOPS) running buffer. NuPAGE antioxidant (Invitrogen) was added to the inner-chamber running buffer. Gels were electroblotted for 50 min at 300 mA onto a 0.45-μm polyvinylidene fluoride (PVDF) (Millipore) membrane, using the semidry Biometra Fastblot B33 apparatus and transfer buffer containing 25 mM Tris (pH 8.3), 150 mM glycine, and 10% (vol/vol) methanol. Following the transfer, the PVDF membrane was fixed for 20 seconds in 100% methanol, air dried, and then immunoblotted or stored dry overnight. PVDF membranes were blocked in casein blocker (Pierce) with 0.05% (vol/vol) Tween 20 for 1 hour at room temperature. The prion protein was detected by a 1-hour incubation at room temperature with the monoclonal antibody F99/97.6.1 (produced in our laboratory; also available from VMRD, Inc.) at a concentration of 3.5 μg/ml in blocking buffer. The membrane was then washed three times in TBST (150 mM NaCl and 0.05% Tween 20 in 10 mM Tris [pH 8.0]) and incubated for 30 minutes at room temperature with a goat anti-mouse IgG1 antibody conjugated to horseradish peroxidase (catalog no. 1070-05; Southern Biotechnology Associates) and diluted 1:5,000 in blocking buffer. Following five additional washes in TBST, the membrane was incubated with chemiluminescent substrate (Western Lightning; Perkin Elmer) for 3 min. Signals were visualized by exposing the membrane to radiographic film (ISC BioExpress) and evaluated for the proper banding pattern associated with proteinase K-resistant PrPSc.
For detection of PrPSc within the SV40 large-T-antigen-transformed cell line (B6), the TeSeE Western blot kit (Bio-Rad, France) was used following the manufacturer's instructions. Fifteen to 20 microliters of the final sample suspension was electrophoresed for 60 to 90 min at 250 V using precast 12% Tris-HCl Ready gels (Bio-Rad) in 25 mM Tris (pH 8.3), 192 mM glycine, and 0.1% sodium dodecyl sulfate running buffer. Gels were electroblotted for 60 min at 115 V onto a 0.22-μm nitrocellulose membrane (Bio-Rad), using the a Mini-Protean 3 cell (Bio-Rad) and transfer buffer containing 1× Tris-CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] (Bio-Rad) and 15% ethanol. Following the transfer, the nitrocellulose membrane was fixed for 10 s in 100% ethanol and briefly transferred to distilled water. The immunoblotting procedure for the detection of the prion protein was similar to the method described above for the primary microglia, except for the use of reagents, including antibodies, provided in the TeSeE kit.
Inoculation of PrPSc-accumulating primary microglia with CAEV and evaluation of effect of CAEV on extracellular PrPSc release.
At passage 10 after PrPSc inoculation, primary microgliaSc were plated at a concentration of 1 × 105/well in 24-well plastic tissue culture plates. MicrogliaC were treated similarly and served as negative controls for this experiment. The Cork strain of CAEV, in 0.5 ml of total OMEM, was added to the cells (microgliaSc-CAEV and microgliaC-CAEV) at a concentration sufficient to infect the majority of the cells within 7 days (based on a previous viral infection experiment [data not shown]) as assayed by immunocytochemistry, which is described below. Aliquots of both cell types were also sham inoculated with medium only (microgliaSc-sham and microgliaC-sham) to serve as negative controls for the viral inoculation. All inoculations were performed with six replicates. At days 4, 8, 12, 16, and 19 after virus inoculation, culture supernatants were collected and saved at −80°C for analysis by HerdChek scrapie antigen test kit ELISA (see above) and for transmission studies (see below). A standard curve prepared from diluted Rov9Sc inoculum (1/50, 1/100, 1/400, 1/800, and 1/1,600) was used to normalize corrected optical density results between ELISA plates. The PrPSc signal of microgliaSc-CAEV supernatants was compared to the PrPSc signal of microgliaSc-sham supernatants using an independent t test with a cutoff P value of 0.05 (SigmaPlot).
Demonstration of PrPSc infectivity from microgliaSc-CAEV-derived supernatants and evaluation of effect of CAEV on intracellular PrPSc accumulation.
Primary microgliaC at passage 21 were plated in 24-well plates at 2 × 105 cells per well. One day later, 400 μl of the cell culture supernatant on day 4 after viral inoculation (see previous paragraph) was applied to the cells, in triplicate for each of the four treatment groups (microgliaSc-CAEV, microgliaSc-sham, microgliaC-CAEV, and microgliaC-sham). Once the cells reached confluence, they were expanded sequentially into 12-well plates and into 25-cm2 flasks. At day 26 postinoculation, aliquots of microglial cells were plated into chambered slides to confirm that the cells were infected with CAEV (see below), and four-fifths of the total cells were collected and lysed (as described above) for analysis by HerdChek scrapie antigen test kit ELISA. A standard curve prepared from diluted Rov9Sc inoculum (1/100, 1/400, 1/800, and 1/1,600) was used to normalize corrected optical density results. The PrPSc signal of microgliaSc-CAEV lysates was compared to the PrPSc signal of microgliaSc-Sham lysates using an independent t test with a cutoff P value of 0.05 (SigmaPlot).
Immunocytochemical detection of CAEV in primary microglia.
Cells were fixed with 4% buffered zinc formaldehyde (Z-fix; Anatech Ltd.) for 2 min and then rinsed in 70% ethanol. CAEV antigen was detected using the MAb 10A1 (VMRD, Inc.) directed against 28-kDa capsid protein. Negative controls included the use of isotype-matched antibodies raised against an irrelevant antigen, omission of the primary antibody, and omission of both the primary and secondary antibodies. CAEV-infected goat synovial membrane cells, derived from neonatal goats (29), served as positive controls for the immunocytochemistry.
RESULTS
Establishment and characterization of primary and transformed fetal sheep brain cells.
To study the cellular and molecular changes associated with prion conversion in a natural host-TSE system, primary brain cell cultures were derived from a Suffolk cross domestic sheep fetus that was homozygous for the prnp VRQ allele (Val-136, Arg-154, and Gln-171). Primary cell cultures were adherent and viable for approximately 20 passages. Additionally, a cell line derived from the primary brain cells was created by transformation with a plasmid containing the SV40 virus large T antigen. Increased rate of growth, lack of contact inhibition, and immunoreactivity for the large T antigen confirmed transformation of the cell culture (data not shown). The transformed cell line has demonstrated viability for 60 passages, as of this writing, and shown no evidence of cell pathology. Both the primary brain cultures and the transformed cell line demonstrated expression of several microglial markers by both immunocytochemistry and flow cytometry (Table 1), although the primary microglia cultures lacked CD68 expression. By immunocytochemistry, approximately 90% of cells demonstrated immunoreactivity for the given markers (data not shown). The approximately 10% of cells lacking immunoreactivity were morphologically indistinguishable from those cells that were demonstrating immunoreactivity. On the basis of the phenotypic features, we classified both the primary cell cultures and the transformed cell line as sheep microglia.
TABLE 1.
Phenotypes of primary sheep brain cultures and transformed glial cell line
| Cells | Method | Markera
|
|||
|---|---|---|---|---|---|
| RCA-I | CD14 | CD68 | Nonspecific esterase | ||
| Primary | Immunocytochemistry | + | + | − | + |
| Flow cytometry | + | + | − | NA | |
| Transformed | Immunocytochemistry | + | + | + | + |
| Flow cytometry | + | + | + | NA | |
The presence (+) or absence (−) of markers is shown. The markers are Ricinus communis agglutinin-1 (RCA-I), CD14 (catalog no. MM61A; VMRD, Inc.), CD68 (EBM11; Dako), and nonspecific esterase. NA, not applicable.
PrPSc infection of primary microglia.
To determine the permissiveness of cultured sheep microglia to PrPSc infectivity, primary microglia were exposed to the prnp genotype-matched Rov9Sc-derived PrPSc. Lysates of the inoculated microglia (microgliaSc) were collected at several time points, and commercially available ELISAs were used to screen the lysates for PrPSc. Primary microgliaSc demonstrated accumulation of PrPSc over time, as illustrated by an increasing PrPSc signal in cell lysates, even after dilution of the cells and residual inoculum due to several 1/5 splits (Fig. 1A). The cell growth rate or morphology of microgliaSc and microgliaC were not different from each other, which is consistent with previously reported findings (11).
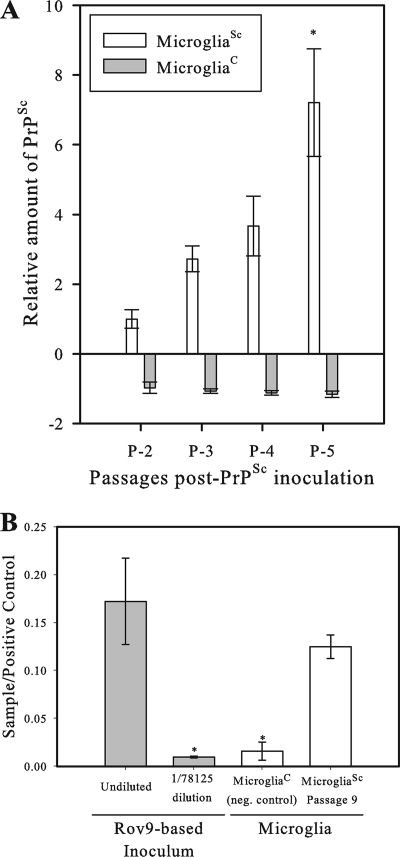
FIG. 1.
PrPSc accumulation in primary microglia. Primary sheep microglia were inoculated in triplicate with either PrPSc-containing (microgliaSc) or PrPSc-lacking (microgliaC) cell lysates derived from the PrPSc-positive or -negative Rov9 cell line (60), respectively, and then serially passaged. Lysates of microgliaSc were assayed for PrPSc by commercial ELISA, and the corrected optical density was determined. The equation for the corrected optical density is OD450 − OD620 (OD450 being the optical density at 450 nm) per the manufacturer's instructions. (A) ELISA results comparing the level of PrPSc in late-passage (five passages after PrPSc inoculation [P-5]) microgliaSc versus early passage (P-2) microgliaSc. Data were normalized using a standard curve. MicrogliaSc results were set at 1, and all other results were normalized to this value (y axis). Each value represents the mean ± 1 standard deviation (error bar). Results for microgliaSc at P-5 and P-2 were compared by a statistical analysis, and the microgliaSc values at P-5 and P-2 were significantly different (P < 0.005) as indicated by an asterisk. (B) ELISA results comparing the relative level of PrPSc in late-passage (P-9) primary microgliaSc to the original inoculum. Data were normalized to the ELISA plate positive control (y axis) to account for plate-to-plate variation between testing repeats performed at different times. Results for microgliaSc at passage 9 were compared to the values for microgliaC and both Rov9Sc-based inoculum data points. The values for P-9 microgliaSc were significantly different (P < 0.05) from those for microgliaC and the diluted RovSc data points, as indicated by an asterisk. neg. control, negative control.
To determine the relative level of PrPSc accumulation compared to the original inoculum and to further verify the detection of newly converted PrPSc versus detection of residual inoculum, the amount of PrPSc in a lysate of microglial cells at passage 9 (representing two expansions and seven 1/5 splits) was compared to the amount of PrPSc in the inoculum. The inoculum was tested at its original concentration and at a 57 (1/78,125) dilution, representing the seven 1/5 splits (and two expansions) of the microgliaSc cells at passage 9. The level of PrPSc in the 1/57-diluted inoculum was below the detection limit of the ELISA, as evidenced by results similar to those for uninoculated microglia cells. The microgliaSc lysates from passage 9 demonstrated a significantly higher PrPSc signal than the 1/57-diluted inoculum did (representing the expected PrPSc signal after seven splits if no accumulation was occurring in the microgliaSc) (Fig. 1B). If the cells lacked the ability to form nascent PrPSc, then the expected result would be that the passage 9 sample would be equal to the 1/57-diluted inoculum. As this was not seen, it was concluded that de novo conversion and accumulation of PrPSc were occurring within the primary sheep microglia in order to compensate for the dilution of PrPSc that occurs during splitting of cells during routine cell culture.
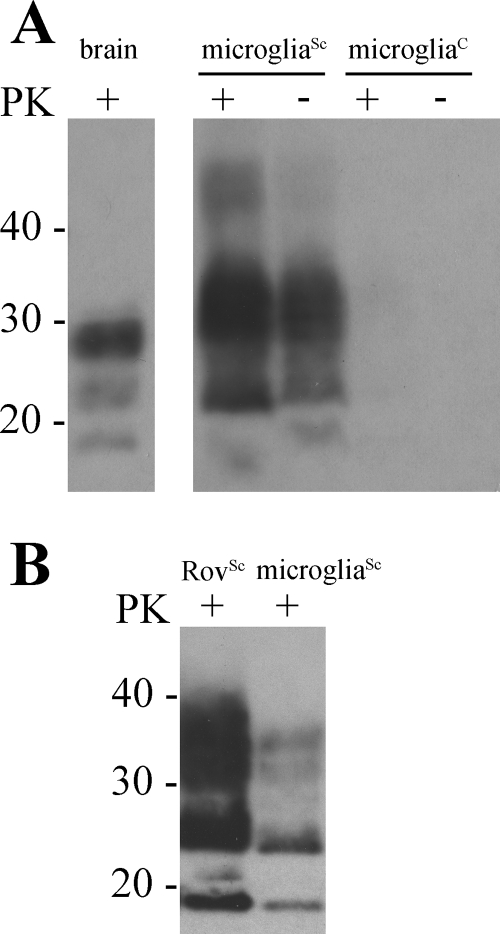
The ELISA that was utilized in this study uses a proprietary chemical, which preferentially binds beta-sheets, to distinguish PrPSc from PrPC. Therefore, immunoblotting of microglial lysates was used to confirm proteinase K resistance, which is the gold standard for PrPSc detection, of the microglia-derived PrPSc and to compare the glycoform profile compared to the profile of Rov9Sc-based inoculum. MicrogliaSc accumulated proteinase K-resistant PrPSc that migrated with a profile different from that of scrapie-positive brain material (Fig. 2A), but with a profile similar to that of the Rov9Sc-based inoculum (Fig. 2B).
FIG. 2.
Immunoblot analysis of primary sheep microglia following exposure to PrPSc (microgliaSc) and unexposed microglia (microgliaC). Microglia were lysed, treated with proteinase K (PK) (+) or not treated with proteinase K (−), precipitated with phosphotungstic acid, and immunoblotted using the monoclonal anti-PrP antibody F99.97.6.1. (A) Comparison of prion immunoreactivity from sheep brain, microgliaSc, and microgliaC. (B) Comparison of proteinase K-resistant glycoform patterns between Rov9Sc and microgliaSc. The positions of molecular mass standards (in kilodaltons) are indicated to the left of the immunoblots.
The infectivity of the intracellular PrPSc produced by the microglia was analyzed by exposing Rov9C cells to three dilutions (1/20, 1/200, and 1/2,000) of microglial cell lysates. The Rov9C cells inoculated with microgliaSc lysates demonstrated accumulation of PrPSc over time as evidenced by an increasing ELISA optical density at each subsequent time point (Fig. 3). Additionally, Rov9C cells exposed to higher concentrations of microgliaSc inoculum demonstrated higher levels of PrPSc accumulation (Fig. 3). The dose responsiveness of the microglia-derived PrPSc is consistent with infectivity.
FIG. 3.
Transmission of microglia-derived PrPSc to Rov9C cells. Rov9C cells were inoculated in triplicate with three dilutions of microgliaSc lysates. The inoculated Rov9 cells were serially passaged, lysed, and analyzed for PrPSc levels by ELISAs. Results for each group of inoculated cells at passage 4 were compared to the results for the corresponding mock-inoculated cells, and the values for inoculated cells at passage 4 that were statistically different (P < 0.05) from the value for the corresponding mock-inoculated cells are indicated by an asterisk. All samples were run on the same ELISA plate; thus, results are reported as the corrected optical density (y axis). Each value represents the mean ± 1 standard deviation (error bar) of the sample corrected optical density. The equation for the corrected optical density is OD450 − OD620 (OD450 being the optical density at 450 nm) per the manufacturer's instructions. Inoc, inoculated; Mock, mock inoculated.
PrPSc infection of transformed microglia and inhibition of PrPSc accumulation.
Transformed microglial cells were cloned and inoculated to test whether sheep microglial cell cultures with a longer culture life span than the primary sheep microglial cell culture support PrPSc accumulation. Clones were inoculated with Rov9-derived PrPSc and then screened by ELISA for evidence of PrPSc accumulation. Twenty clones were inoculated, and 15 of those clones were stable and were able to be expanded and tested for PrPSc accumulation. One of the 15 clones tested positive for PrPSc by ELISA at passage five and later (data not shown). The positive clone, B6, was the clone that was the slowest to grow to confluence during the initial inoculation period, taking 16 days compared to an average of 6 days for the 14 negative clones. The PrPSc signal in the transformed cell line remained consistently low, compared to the signal in the primary sheep microglia, and spontaneous loss of PrPSc was seen after approximately 15 passages.
To further characterize the PrPSc being accumulated within the transformed microglia, the positive clone was treated with the recombinant anti-prion Fab D18. Following a 13-day exposure to this Fab and 4 weeks of culture without the Fab, the transformed microglial cell line demonstrated loss of PrPSc detectable by ELISA (data not shown) and detectable by immunoblotting compared to the cultures treated with the negative-control recombinant anti-prion Fab R72, which maintained detectable levels of PrPSc (Fig. 4). This specific antibody-based inhibition of PrPSc accumulation and the multiple assays used to detect PrPSc increase over time indicate that the PrPSc detected in the microglial cells represents PrPSc molecules that are newly formed in microglia.
FIG. 4.
Immunoblot analysis of PrPSc-infected sheep microglial cell line after incubation with the recombinant anti-prion Fab D18. Primary sheep cultures were transformed with the SV40 virus large T antigen, cloned by limiting dilutions, and then inoculated with PrPSc. The clone that was positive for PrPSc was incubated with Fab D18 for 13 days and then cultured in the absence of the Fab for 4 weeks. Cells were lysed, treated with proteinase K (PK) (+) or not treated with proteinase K (−), and immunoblotted using the TeSeE Western blot kit (Bio-Rad, France). Lysates of replicate PrPSc-infected B6 cells that were treated with the control anti-prion Fab R72, which does not inhibit PrPSc accumulation, are included as a control for spontaneous loss of detectable PrPSc. The positions of molecular mass standards (in kilodaltons) are indicated to the left of the immunoblot.
Enhancement of PrPSc levels by CAEV coinfection.
Previous studies with ScN2a cells and MLV, a gammaretrovirus, have demonstrated enhancement of prion infectivity release into the culture supernatant (39). While no gammaretroviruses of small ruminants have been identified (17), sheep and goats are susceptible to the SRLVs VISNA and CAEV, which are closely related to one another and are considered members of the same lentivirus group (45). While gammaretroviruses and lentiviruses have unique properties, much of the basic biology between these two groups of retroviruses is shared. To determine whether SRLV coinfection enhances the release of PrPSc into the culture supernatant, similar to the results of MLV in ScN2a cells, late-passage primary microgliaSc and microgliaC were inoculated with the Cork strain of CAEV. CAEV-Cork induced less cytotoxicity than the other tested SRLV strains (VISNA 84-28, VISNA LMH11, and VISNA WLC-1) that each resulted in nearly 100% cell lysis within 1 week (data not shown). The culture supernatants were collected regularly and assayed for relative levels of PrPSc concentration by normalized ELISA optical densities. There was no detectable difference between cultures infected and uninfected with SRLV in cell growth, as determined by rate to confluence, or phenotype. The results demonstrate that following subsequent infection by CAEV, microglia cells that were already PrPSc infected have significantly enhanced (up to 1.5-fold) PrPSc release into the culture supernatant compared to that of microgliaSc-Sham (Fig. 5A). The supernatants from cultures of microgliaC-CAEV and microgliaC-Sham remained free from detectable levels of PrPSc, indicating that CAEV does not result in detectable levels of spontaneous conversion of PrPC into PrPSc.
FIG. 5.
Effects of CAEV coinfection on PrPSc accumulation in primary sheep microglia and transmission of microglia-derived PrPSc to primary microglial cells. (A) PrPSc accumulation in the culture supernatant of primary microglia infected with CAEV after establishment of PrPSc accumulation. MicrogliaSc and microgliaC were inoculated with CAEV, and cell culture supernatants were collected at 4, 8, 12, 16, and 19 days after CAEV inoculation (dpi). Aliquots of supernatant were analyzed by an ELISA for PrPSc levels, which were normalized via a standard curve. Normalized 4 dpi microgliaSc-CAEV levels were set at 1, and all other results were normalized based on this value (y axis). Six replicates for each treatment group were used. At each time point, results for microgliaSc-CAEV were compared to the results for microgliaSc-Sham in a statistical analysis; microgliaSc-CAEV values that were significantly different from microgliaSc-Sham values are indicated as follows: *, P < 0.05; †, P < 0.0005. (B) PrPSc levels in the lysates of primary microglia simultaneously coinfected with PrPSc and CAEV. MicrogliaC were inoculated with the 4 dpi culture supernatants from the previous experiment: Sc-CAEV, Sc-Sham, C-CAEV, and C-Sham. At 26 dpi, cell lysates were analyzed by an ELISA for PrPSc levels, which were normalized via a standard curve. Normalized microgliaSc-Sham levels were set at 1, and all other results were normalized to this value (y axis). Three replicates for each treatment group were used. Results for microgliaSc-CAEV were compared to the results for microgliaSc-Sham and evaluated statistically; the microgliaSc-CAEV value that was significantly different (P < 0.01) from the microgliaSc-Sham value is indicated by an asterisk. Each error bar represents 1 standard deviation.
It is known that the mechanism of PrPSc transmission within one cell culture system may vary compared to another cell culture system. For instance, in ScN2a cells, the spread of PrPSc is mainly vertical (i.e., from mother to daughter cells) (22), whereas spread within Rov9 cells is horizontal to nearby cells, with rare spreading to spatially distant cells (44). To determine whether supernatant-derived PrPSc, which was enhanced by SRLV coinfection in primary sheep microglia, is associated with infectivity toward primary microglia, microgliaC were inoculated with supernatants from the previous primary microglia experiment that contained both PrPSc and CAEV. Additionally, this experiment also served three additional purposes: to repeat the observed enhancement of PrPSc accumulation following CAEV coinfection, to determine whether the increase in PrPSc also applied to intracellular PrPSc, and to determine whether simultaneous PrPSc-CAEV coinfection (versus CAEV infection of established PrPSc-infected microglia as shown above) yielded a similar enhancement of relative PrPSc levels. Lysates were analyzed by ELISA to determine the relative amounts of PrPSc. Both microgliaSc-CAEV and microgliaSc-Sham contain detectable levels of PrPSc, thus confirming that primary microgliaSc-derived PrPSc is infectious to microglia and suggesting that horizontal spread within microglial cultures is likely (Fig. 5b). Additionally, microgliaSc-CAEV lysates demonstrated an approximately twofold-higher PrPSc signal compared to the signal of microgliaSc-Sham (Fig. 5b). Thus, these results confirm the repeatability of CAEV-induced enhancement of PrPSc accumulation in microglia, show that the intracellular PrPSc levels are also elevated, and demonstrate that the PrPSc enhancement is evident with simultaneous PrPSc and CAEV coinfections. These two coinfection experiments (Fig. 5) indicate an overall increase, up to twofold, in PrPSc within primary sheep microglia that are coinfected with CAEV. As expected, primary microglia not infected with PrPSc (microgliaC-CAEV and microgliaC-Sham treatment groups) did not demonstrate detectable levels of PrPSc. CAEV infection was verified by immunocytochemistry for the CAEV 28-kDa capsid protein (data not shown).
DISCUSSION
Scrapie is an important disease in the United States and has been targeted for eradication (47). While scrapie has been recognized since the early 18th century (48), many questions still remain including the details of transmission between animals and the identification of factors that promote transmission and infection. Recent studies in mice and mouse-derived cell cultures have shown that gammaretroviral infections enhance the amount of prion infectivity produced by prion-infected cells (39). Our results have extended this connection to sheep-derived microglia cultures coinfected with an SRLV. While the VISNA strains that were most readily available were too cytopathic for these studies, we have shown that coinfection with the very closely related SRLV CAEV-Cork strain results in increased PrPSc accumulation in sheep brain macrophages (microglia), as evidenced by the increase in PrPSc signal both within the supernatant and within the microglial cells. Additionally, we have demonstrated that the enhancement of PrPSc accumulation occurs following either sequential PrPSc/CAEV infection or simultaneous coinfection and that both the intracellular microglia-derived PrPSc and extracellular microglia-derived PrPSc are infectious.
Studies analyzing prion and retroviral coinfections in natural hosts are limited. A previous report has demonstrated PrPSc in macrophages within lesions of SRLV-induced lymphofollicular mastitis in sheep (41). However, the mechanism of colocalization was not determined. One possible mechanism is recruitment of macrophages and follicular dendritic cells, unrelated to prion pathogenesis, to tissues with SRLV-induced inflammation, which then increased the number of PrPSc-permissive cells in the mammary gland. A second possible mechanism is that the SRLV infection specifically enhances PrPSc accumulation in the macrophages and follicular dendritic cells. While these two mechanisms are not mutually exclusive, our results demonstrating SRLV-induced enhancement of PrPSc in microglia (brain-derived macrophages) suggest that a specific interaction may be at least partially responsible for the presence of PrPSc in SLRV-induced mastitis.
The SLRV-induced enhancement of PrPSc in these microglia cultures is modest; thus, the in vivo relevance of any direct interaction between SRLV and PrPSc is not known. However, prion diseases are extremely chronic diseases that take months to years to fully develop (61); thus, small changes in the rate of PrPSc accumulation can potentially be significant following amplification over that extended time period. Furthermore, both PrPSc and SRLV are present independently in the same cell types, e.g., macrophages (20, 23, 24, 43, 49), dendritic cells (24, 31, 53), and microglia (5, 7, 16, 33). Additionally, nearly one in four domestic sheep within the United States is infected with VISNA (56). Thus, given the increased PrPSc accumulation associated with SRLV coinfection that is reported herein, the overlapping cellular tropism of PrPSc and SRLVs, and the prevalence of SRLV, it is important to determine the extent of interaction between these two agents in vivo and the effects, if any, on any scrapie eradication measures.
This study does not investigate the mechanism of interaction between SRLVs and PrPSc. Possible mechanisms for the enhanced PrPSc accumulation in the sheep microglia include but are not limited to the coinfecting SRLV increasing PrPC protein expression, enhancing access to conversion accessory molecules, increasing colocalization of PrPC and PrPSc during viral assembly, and increasing transmission of PrPSc by cotransmission with the virus. The study using MLV and ScN2a cells demonstrated that PrPC and PrPSc are recruited to virions (39), suggesting that increased PrPSc transmission is likely to play at least some role in the enhancement of PrPSc accumulation. Another recent study investigated the interaction of PrPSc and minute virus of mice, a parvovirus, and demonstrated that the parvovirus-infected cells internalize exogenous PrPSc more efficiently than the virus-free cells, most likely due to virus-induced changes in the cellular membranes (4). However, it remains to be determined whether similar mechanisms are induced by infections with retroviruses and other viruses. Another possible mechanism for the enhanced prion accumulation is that there are virus-induced changes in the abundance and/or character of RNA, which has been suggested as a possible cofactor for the conversion of PrPC to PrPSc (2, 15, 19, 21). Determining the mechanism(s) of the virus-induced enhancement of PrPSc accumulation warrants further study, which may also help to elucidate some of the basic cellular mechanisms of PrPSc conversion.
During the course of scrapie infection, numerous cell types, including glia, macrophages, and follicular dendritic cells, demonstrate accumulation of PrPSc (58), but the cell-specific mechanisms that allow for prion conversion are not known. The ability to investigate these mechanisms requires the tools to study PrPSc infection in these various cell types, mainly having permissive cell culture systems from a variety of cell types. Herein we describe the second cell line derived from a natural TSE host and the first microglial cell culture derived from a natural TSE host (59).
These microglial cells are of the highly susceptible VRQ genotype (Val-136, Arg-154, and Gln-171). Acquiring PrPSc-containing VRQ/VRQ sheep brain for research material can be difficult in the United States due to the rarity (approximately 0.5%) of this genotype in the United States (57). While it is recognized that this study used an inoculum derived from lagomorph cells, it should be noted that the inoculum's PrPSc has an ovine, and not lagomorph, amino acid sequence (prnp in Rov cells is the ovine insert). Thus, the disadvantage of utilizing material derived from a leporine source is outweighed in this particular case by the availability of Rov9-derived PrPSc and its matched ovine amino acid sequence. Furthermore, now that the concept of PrPSc-susceptible sheep microglia cultures has been demonstrated, these cells can be utilized in further studies to determine the relevance of prion allelic usage, prion allelic susceptibility, and other sheep-specific prion questions.
Cell culture models of PrPSc accumulation have been used for numerous studies to screen for inhibitors of PrPSc accumulation (59). However, recent work has indicated one of the pitfalls of using cell culture models, namely, that the amount of PrPSc accumulating in cells is inversely related to the mitotic rate of the cell cultures (22). Thus, the ability to cure cell culture systems of PrPSc differs significantly compared to mitotically inactive neurons, which are infected with PrPSc in clinical patients. Our results support this conclusion, as the only positive cell line derived from the transformed microglia was the clone that was slow growing during the initial inoculation phase and even this clone lost the ability to maintain detectable levels of PrPSc after the cultures began achieving confluence at a faster rate. While the mitotic activity of cell culture systems may alter the biological relevance of using these systems to screen for antiprion compounds, cell culture systems do have the ability to screen for factors that promote PrPSc accumulation. Multiple lines of evidence suggest a role for an accessory molecule(s) to enable PrPSc accumulation (35, 54, 55). The coinfection model of PrPSc and SRLV in primary sheep microglia is one example of using cell cultures to identify factors that enhance PrPSc accumulation. Further studies, including screening for additional factors that promote PrPSc accumulation in microglia and studying those cofactors in a variety of PrPSc-permissive cell lines, are warranted.
Acknowledgments
This work was supported by NIH grant K08 AI064729 and USDA/ARS specific cooperative agreement 58-5348-577.
We thank Didier Vilette for permission to use the Rov9 cells, Byron Caughey for supplying the Rov9 cells and methods for culturing Rov9 cells, and Dongyue Zhuang for assistance with the phosphotungstic acid precipitation and immunoblotting.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Ackermann, M. R., B. M. DeBey, T. J. Stabel, J. H. Gold, K. B. Register, and J. T. Meehan. 1994. Distribution of anti-CD68 (EBM11) immunoreactivity in formalin-fixed, paraffin-embedded bovine tissues. Vet. Pathol. 31340-348. [DOI] [PubMed] [Google Scholar]
- 2.Adler, V., B. Zeiler, V. Kryukov, R. Kascsak, R. Rubenstein, and A. Grossman. 2003. Small, highly structured RNAs participate in the conversion of human recombinant PrPSen to PrPRes in vitro. J. Mol. Biol. 33247-57. [DOI] [PubMed] [Google Scholar]
- 3.Alverson, J., K. I. O'Rourke, and T. V. Baszler. 2006. PrPSc accumulation in fetal cotyledons of scrapie-resistant lambs is influenced by fetus location in the uterus. J. Gen. Virol. 871035-1041. [DOI] [PubMed] [Google Scholar]
- 4.Avrahami, D., Y. Dayan-Amouyal, S. Tal, M. Mincberg, C. Davis, O. Abramsky, and R. Gabizon. 13 May 2008. Virus-induced alterations of membrane lipids affect the incorporation of PrPSc into cells. J. Neurosci. Res. doi: 10.1002/jnr.21720. [DOI] [PubMed]
- 5.Baker, C. A., D. Martin, and L. Manuelidis. 2002. Microglia from Creutzfeldt-Jakob disease-infected brains are infectious and show specific mRNA activation profiles. J. Virol. 7610905-10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler, K., B. Oesch, M. Scott, D. Westaway, M. Walchli, D. F. Groth, M. P. McKinley, S. B. Prusiner, and C. Weissmann. 1986. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell 46417-428. [DOI] [PubMed] [Google Scholar]
- 7.Baszler, T. V., W. G. Harwood, K. L. Lester, W. C. Davis, and D. P. Knowles. 1994. Characterization of caprine microglial cells and in vitro infection with caprine arthritis-encephalitis lentivirus. Lab. Investig. 70933-943. [PubMed] [Google Scholar]
- 8.Bate, C., S. Reid, and A. Williams. 2001. Killing of prion-damaged neurons by microglia. Neuroreport 122589-2594. [DOI] [PubMed] [Google Scholar]
- 9.Beringue, V., P. Couvreur, and D. Dormont. 2002. Involvement of macrophages in the pathogenesis of transmissible spongiform encephalopathies. Dev. Immunol. 919-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borchelt, D. R., M. Scott, A. Taraboulos, N. Stahl, and S. B. Prusiner. 1990. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J. Cell Biol. 110743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosque, P. J., and S. B. Prusiner. 2000. Cultured cell sublines highly susceptible to prion infection. J. Virol. 744377-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carp, R. I., H. C. Meeker, V. Caruso, and E. Sersen. 1999. Scrapie strain-specific interactions with endogenous murine leukaemia virus. J. Gen. Virol. 805-10. [DOI] [PubMed] [Google Scholar]
- 13.Caughey, B., and G. J. Raymond. 1991. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 26618217-18223. [PubMed] [Google Scholar]
- 14.Cordeiro, Y., F. Machado, L. Juliano, M. A. Juliano, R. R. Brentani, D. Foguel, and J. L. Silva. 2001. DNA converts cellular prion protein into the beta-sheet conformation and inhibits prion peptide aggregation. J. Biol. Chem. 27649400-49409. [DOI] [PubMed] [Google Scholar]
- 15.Deleault, N. R., R. W. Lucassen, and S. Supattapone. 2003. RNA molecules stimulate prion protein conversion. Nature 425717-720. [DOI] [PubMed] [Google Scholar]
- 16.Ebrahimi, B., T. E. Allsopp, J. K. Fazakerley, and G. D. Harkiss. 2000. Phenotypic characterisation and infection of ovine microglial cells with Maedi-Visna virus. J. Neurovirol. 6320-328. [DOI] [PubMed] [Google Scholar]
- 17.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 18.Gabus, C., S. Auxilien, C. Pechoux, D. Dormont, W. Swietnicki, M. Morillas, W. Surewicz, P. Nandi, and J. L. Darlix. 2001. The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J. Mol. Biol. 3071011-1021. [DOI] [PubMed] [Google Scholar]
- 19.Gabus, C., E. Derrington, P. Leblanc, J. Chnaiderman, D. Dormont, W. Swietnicki, M. Morillas, W. K. Surewicz, D. Marc, P. Nandi, and J. L. Darlix. 2001. The prion protein has RNA binding and chaperoning properties characteristic of nucleocapsid protein NCP7 of HIV-1. J. Biol. Chem. 27619301-19309. [DOI] [PubMed] [Google Scholar]
- 20.Gendelman, H. E., O. Narayan, S. Kennedy-Stoskopf, P. G. Kennedy, Z. Ghotbi, J. E. Clements, J. Stanley, and G. Pezeshkpour. 1986. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. J. Virol. 5867-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geoghegan, J. C., P. A. Valdes, N. R. Orem, N. R. Deleault, R. A. Williamson, B. T. Harris, and S. Supattapone. 2007. Selective incorporation of polyanionic molecules into hamster prions. J. Biol. Chem. 28236341-36353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghaemmaghami, S., P. W. Phuan, B. Perkins, J. Ullman, B. C. May, F. E. Cohen, and S. B. Prusiner. 2007. Cell division modulates prion accumulation in cultured cells. Proc. Natl. Acad. Sci. USA 10417971-17976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilch, S., F. Schmitz, Y. Aguib, C. Kehler, S. Bulow, S. Bauer, E. Kremmer, and H. M. Schatzl. 2007. CpG and LPS can interfere negatively with prion clearance in macrophage and microglial cells. FEBS J. 2745834-5844. [DOI] [PubMed] [Google Scholar]
- 24.Gorrell, M. D., M. R. Brandon, D. Sheffer, R. J. Adams, and O. Narayan. 1992. Ovine lentivirus is macrophagetropic and does not replicate productively in T lymphocytes. J. Virol. 662679-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haldorson, G. J., J. B. Stanton, B. A. Mathison, C. E. Suarez, and T. V. Baszler. 2006. Neospora caninum: antibodies directed against tachyzoite surface protein NcSRS2 inhibit parasite attachment and invasion of placental trophoblasts in vitro. Exp. Parasitol. 112172-178. [DOI] [PubMed] [Google Scholar]
- 26.Heikenwalder, M., N. Zeller, H. Seeger, M. Prinz, P. C. Klohn, P. Schwarz, N. H. Ruddle, C. Weissmann, and A. Aguzzi. 2005. Chronic lymphocytic inflammation specifies the organ tropism of prions. Science 3071107-1110. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann, L. M., T. V. Baszler, K. I. O'Rourke, C. E. Suarez, M. Bakko, J. Alverson, and D. P. Knowles. 2006. Fewer PrPc myeloid-based cells in sheep with the prion-resistant genotype. Neuroreport 17125-129. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann, L. M., W. P. Cheevers, W. C. Davis, D. P. Knowles, and K. I. O'Rourke. 2003. CD21-positive follicular dendritic cells: a possible source of PrPSc in lymph node macrophages of scrapie-infected sheep. Am. J. Pathol. 1621075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrmann, L. M., T. C. McGuire, I. Hotzel, G. S. Lewis, and D. P. Knowles. 2005. Surface envelope glycoprotein is B-lymphocyte immunodominant in sheep naturally infected with ovine progressive pneumonia virus. Clin. Diagn. Lab. Immunol. 12797-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holness, C. L., and D. L. Simmons. 1993. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood 811607-1613. [PubMed] [Google Scholar]
- 31.Huang, F. P., C. F. Farquhar, N. A. Mabbott, M. E. Bruce, and G. G. MacPherson. 2002. Migrating intestinal dendritic cells transport PrPSc from the gut. J. Gen. Virol. 83267-271. [DOI] [PubMed] [Google Scholar]
- 32.Huang, H., J. Rendulich, D. Stevenson, K. O'Rourke, and A. Balachandran. 2005. Evaluation of Western blotting methods using samples with or without sodium phosphotungstic acid precipitation for diagnosis of scrapie and chronic wasting disease. Can. J. Vet. Res. 69193-199. [PMC free article] [PubMed] [Google Scholar]
- 33.Iwamaru, Y., T. Takenouchi, K. Ogihara, M. Hoshino, M. Takata, M. Imamura, Y. Tagawa, H. Hayashi-Kato, Y. Ushiki-Kaku, Y. Shimizu, H. Okada, M. Shinagawa, H. Kitani, and T. Yokoyama. 2007. Microglial cell line established from prion protein-overexpressing mice is susceptible to various murine prion strains. J. Virol. 811524-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janeway, C. 2001. Immunobiology 5: the immune system in health and disease, 5th ed., p.67-68. Garland Publishers, New York, NY.
- 35.Kaneko, K., L. Zulianello, M. Scott, C. M. Cooper, A. C. Wallace, T. L. James, F. E. Cohen, and S. B. Prusiner. 1997. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. USA 9410069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klatzo, I., D. C. Gajdusek, and V. Zigas. 1959. Pathology of Kuru. Lab. Investig. 8799-847. [PubMed] [Google Scholar]
- 37.Kovacs, G. G., E. Lindeck-Pozza, L. Chimelli, A. Q. Araujo, A. A. Gabbai, T. Strobel, M. Glatzel, A. Aguzzi, and H. Budka. 2004. Creutzfeldt-Jakob disease and inclusion body myositis: abundant disease-associated prion protein in muscle. Ann. Neurol. 55121-125. [DOI] [PubMed] [Google Scholar]
- 38.Lasmezas, C. I. 2003. The transmissible spongiform encephalopathies. Rev. Sci. Tech. 2223-36. [PubMed] [Google Scholar]
- 39.Leblanc, P., S. Alais, I. Porto-Carreiro, S. Lehmann, J. Grassi, G. Raposo, and J. L. Darlix. 2006. Retrovirus infection strongly enhances scrapie infectivity release in cell culture. EMBO J. 252674-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, K. H., B. H. Jeong, J. K. Jin, H. C. Meeker, J. I. Kim, R. I. Carp, and Y. S. Kim. 2006. Scrapie infection activates the replication of ecotropic, xenotropic, and polytropic murine leukemia virus (MuLV) in brains and spinal cords of senescence-accelerated mice: implication of MuLV in progression of scrapie pathogenesis. Biochem. Biophys. Res. Commun. 349122-130. [DOI] [PubMed] [Google Scholar]
- 41.Ligios, C., C. J. Sigurdson, C. Santucciu, G. Carcassola, G. Manco, M. Basagni, C. Maestrale, M. G. Cancedda, L. Madau, and A. Aguzzi. 2005. PrPSc in mammary glands of sheep affected by scrapie and mastitis. Nat. Med. 111137-1138. [DOI] [PubMed] [Google Scholar]
- 42.Moscardini, M., M. Pistello, M. Bendinelli, D. Ficheux, J. T. Miller, C. Gabus, S. F. Le Grice, W. K. Surewicz, and J. L. Darlix. 2002. Functional interactions of nucleocapsid protein of feline immunodeficiency virus and cellular prion protein with the viral RNA. J. Mol. Biol. 318149-159. [DOI] [PubMed] [Google Scholar]
- 43.Narayan, O., S. Kennedy-Stoskopf, D. Sheffer, D. E. Griffin, and J. E. Clements. 1983. Activation of caprine arthritis-encephalitis virus expression during maturation of monocytes to macrophages. Infect. Immun. 4167-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paquet, S., C. Langevin, J. Chapuis, G. S. Jackson, H. Laude, and D. Vilette. 2007. Efficient dissemination of prions through preferential transmission to nearby cells. J. Gen. Virol. 88706-713. [DOI] [PubMed] [Google Scholar]
- 45.Pasick, J. 1998. Maedi-visna virus and caprine arthritis-encephalitis virus: distinct species or quasispecies and its implications for laboratory diagnosis. Can. J. Vet. Res. 62241-244. [PMC free article] [PubMed] [Google Scholar]
- 46.Peretz, D., R. A. Williamson, K. Kaneko, J. Vergara, E. Leclerc, G. Schmitt-Ulms, I. R. Mehlhorn, G. Legname, M. R. Wormald, P. M. Rudd, R. A. Dwek, D. R. Burton, and S. B. Prusiner. 2001. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature 412739-743. [DOI] [PubMed] [Google Scholar]
- 47.Peterhans, E., T. Greenland, J. Badiola, G. Harkiss, G. Bertoni, B. Amorena, M. Eliaszewicz, R. A. Juste, R. Krassnig, J. P. Lafont, P. Lenihan, G. Petursson, G. Pritchard, J. Thorley, C. Vitu, J. F. Mornex, and M. Pepin. 2004. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 35257-274. [DOI] [PubMed] [Google Scholar]
- 48.Poser, C. M. 2002. Notes on the history of the prion diseases. Part II. Clin. Neurol. Neurosurg. 10477-86. [DOI] [PubMed] [Google Scholar]
- 49.Prinz, M., F. Montrasio, M. A. Klein, P. Schwarz, J. Priller, B. Odermatt, K. Pfeffer, and A. Aguzzi. 2002. Lymph nodal prion replication and neuroinvasion in mice devoid of follicular dendritic cells. Proc. Natl. Acad. Sci. USA 99919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prusiner, S. B. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216136-144. [DOI] [PubMed] [Google Scholar]
- 51.Raymond, G. J., E. A. Olsen, K. S. Lee, L. D. Raymond, P. K. Bryant III, G. S. Baron, W. S. Caughey, D. A. Kocisko, L. E. McHolland, C. Favara, J. P. Langeveld, F. G. van Zijderveld, R. T. Mayer, M. W. Miller, E. S. Williams, and B. Caughey. 2006. Inhibition of protease-resistant prion protein formation in a transformed deer cell line infected with chronic wasting disease. J. Virol. 80596-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rock, R. B., G. Gekker, S. Hu, W. S. Sheng, M. Cheeran, J. R. Lokensgard, and P. K. Peterson. 2004. Role of microglia in central nervous system infections. Clin. Microbiol. Rev. 17942-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan, S., L. Tiley, I. McConnell, and B. Blacklaws. 2000. Infection of dendritic cells by the Maedi-visna lentivirus. J. Virol. 7410096-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Telling, G. C., M. Scott, K. K. Hsiao, D. Foster, S. L. Yang, M. Torchia, K. C. Sidle, J. Collinge, S. J. DeArmond, and S. B. Prusiner. 1994. Transmission of Creutzfeldt-Jakob disease from humans to transgenic mice expressing chimeric human-mouse prion protein. Proc. Natl. Acad. Sci. USA 919936-9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Telling, G. C., M. Scott, J. Mastrianni, R. Gabizon, M. Torchia, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 1995. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 8379-90. [DOI] [PubMed] [Google Scholar]
- 56.U.S. Department of Agriculture. 2003. Ovine progressive pneumonia: awareness, management, and seroprevalence. Info sheet. Centers for Epidemiology and Animal Health, Veterinary Services, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Fort Collins, CO. http://nahms.aphis.usda.gov/sheep/sheep01/OPP.pdf.
- 57.U.S. Department of Agriculture. 2003. Phase II: scrapie: ovine slaughter surveillance study 2002-2003. U.S. Department of Agriculture document no. N419.0104. National Animal Health Monitoring System, Centers for Epidemiology and Animal Health, Veterinary Services, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Fort Collins, CO. http://nahms.aphis.usda.gov/sheep/SOSSphase2.pdf.
- 58.van Keulen, L. J., A. Bossers, and F. van Zijderveld. 8 February 2008. TSE pathogenesis in cattle and sheep. Vet. Res. doi: 10.1051/vetres:2007061. (Subsequently published, Vet. Res. 39:24, 2008.) [DOI] [PubMed]
- 59.Vilette, D. 27 November 2007. Cell models of prion infection. Vet. Res. doi: 10.1051/vetres:2007049. (Subsequently published, Vet. Res. 39:10, 2008.) [DOI] [PubMed]
- 60.Vilette, D., O. Andreoletti, F. Archer, M. F. Madelaine, J. L. Vilotte, S. Lehmann, and H. Laude. 2001. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc. Natl. Acad. Sci. USA 984055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weissmann, C. 2005. Birth of a prion: spontaneous generation revisited. Cell 122165-168. [DOI] [PubMed] [Google Scholar]
- 62.Westaway, D., S. J. DeArmond, J. Cayetano-Canlas, D. Groth, D. Foster, S. L. Yang, M. Torchia, G. A. Carlson, and S. B. Prusiner. 1994. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell 76117-129. [DOI] [PubMed] [Google Scholar]
- 63.Williams, A. E., L. J. Lawson, V. H. Perry, and H. Fraser. 1994. Characterization of the microglial response in murine scrapie. Neuropathol. Appl. Neurobiol. 2047-55. [DOI] [PubMed] [Google Scholar]
- 64.Zou, W. Q., J. Zheng, D. M. Gray, P. Gambetti, and S. G. Chen. 2004. Antibody to DNA detects scrapie but not normal prion protein. Proc. Natl. Acad. Sci. USA 1011380-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]