Abstract
The NF-κB signaling pathway has previously been shown to be required for efficient influenza A virus replication, although the molecular mechanism is not well understood. In this study, we identified a specific step of the influenza virus life cycle that is influenced by NF-κB signaling by using two known NF-κB inhibitors and a variety of influenza virus-specific assays. The results of time course experiments suggest that the NF-κB inhibitors Bay11-7082 and ammonium pyrrolidinedithiocarbamate inhibited an early postentry step of viral infection, but they did not appear to affect the nucleocytoplasmic trafficking of the viral ribonucleoprotein complex. Instead, we found that the levels of influenza virus genomic RNA (vRNA), but not the corresponding cRNA or mRNA, were specifically reduced by the inhibitors in virus-infected cells, indicating that NF-κB signaling is intimately involved in the vRNA synthesis. Furthermore, we showed that the NF-κB inhibitors specifically diminished influenza virus RNA transcription from the cRNA promoter but not from the vRNA promoter in a reporter assay, a result which is consistent with data obtained from virus-infected cells. The overexpression of the p65 NF-κB molecule could not only eliminate the inhibition but also activate influenza virus RNA transcription from the cRNA promoter. Finally, using p65-specific small interfering RNA, we have shown that p65 knockdown reduced the levels of influenza virus replication and vRNA synthesis. In summary, we have provided evidence showing, for the first time, that the NF-κB host signaling pathway can differentially regulate influenza virus RNA synthesis, which may also offer some new perspectives into understanding the host regulation of RNA synthesis by other RNA viruses.
Influenza A virus causes acute respiratory infections in humans, with severity ranging from morbidity to mortality. Annual flu epidemics generally cause ∼36,000 deaths in the United States alone. The death toll can be much higher during occasional pandemics. For example, the 1918 Spanish flu has been estimated to have claimed up to 100 million lives worldwide.
Influenza A virus is an enveloped RNA virus whose genome consists of eight negative single-strand RNA segments, each encoding one or two viral proteins in negative sense. Upon binding to the host receptors, influenza A virus enters cells via receptor-mediated endocytosis and fuses with the endosomal membrane to release the uncoated virus ribonucleoprotein (vRNP) complex, which then translocates into the nucleus for viral RNA transcription and replication. Influenza virus RNA synthesis consists of three steps: (i) the transcription of virus genomic RNA (vRNA) into mRNA, (ii) the replication of vRNA into cRNA, and (iii) the replication of cRNA into vRNA. The newly synthesized vRNA is encapsidated with the nucleoprotein (NP) and also associated with viral polymerase components to form vRNPs, which are then exported out of the nucleus and incorporated into budding virion particles at the plasma membrane (reviewed in reference 22).
Multiple host signaling pathways have been implicated in influenza virus replication. Pleschka and colleagues have reported that the inhibition of Raf/MEK/extracellular signal-regulated kinase signaling by the MEK inhibitor U0126 or by dominant negative mutant forms of extracellular signal-regulated kinase or Raf results in the inhibition of influenza virus production (31). They have further shown that this inhibition is due to the impaired function of the nuclear export protein NEP/NS2, which leads to the nuclear retention of vRNPs (31). The phosphatidylinositol 3-kinase pathway has been shown previously to be activated by the viral NS1 protein upon influenza A virus infection and to play important roles in the influenza A virus life cycle (6, 7, 13). Using a caspase 3 inhibitor and RNA interference reagents, Wurzer and colleagues (41) have demonstrated a crucial role for caspase 3 activation in influenza virus propagation, particularly in the nuclear export of the vRNPs.
A major host signaling pathway implicated in influenza virus replication is the NF-κB pathway. Infection with influenza A viruses has been shown to activate the NF-κB pathway (11, 35). This activation may be caused by the overexpression of viral proteins such as hemagglutinin (HA), NP, and M1 during virus infection (8, 29). Using stable cell lines expressing inhibitors (a dominant negative mutant form of IκB kinase 2 [IKK2] and a nondegradable phosphorylation site mutant form of IκBα) and an activator (an active mutant form of IKK2) of NF-κB signaling, Wurzer and colleagues (40) found that NF-κB activity promotes efficient influenza virus production, an observation made with different cell lines and different virus subtypes. This effect, according to the authors, is due to NF-κB-dependent viral induction of the proapoptotic factors tumor necrosis factor-related apoptosis-inducing ligand and FasL, which can enhance virus propagation in autocrine and paracrine fashions. However, this study did not address which step(s) of influenza virus replication involves the NF-κB pathway and how tumor necrosis factor-related apoptosis-inducing ligand and FasL enhance influenza virus replication at the molecular level. In a separate study, Nimmerjahn et al. (28) have shown that an active NF-κB signaling pathway is a general prerequisite for a productive influenza virus infection. They noticed that Epstein-Barr virus-negative Burkitt's lymphoma cell lines, which have low levels of NF-κB activity, are resistant to influenza virus infection but become susceptible upon the activation of NF-κB signaling. They also reported that the inhibition of NF-κB by the chemical inhibitor Bay11-7082 or Bay11-7085 severely impairs influenza virus infection in a variety of cell lines. Again, this study did not identity the major step(s) of the influenza virus life cycle that is affected by an NF-κB inhibitor, except to note that receptor expression and virus attachment to cells are not affected.
The NF-κB signaling pathway plays indispensable roles in mediating inflammation, immune responses to pathogen infection, proliferation, apoptosis, and other cellular activities (reviewed in references 1 and 16). The mammalian Rel/NF-κB family of transcription factors consists of five members, p65 (RelA), RelB, c-Rel, p50/p105 (NF-κB1), and p52/p100 (NF-κB2). In the canonical pathway, the IKK complex is activated and mediates the phosphorylation and degradation of IκB molecules and the release of p50/RelA and p50/c-Rel dimers (reviewed in reference 20). In the noncanonical, or alternative, pathway, NF-κB-inducing kinase is activated (43) and in association with IκBα binds to the C terminus of p100, leading to p100 processing into p52 and the preferential release of p52/RelB dimers (4, 42). In both pathways, the freed NF-κB dimers then translocate to the nucleus, where they bind to specific κB sequences in the promoter or enhancer regions of multiple target genes to induce the expression of these genes, including those encoding proinflammatory cytokines, adhesion molecules, interferon (IFN), and proapoptotic molecules.
Despite compelling evidence suggesting that an active NF-κB pathway is required for influenza virus replication, its exact roles in influenza virus replication and the molecular mechanism involved remain unclear. In this study, we employed multiple methods to identify the specific step(s) of the influenza virus life cycle that is affected by NF-κB chemical inhibitors. We have further shown that NF-κB inhibitors specifically decrease (i) the level of vRNA in virus-infected cells and (ii) the level of RNA transcription from the cRNA promoter in a reporter assay. In addition, we have provided evidence to suggest that the NF-κB molecule p65 appears to be responsible for the differential regulation of vRNA synthesis.
MATERIALS AND METHODS
Cells and viruses.
293T cells (human kidney epithelial cells) and A549 cells (human lung epithelial cells) were grown in Dulbecco's modified Eagle's medium (with a low level of glucose) supplemented with 10% heat-inactivated fetal bovine serum. Madin-Darby canine kidney (MDCK) cells were maintained in Eagle's minimal essential medium supplemented with 5% fetal bovine serum. After infection with influenza A virus, MDCK cells were grown in L-15 medium containing 15 mM HEPES, pH 7.5, nonessential amino acids, 0.75 g of NaHCO3 per liter, and 0.125% (wt/vol) bovine serum albumin. Influenza A viruses WSN/H1N1 and PR8/H1N1 were grown in MDCK cells, and virus titers were determined by plaque assays of MDCK cells. The WSN-LUC virus was generated essentially as described before for the WSN-GFP virus (26). In brief, 293T cells were transfected with the eight plasmids that are required to recover infectious influenza A virus WSN (19), along with a plasmid carrying viral RNA in which most of the influenza virus PB2 coding sequence was replaced with a firefly luciferase (LUC) gene (pHH21-PB2LUC). Viruses in the supernatant of the transfected cells were harvested at 48 h posttransfection.
Plasmids and reagents.
The 17-plasmid and the 8-plasmid influenza A virus reverse genetic systems were obtained from Y. Kawaoka (University of Wisconsin) and G. Hobom (Justus Liebig University, Germany), respectively. The plasmids expressing NF-κB molecules p50, p65, and c-Rel were obtained from W. Greene (University of California, San Francisco). The generation of the LUC-encoding reporter constructs vNA-LUC and cNA-LUC was described previously (34); each construct contained the LUC gene flanked by the 5′ and 3′ untranslated regions (UTRs) of the genomic (vNA-LUC) or antigenomic (cNA-LUC) NA segment. The pHH21-PB2LUC plasmid was constructed by replacing the green fluorescent protein gene with the LUC gene in the PB2 0-G-100 construct that was described previously (26). Bay11-7082, ammonium pyrrolidinedithiocarbamate (PDTC), and ribavirin were purchased from Sigma. U0126 was purchased from Promega. The control and p65-specific small interfering RNA (siRNA) reagents were purchased from Dharmacon. Anti-p65 mouse antibody and the NF-κB-specific oligonucleotides used in the gel shift assay were purchased from Santa Cruz.
NF-κB gel shift assay.
A549 cells were infected with influenza A virus WSN at a multiplicity of infection (MOI) of 5. At different time points postinfection, nuclear extracts were prepared using a nuclear extract kit (Active Motif, Carlsbad, CA) and total proteins were quantified by the Bradford assay with a kit from Bio-Rad. The NF-κB-specific oligonucleotides (Santa Cruz) were end labeled with [γ-32P]ATP by using T4 polynucleotide kinase and purified through a G-25 microspin column (GE Healthcare). The gel shift assay was performed as described previously (25). In brief, radiolabeled NF-κB oligonucleotides were incubated for 30 min at room temperature with 6-μg aliquots of nuclear extracts obtained at different time points post-viral infection (see Fig. 1). The mixtures were separated on a 4% nondenaturing polyacrylamide gel, which was dried and exposed to a piece of film.
FIG. 1.

Influenza A virus infection strongly activates NF-κB signaling in human lung epithelial cells early in the infection and throughout the infection. A549 cells were infected with influenza A virus (WSN) at an MOI of 5. At different time points postinfection, nuclear extracts were prepared and subjected to gel shift assays with NF-κB-specific probes. c, oligonucleotide only; m, mock infection; 15′, 15 min.
vRNP nucleocytoplasmic trafficking.
To monitor the nucleocytoplasmic trafficking of vRNP in infected cells, we seeded A549 cells onto coverslips and infected them with WSN virus at an MOI of 5 for 1 h at 37°C. After the infected cells were washed with phosphate-buffered saline (PBS) three times, the culture medium was replaced with medium containing chemicals or controls. At different time points postinfection, cells were fixed in 4% paraformaldehyde for 15 min and blocked by 1% bovine serum albumin for 30 min at room temperature. After being washed with PBS, cells were stained with mouse anti-influenza A virus NP antibody (Serotec) for 30 min in the presence of 0.2% saponin. Cells were washed three times with PBS and incubated with a secondary fluorescein isothiocyanate-conjugated anti-mouse antibody (Santa Cruz) in the presence of 0.2% saponin for 30 min. After being washed again with PBS, the cells were mounted with medium containing 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories) and observed by fluorescence microscopy.
Quantitative real-time RT-PCR.
As described previously (26), total RNA was extracted from cells by using RNAbee reagent according to the instructions of the manufacturer (TEL-TEST, Inc.). The total RNA was cleared of possible plasmid DNA contamination by incubation for 30 min at 37°C with DNase I, which was inactivated by incubation at 85°C for 15 min. Reverse transcription (RT) was conducted using strand- and sense-specific oligonucleotides for vRNA (5′ AGCGAAAGCAGG 3′ and 5′ AGCAAAAGCAGG 3′), cRNA (5′ AGTAGAAACAAGG 3′), and mRNA [oligo(dT)]. A glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primer (5′ GAAGATGGTGATGGGATTTC 3′) was also included in the RT reaction mixture for vRNA or cRNA analysis. The quantitative real-time PCR was carried out with a 20-μl reaction mixture containing primers specific for each of the eight RNA segments (oligonucleotide sequences will be provided upon request) or for GAPDH RNA (5′ GAAGGTGAAGGTCGGAGTC 3′ and 5′ GAAGATGGTGATGGGATTTC 3′) by using Sybr green DNA dye (Invitrogen) in the reaction mixture. The PCR conditions were 50°C for 2 min, 95°C for 2 min, and 45 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The viral RNA levels, expressed as threshold cycle (CT) values, were normalized by the GAPDH RNA level.
Reporter-based influenza virus RNA transcription assay.
Influenza virus RNA replication and transcription were analyzed by a LUC assay using the five-plasmid system. 293T cells were transfected with 0.1 μg of the LUC reporter construct (vNA-LUC or cNA-LUC) and 0.25 μg of each of the protein expression vectors encoding PB2, PB1, PA, and NP. A β-galactosidase (β-gal) expression plasmid was included as an internal control to normalize transfection efficiency. Chemical or control treatments were applied at 8 h posttransfection. At 24 h posttransfection, cells were harvested for the LUC assay, and the LUC activity was normalized by β-gal activity.
siRNA transfection.
A549 cells in six-well plates were transfected with 50 nM p65-specific and control siRNAs by using the Lipofectamine 2000 reagent (Invitrogen) twice on two consecutive days. On the fourth day, cells were infected with influenza A virus (WSN). Viral RNA synthesis and virus titers were determined as described above. The expression level of p65 was evaluated by Western blot analysis with an anti-p65 antibody.
RESULTS
Influenza A virus infection activates NF-κB signaling.
Garoufalis et al. have shown previously that infection with influenza A viruses activates the NF-κB pathway in macrophages, even at 1 h postinfection (hpi) (11). To determine whether and when the NF-κB pathway is activated in the A549 human lung epithelial cell line by influenza A virus infection, we prepared nuclear extracts from WSN-infected A549 cells at different time points postinfection and performed a gel shift assay using NF-κB-specific DNA oligonucleotides as probes. As shown in Fig. 1, a basal level of an active p65/p50 complex was present in the mock-infected A549 cells (Fig. 1, lane 2), forming a specifically shifted band, as seen in the sample at 15 min post-viral infection (Fig. 1, lane 3). This level increased significantly at 30 min after infection and remained high during the course of infection (Fig. 1, lanes 4 to 9). These data clearly show that influenza A virus infection strongly activates NF-κB signaling in human lung epithelial cells as early as 30 min postinfection and throughout the infection, which correlates well with published data on macrophages infected with influenza A virus (11).
NF-κB inhibitors decrease influenza virus production and viral gene expression.
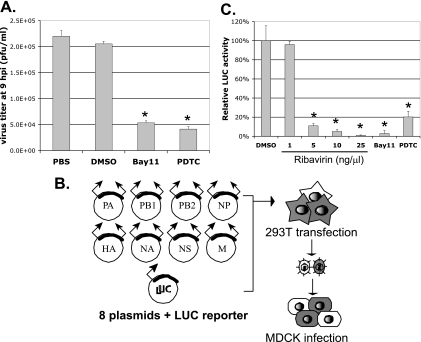
We used two well-known chemical inhibitors of NF-κB signaling, PDTC and Bay11-7082, throughout this study. PDTC inhibits the IκB-ubiquitin ligase activity to prevent the degradation of IκB (15). Bay11-7082 inhibits IKK activity to prevent the phosphorylation and subsequent degradation of IκB (30). Both chemicals therefore block NF-κB activation by stabilizing IκB in order to sequester NF-κB molecules in the cytoplasm of cells. We have examined the potential cytotoxicities of Bay11-7082 and PDTC at different concentrations by the established MTT assay (data not shown). No cytotoxicity of Bay11-7082 at 10 μM and PDTC at 100 μM was observed. Therefore, we decided to use 10 μM Bay11-7082 and 50 to 100 μM PDTC throughout this work. A549 human lung epithelial cells were infected with influenza A virus WSN/33 at an MOI of 5 for 1 h, after which Bay11-7082, PDTC, or a vehicle control consisting of PBS or dimethyl sulfoxide (DMSO) was added. Virus titers in the supernatants at 9 hpi were determined by plaque assays of MDCK cells. As shown in Fig. 2A, Bay11-7082 and PDTC effectively inhibited virus production by ∼75% compared to that in samples treated with vehicle controls. Similar results were obtained using the epithelial MDCK cells or a different influenza A virus strain, PR8 (data not shown). Our results corroborate the findings in previously published works (28, 40) in showing that blocking NF-κB activation decreases influenza virus replication, consistent with the idea that active NF-κB signaling is required for efficient influenza virus replication.
FIG. 2.
NF-κB inhibitors decrease influenza A virus production and virus gene expression in virus-infected cells. (A) NF-κB inhibitors decrease influenza A virus production. A549 cells were infected with influenza A virus WSN at an MOI of 5 for 1 h, after which medium containing either a vehicle control (PBS or DMSO) or an NF-κB inhibitor (10 μM Bay11 or 50 μM PDTC) was added to the infected cells. The virus titers in the supernatants at 9 hpi were determined by plaque assays in three independent experiments. (B) Schematic illustration of the generation of the WSN-LUC virus. As detailed in Materials and Methods, 293T cells were transfected with eight plasmids required to generate infectious influenza A virus WSN, together with a plasmid carrying a LUC-encoding viral reporter PB2-LUC RNA segment. Viruses were harvested from the supernatants at 48 h posttransfection. (C) Ribavirin and NF-κB inhibitors inhibit LUC expression in the WSN-LUC-infected cells. WSN-LUC viruses were used to infect fresh MDCK cells at an MOI of 1 for 1 h. DMSO, different concentrations of ribavirin, or an NF-κB inhibitor (10 μM Bay11 or 50 μM PDTC) was applied at 1 hpi, and a LUC assay was conducted at 8 hpi. * represents statistical significance (P < 0.05).
Because previous studies did not provide a detailed mechanism for how NF-κB inhibitors affect influenza virus replication, we wished to determine whether these inhibitors can directly block influenza virus gene expression. To conveniently and quantitatively monitor viral gene expression, we employed a recombinant influenza virus (WSN-LUC) that expresses the LUC reporter gene during virus infection. This virus was generated by transfecting 293T cells with a plasmid that contains a LUC-encoding influenza virus RNA segment that retains sufficient cis-acting elements for viral RNA transcription/replication and packaging (26), together with the eight plasmids of the influenza virus reverse genetics system (19) (Fig. 2B). Since both full-length vRNA and its LUC-encoding counterpart segment were present in the cells, the released recombinant viruses were a mixture of wild-type influenza A virus virions and viral particles that packaged LUC reporter vRNA. Upon the infection of fresh target cells, the wild-type influenza A viruses served as helper viruses for the efficient replication and expression of the LUC reporter vRNA. We used these recombinant WSN-LUC virus mixtures to track viral infection by monitoring the LUC expression levels. MDCK cells were infected with these recombinant WSN-LUC viruses and treated with the vehicle control DMSO, ribavirin, or NF-κB inhibitors (PDTC and Bay11-7082), and LUC activities were measured at 8 hpi. Ribavirin, a known inhibitor of influenza virus RNA synthesis (36), considerably inhibited LUC gene expression in a dose-dependent manner (Fig. 2C). NF-κB inhibitors PDTC and Bay11-7082 also significantly decreased LUC gene expression in the WSN-LUC-infected cells (Fig. 2C). These data suggest that blocking NF-κB activation can effectively inhibit virus-specific gene expression in influenza virus-infected cells.
NF-κB inhibitors block early stages of the influenza virus life cycle.
The results shown in Fig. 2C also indicate that NF-κB inhibitors appear to block an early step of influenza virus infection, prior to viral protein expression. To further determine which step(s) of the influenza virus life cycle is affected by NF-κB inhibitors, a vehicle control (PBS or DMSO) or a chemical inhibitor (PDTC or Bay11-7082) was added to influenza virus-infected A549 cells at different times post-viral infection and the infectious viruses released into the supernatants at 9 hpi were quantified. As shown in Fig. 3, inhibitors applied at a preinfection time point (1 h before infection) were as effective at inhibiting infectious virus production as those applied at 1, 2, or 3 hpi, indicating that the steps of virus entry and uncoating, which are largely completed by 1 hpi, are not affected by the NF-κB inhibitors. This result is also consistent with the findings of an earlier study by Nimmerjahn et al. showing that receptor expression and virus attachment to cells are not affected by NF-κB inhibition (28). On the other hand, when applied at or after 5 hpi, NF-κB inhibitors did not affect virus production, suggesting that they were ineffective at late stages of the viral life cycle. Taken together, these data suggest that NF-κB inhibitors block an early stage(s) of the influenza virus life cycle preceding the viral gene expression that occurs between approximately 3 and 5 hpi.
FIG. 3.
Time course analysis of the effects of NF-κB inhibitors on virus production. A549 cells were infected with influenza A virus WSN at an MOI of 5. Chemical treatment (10 μM Bay11 or 50 μM PDTC) or a vehicle control (PBS or DMSO) was applied at different times postinfection. The virus titers in the supernatants at 9 hpi were determined by plaque assays. −1 h, 1 h before infection; 1, 2, 3, 5, and 7 h, different times postinfection.
NF-κB inhibitors do not affect vRNP nucleocytoplasmic trafficking.
After uncoating, the vRNP complex, which is composed of viral RNA, NP protein, and the three viral polymerase components (PA, PB1, and PB2), needs to be imported into the nuclei of the infected cells for virus RNA replication and transcription and then exported out of the nuclei for packaging into newly formed virions at the cellular plasma membranes. To study the effects of NF-κB inhibitors on the nucleocytoplasmic trafficking of vRNPs, we infected A549 cells with wild-type influenza A virus WSN for 1 h and then treated cells with DMSO, Bay11-7082, PDTC, or the MEK inhibitor U0126. Influenza virus NP protein was detected by indirect immunofluorescence at different time points postinfection in order to study the intracellular localization of vRNP in an infected cell. As shown in Fig. 4, in vehicle control DMSO-treated samples, the vRNP complexes were localized mostly in the cytoplasm at 1.5 hpi but were present in the nucleus at 3 hpi. By 4 hpi, vRNPs were localized predominantly in the nucleus, and by 6 hpi, most of the presumably newly generated vRNPs were exported out of the nucleus. As a control, we used the MEK inhibitor U0126, which has been shown by Pleschka et al. to cause vRNP nuclear retention in influenza virus-infected cells (31). Consistent with this report, we showed that U0126 prevented the export of vRNPs, even at the late stage of infection (at 9 hpi), whereas their nuclear import was not affected (Fig. 4, compare images from the control and the U0126-treated samples at 2, 3, and 4 hpi). In contrast, neither PDTC nor Bay11-7082 showed any effect on the vRNP trafficking (Fig. 4). These data strongly suggest that NF-κB inhibitors do not affect the nuclear import and export of vRNPs in influenza virus-infected cells.
FIG. 4.
NF-κB inhibitors do not affect the nucleocytoplasmic trafficking of vRNPs. A549 cells were infected with influenza A virus WSN at an MOI of 5 in the presence of either a vehicle control (PBS or DMSO) or a chemical compound (10 μM U0126, 10 μM Bay11, or 50 μM PDTC). At different times postinfection, cells were stained with anti-NP antibody and observed via microscopy.
NF-κB inhibitors specifically decrease the vRNA level during influenza virus infection.
We then asked whether NF-κB inhibitors affect viral RNA synthesis. To address this question, we infected A549 cells with the wild-type WSN virus at an MOI of 5 for 1 h, by which time viral entry and uncoating are believed to have occurred. An NF-κB inhibitor (PDTC) or vehicle control (PBS) was then added to the infected cultures. At 5 hpi, when viral RNA synthesis has already been completed, total RNA from the infected cells was prepared and the levels of individual segments of influenza virus-specific vRNA, cRNA, and mRNA were determined by quantitative real-time RT-PCR. The RNA amounts are expressed as CT values, so that a lower value indicates a larger RNA quantity. Table 1 shows the levels of each of the eight viral segments of vRNA, cRNA, and mRNA in PDTC- or PBS-treated virus-infected cells. It also shows the respective differences in the CT values and the results of the statistical analysis (P values) for each of the RNA species. The levels of each segment of the vRNA in PDTC-treated samples were significantly lower than those in PBS-treated ones (the highest P values were <0.01); the differences in the CT values were 1.5 to 2 for most of the segments and 1.1 for the NS segment. In contrast, the differences in the CT values for cRNA and mRNA were 0.1 to 0.75 in most cases, except for the PA segment of cRNA (0.9). Moreover, most of the CT differences in cRNA and mRNA levels were not statistically significant. Only the CT differences in PB2 cRNA, PB1 cRNA, PA cRNA, and HA mRNA were significant at a 5% level of confidence (P < 0.05). Collectively, our data suggest that NF-κB inhibitors specifically decrease the level of vRNA, but not necessarily those of cRNA and mRNA, in virus-infected cells.
TABLE 1.
Levels of all eight segments of vRNA, cRNA, and mRNA in virus-infected cells treated with either PBS or PDTC
| RNA species | RNA segment | Levela after treatment with:
|
CT difference | P value | |
|---|---|---|---|---|---|
| PBS | PDTC | ||||
| vRNA | PB2 | 14.52 ± 0.36 | 16.58 ± 0.17 | 2.06 | <0.005 |
| PB1 | 13.83 ± 0.08 | 15.7 ± 0.12 | 1.87 | <0.001 | |
| PA | 14.67 ± 0.33 | 16.41 ± 0.09 | 1.74 | <0.01 | |
| HA | 12.64 ± 0.23 | 14.14 ± 0.3 | 1.50 | <0.005 | |
| NP | 15.13 ± 0.31 | 16.83 ± 0.02 | 1.70 | <0.01 | |
| NA | 16.02 ± 0.2 | 18.01 ± 0.19 | 1.99 | <0.001 | |
| M | 15.9 ± 0.11 | 17.7 ± 0.04 | 1.80 | <0.001 | |
| NS | 17.04 ± 0.21 | 18.18 ± 0.14 | 1.14 | <0.005 | |
| cRNA | PB2 | 19.27 ± 0.1 | 20.02 ± 0.27 | 0.75 | <0.05 |
| PB1 | 15.6 ± 0.22 | 16.18 ± 0.22 | 0.58 | <0.05 | |
| PA | 17.65 ± 0.29 | 18.56 ± 0.26 | 0.91 | <0.05 | |
| HA | 15.63 ± 0.44 | 16.36 ± 0.83 | 0.73 | >0.05 | |
| NP | 15.49 ± 0.37 | 15.58 ± 0.15 | 0.09 | >0.05 | |
| NA | 19.92 ± 0.26 | 20.6 ± 0.59 | 0.68 | >0.05 | |
| M | 20.47 ± 0.44 | 20.74 ± 0.11 | 0.27 | >0.05 | |
| NS | 20.49 ± 0.41 | 20.84 ± 0.37 | 0.35 | >0.05 | |
| mRNA | PB2 | 14.05 ± 0.16 | 14.3 ± 0.3 | 0.25 | >0.05 |
| PB1 | 14.07 ± 0.52 | 14.69 ± 0.12 | 0.62 | >0.05 | |
| PA | 16.07 ± 0.42 | 16.22 ± 0.14 | 0.15 | >0.05 | |
| HA | 16.21 ± 0.13 | 16.88 ± 0.24 | 0.67 | <0.05 | |
| NP | 15.66 ± 0.32 | 16.22 ± 0.32 | 0.56 | >0.05 | |
| NA | 13.44 ± 0.08 | 13.68 ± 0.45 | 0.24 | >0.05 | |
| M | 13.09 ± 0.42 | 13.72 ± 0.21 | 0.63 | >0.05 | |
| NS | 11.67 ± 0.4 | 12.06 ± 0.16 | 0.39 | >0.05 | |
The RNA levels were normalized to GAPDH RNA levels and are shown as mean CT values ± standard deviations.
NF-κB inhibitors differentially decrease influenza virus RNA transcription from the cRNA promoter.
vRNA is synthesized from the cRNA promoter, whereas both cRNA and mRNA are transcribed from the vRNA promoter. We wondered whether the reduced level of vRNA in virus-infected cells (Table 1) was due to the inhibition of RNA transcription by NF-κB inhibitors. We therefore analyzed the effects of NF-κB inhibitors on RNA transcription from the different promoters by using a LUC-based RNA transcription assay. Two reporter constructs, each carrying the LUC reporter gene in negative sense and either the NA cRNA UTR (cRNA-LUC) or the NA vRNA UTR (vRNA-LUC) (34), were used. 293T cells were transfected with each reporter construct, along with four protein expression plasmids (i.e., the PB2, PB1, PA, and NP expression plasmids) required for influenza virus RNA synthesis. At 8 h posttransfection, a chemical (PDTC or ribavirin) or a vehicle control (PBS) was applied, and the LUC assay was conducted 16 h later. The LUC activity of each chemically treated sample was compared to that of PBS-treated samples transfected with either the cRNA-LUC or the vRNA-LUC construct. As shown in Fig. 5, PDTC decreased LUC expression from the cRNA promoter (cRNA-LUC) to ∼20% of that in the PBS-treated samples, whereas it seemed to have a much less inhibitory effect on the vRNA promoter (vRNA-LUC), which still retained ∼65% of the reporter activity. Furthermore, the statistical analysis revealed that the inhibitory effect on the cRNA promoter was significant (P < 0.05) but that the P value for the inhibition of the vRNA promoter was around the borderline of 0.05. In contrast, ribavirin, a known inhibitor of influenza virus RNA synthesis (36), significantly decreased LUC expression from the vRNA and cRNA promoters to similar extents (to ∼10% of that in PBS-treated samples). These results were not restricted to NA segment-based reporter constructs, as we have obtained similar results with different reporter constructs that contained the UTRs from different RNA segments (data not shown). Excluding the possibility that these inhibitors may affect plasmid transfection and general RNA/protein expression efficiency in the reporter assay, we have observed a differential reduction of RNA transcription from the cRNA promoter by NF-κB inhibitors, which correlates well with the viral RNA levels in virus-infected cells (Table 1).
FIG. 5.
NF-κB inhibitors differentially decrease influenza virus RNA transcription from the cRNA promoter compared to that from the vRNA promoter. As described in Materials and Methods, 293T cells were transfected with the five plasmids to recreate the LUC-based influenza virus RNA transcription assay, along with a reporter construct based on either the cRNA (cRNA-LUC) or the vRNA (vRNA-LUC) promoter. Treatment with a control (PBS) or chemical (50 μM PDTC or 10 ng of ribavirin/μl) was applied at 8 h posttransfection. LUC activity was determined at 24 h posttransfection, normalized by the β-gal level, and compared to the activity in the cells subjected to the control PBS treatment (set as 100%). * represents statistical significance (P < 0.05).
Potential function of p65 in influenza virus RNA synthesis.
Our results obtained from studying the chemical inhibition of the NF-κB signaling pathway as described above indicated that active NF-κB signaling might participate in regulating vRNA synthesis from the cRNA promoter. We sought to directly examine the effect of individual NF-κB molecules on vRNA synthesis. First, we asked whether the transient expression of each of the NF-κB molecules could abolish the inhibitory effect of the NF-κB inhibitors on vRNA synthesis. To address this issue, we transfected 293T cells with the reporter plasmid cRNA-LUC and the four influenza virus protein expression plasmids encoding PB2, PB1, PA, and NP, along with either an empty vector control or an individual plasmid expressing p50, p65, or c-Rel (kindly provided by Warner Greene at the University of California, San Francisco). PBS or PDTC was added at 8 h posttransfection. The LUC assay was conducted at 24 h posttransfection. Similar to the results shown in Fig. 5, PDTC decreased LUC expression from the influenza virus cRNA promoter in the sample with the empty vector control (Fig. 6A). This PDTC-induced inhibitory effect could be abolished by the transient expression of p65 but not of the p50 or c-Rel factor (Fig. 6A), indicating that p65 plays an important role in regulating vRNA synthesis from the cRNA promoter. We next determined whether the overexpression of p65 alone could increase vRNA synthesis from the cRNA promoter in the absence of any chemical treatment. To examine this issue, we transfected 293T cells with a set of plasmids similar to that described above in order to recreate the LUC-based vRNA transcription from the cRNA promoter in the presence of either an empty vector or increasing amounts of a p50, p65, or c-Rel expression plasmid. As shown in Fig. 6B, while p50 had no effect on LUC expression, p65 significantly increased the reporter gene expression from the influenza virus cRNA promoter in a dose-dependent manner (Fig. 6B). Interestingly, c-Rel appeared to exhibit a dose-dependent inhibitory effect on vRNA transcription from the cRNA promoter (Fig. 6B).
FIG. 6.
Effect of individual NF-κB molecules on influenza virus RNA transcription. (A) The overexpression of p65 abolishes the inhibition of influenza virus RNA transcription from the cRNA promoter by the NF-κB inhibitors. 293T cells were transfected with five plasmids to recreate influenza virus RNA transcription from the cRNA-LUC reporter, together with increasing amounts of either an empty vector or a vector expressing p50, p65, or c-Rel. Control treatment (PBS, indicated by −) or chemical treatment (50 μM PDTC, indicated by +) was applied at 8 h posttranfection. LUC activity was measured at 24 h posttransfection and normalized by the β-gal level. (B) The overexpression of p65 increases influenza virus RNA transcription from the cRNA promoter. 293T cells were transfected with the five plasmids to recreate influenza virus RNA transcription from the cRNA promoter, together with increasing amounts (indicated by triangles) of either an empty vector or a vector expressing p50, p65, or c-Rel. LUC activity was measured at 24 h posttransfection and normalized by the β-gal enzymatic activity. (C) A549 cells were transfected with p65 or control siRNA as described in Materials and Methods. Cell lysates were analyzed by Western blotting with antibodies against p65 and β-actin. (D) A549 cells were transfected with p65 or control siRNA and then infected with influenza A virus (WSN). The virus titer was determined at 9 hpi. (E) A549 cells were transfected with p65 or control siRNA and then infected with influenza A virus. Viral RNA was prepared at 5 hpi and quantified by real-time RT-PCR as described in Materials and Methods. The differences in CT values [ΔC(t)] between control and p65 siRNA-treated samples and the results of the statistical analysis of significance (P values) are shown. * represents statistical significance (P < 0.05).
Since most of the studies described above utilized chemical inhibitors of NF-κB signaling, their specificity in determining the role of NF-κB signaling and, more specifically, of the p65 molecule in influenza A virus infection can be a subject for debate. Instead, we chose the approach of knocking down the level of p65 expression by transfection with siRNA and used the transfected cells to analyze the effect of a reduced p65 level on influenza A virus replication and virus RNA synthesis. As shown in Fig. 6C, p65-specific siRNA effectively reduced the endogenous level of p65 in A549 cells by ∼80 to 90% compared to the level in cells transfected with control siRNA. Influenza A virus replication in the presence of p65 siRNA decreased by 50% compared to that in cells transfected with control siRNA (Fig. 6D). We have also quantified the levels of most segments of vRNA and cRNA (except for the PB1 and PA segments). With the exception of the NP segment, the levels of most vRNA segments were significantly reduced by p65 knockdown (Fig. 6E), although the differences in CT values were smaller than those for chemical inhibitor-treated samples (Table 1). In contrast, none of the cRNA levels were significantly affected (Fig. 6E). In general, the inhibitory effect of siRNA-mediated p65 knockdown was smaller than those of chemical inhibitors. This result may be due to the incomplete knockdown of active p65 molecules by the siRNA and/or the potential contributions of other unknown cellular factors. Nonetheless, siRNA-mediated p65 knockdown significantly reduced influenza A virus replication and the synthesis of most vRNA segments, indicating that NF-κB signaling, specifically that of the NF-κB molecule p65, may play an important role in regulating influenza virus RNA synthesis.
DISCUSSION
The NF-κB signaling pathway plays a dual role in influenza virus infection. On one hand, the NF-κB pathway is important in inducing host innate immune responses against viral infection by activating IFN expression (17). Virus infection generally can trigger multiple signaling cascades through the Toll-like-receptor (TLR)-dependent (TLR3, TLR7, and TLR9) and TLR-independent (RIG-I and Mda-5) pathways that lead to the downstream activation of the NF-κB pathway, ATF2-cJun, or the IFN regulatory factors (IRF-3 and IRF-7), which coordinate to induce the transcription of the IFN-β factor (27). Because of the critical role of the NF-κB pathway in mediating antiviral responses, many viruses evolve to target specific aspects of this pathway and the innate immune responses (reviewed in reference 18). For example, vaccinia virus produces the TIR domain-containing proteins A46R and A52R in order to suppress TLR- or interleukin-1 receptor-induced NF-κB activation (2, 37). On the other hand, some viruses are known to incorporate the NF-κB pathway into their own life cycles and pathogenesis (reviewed in reference 18). Perhaps the most well known example is that of the human immunodeficiency virus type 1 long terminal repeat containing multiple NF-κB binding sites, through which the activated host NF-κB signaling can promote long terminal repeat-driven viral transcription. Both Kaposi's sarcoma-associated herpesvirus (or human herpesvirus 8) and Epstein-Barr virus, two members of the gammaherpesvirus family, activate the NF-κB signaling during latent infections, which are associated with virus-induced transformation and tumorigenesis (reviewed in reference 3). Human T-cell leukemia virus type 1 Tax is a well-studied oncoprotein that targets multiple components of the NF-κB signaling pathway in order to enhance its oncogenic activity (reviewed in reference 38). NF-κB signaling has been shown to be strongly activated during influenza virus infection (this work and references 11 and 35). We and other groups have demonstrated that active NF-κB signaling is required for efficient influenza virus replication (this work and references 28 and 40). To combat innate immune responses induced by activated NF-κB signaling during virus infection, influenza virus encodes a multifunctional NS1 protein that serves as an antagonist against cellular IFN responses (10). Thus, influenza virus is able to counteract the antiviral functions of the NF-κB pathway while at the same time utilizing the active NF-κB signaling for its own replication purpose.
Our work suggests that NF-κB signaling contributes to efficient influenza virus replication via the preferential regulation of the synthesis of vRNA but not of the complementary positive-strand cRNA or of the mRNA (Table 1). This is, to the best of our knowledge, the first evidence of a novel function played by a host signaling pathway in regulating viral replication. Influenza virus RNA synthesis consists of three steps: (i) the transcription of vRNA into mRNA, (ii) the replication of vRNA into cRNA, and (iii) the replication of cRNA into vRNA (reviewed in reference 22). The transcription of influenza virus mRNA differs from the replication of vRNA or cRNA in both its initiation and termination steps. Influenza virus mRNA transcription is initiated by short capped RNAs that are stolen from host pre-mRNAs (12, 23, 24) and is terminated by a poly(A) signal located near the 5′ end of the vRNA template (22, 33). Both the vRNA and cRNA of influenza virus are full-length transcripts which are synthesized by primer-independent de novo initiation and NP-mediated antitermination mechanisms. The mechanisms by which the synthesis of the three different viral RNA species is regulated in virus-infected cells are not well understood. It has been hypothesized previously that the form of the polymerase required for replication (i.e., replicase) is different from that required for transcription (i.e., transcriptase) (21). Some evidence to support such a hypothesis exists. For example, the replicase seems to require the NP protein, which is consistent with the facts that vRNA and cRNA are encapsidated along with the NP proteins and that virus mRNAs are not (14, 21, 32). On the other hand, the transcriptase was previously proposed to require PB2 but not PA protein. However, recent mutagenesis studies of the PA protein have demonstrated that PA is also involved in transcription (9, 34). Therefore, definitive experimental evidence for the differences between a replicase and a transcriptase is still lacking. In addition, both vRNA and cRNA should be generated by a replicase, yet their syntheses also differ. A recent study by Deng et al. has shown that vRNA synthesis is initiated internally and that the developing vRNA is then realigned to the terminus of the cRNA promoter but that cRNA synthesis is initiated at the terminus of the vRNA promoter (5). However, the mechanism to regulate the synthesis of vRNA versus that of cRNA is less well characterized. In the present study, we have shown for the first time that NF-κB signaling, an essential host signaling pathway, preferentially enhances the synthesis of vRNA but not cRNA or mRNA of influenza virus. The differential regulation of influenza virus RNA synthesis by NF-κB signaling may play a significant role in controlling the levels of the three viral RNA species in infected cells in order to optimize viral RNA and protein syntheses for the purpose of enhancing virus production.
How might NF-κB signaling specifically regulate vRNA synthesis? vRNA is synthesized from the cRNA promoter, while both cRNA and mRNA are synthesized from the vRNA promoter. We have shown that NF-κB inhibitors strongly affect RNA transcription from the cRNA promoter, in contrast to that from the vRNA promoter (Fig. 5), indicating that NF-κB signaling may specifically target the cRNA promoter. The cRNA promoter differs from the vRNA promoter in several ways (Fig. 7): in the cRNA promoter, (i) the position 3′-position 8′ base pair of the 5′ hairpin loop is C-G, (ii) the nucleotide at position 5′ in the 5′ tetraloop sequence is an adenine, (iii) there is no hinge in the 5′ hairpin loop, (iv) there is a hinge U at the 3′ hairpin loop, (v) the position 2-position 9 base pair of the 3′ hairpin loop is A-U, and (vi) the nucleotide at position 5 in the 3′ tetraloop sequence is a cytosine. These distinct features of the cRNA promoter relative to the vRNA promoter have been shown to be essential to induce endonuclease activity (24), to serve as the nuclear export signals for the selective packaging of the vRNA rather than the cRNA into virion particles (39), and to mediate the internal pppApG initiation of cRNA synthesis (5). It is likely that the distinct sequence and/or structural features of the cRNA promoter may also determine its differential regulation by the NF-κB signaling. This hypothesis still needs to be examined experimentally, however.
FIG. 7.
Diagram of the corkscrew structures of cRNA and vRNA promoters. The differences between the cRNA and vRNA promoters are highlighted by circles and arrows.
What components of the NF-κB signaling pathway are involved in regulating influenza virus RNA synthesis? We have shown that p65 alone could block the inhibition of the influenza virus cRNA promoter by NF-κB inhibitors (Fig. 6A) and that the overexpression of p65 alone increased influenza virus cRNA promoter activity (Fig. 6B). Consistently, p65 reduction by the corresponding specific siRNA molecule reduced influenza virus replication and the levels of most vRNA segments in the infected cells (Fig. 6D and E). Taken together, these data strongly implicate p65 in regulating vRNA synthesis from the cRNA promoter. It is noteworthy that c-Rel was also found to inhibit the influenza virus cRNA promoter in a dose-dependent manner (Fig. 6B). Further studies are needed in order to characterize the molecular mechanisms by which p65 and c-Rel contribute to vRNA synthesis.
In summary, we have provided for the first time a molecular mechanism to account for the effect of the important NF-κB signaling pathway on influenza virus RNA synthesis. Specifically, the p65 molecule of this signaling event may directly participate in regulating efficient vRNA synthesis from the cRNA promoter. Information gained from these studies may offer new cellular targets, some that cannot be altered as easily as the viral genomic sequence, for the development of novel antiviral agents against influenza virus.
Acknowledgments
We thank Y. Kawaoka and G. Hobom for the influenza virus plasmids and Warner Greene for the NF-κB molecule plasmids.
This work was supported in part by funds from the Southeast Regional Center of Excellence for Emerging Infections and Biodefense (SERCEB) to Yuying Liang and Hinh Ly, the pilot component of the U19 grant (RFA-AI-02-042) to Yuying Liang, the Emory Center for AIDS Research seed grant (CFAR P30 AI050409) to Hinh Ly, and grant R01 AI-40317 to T. G. Parslow and Yuying Liang.
Footnotes
Published ahead of print on 13 August 2008.
REFERENCES
- 1.Bonizzi, G., and M. Karin. 2004. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25280-288. [DOI] [PubMed] [Google Scholar]
- 2.Bowie, A., E. Kiss-Toth, J. A. Symons, G. L. Smith, S. K. Dower, and L. A. O'Neill. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 9710162-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkmann, M. M., and T. F. Schulz. 2006. Regulation of intracellular signalling by the terminal membrane proteins of members of the Gammaherpesvirinae. J. Gen. Virol. 871047-1074. [DOI] [PubMed] [Google Scholar]
- 4.Dejardin, E., N. M. Droin, M. Delhase, E. Haas, Y. Cao, C. Makris, Z. W. Li, M. Karin, C. F. Ware, and D. R. Green. 2002. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 17525-535. [DOI] [PubMed] [Google Scholar]
- 5.Deng, T., F. T. Vreede, and G. G. Brownlee. 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 802337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrhardt, C., H. Marjuki, T. Wolff, B. Nurnberg, O. Planz, S. Pleschka, and S. Ludwig. 2006. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 81336-1348. [DOI] [PubMed] [Google Scholar]
- 7.Ehrhardt, C., T. Wolff, S. Pleschka, O. Planz, W. Beermann, J. G. Bode, M. Schmolke, and S. Ludwig. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 813058-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flory, E., M. Kunz, C. Scheller, C. Jassoy, R. Stauber, U. R. Rapp, and S. Ludwig. 2000. Influenza virus-induced NF-κB-dependent gene expression is mediated by overexpression of viral proteins and involves oxidative radicals and activation of IκB kinase. J. Biol. Chem. 2758307-8314. [DOI] [PubMed] [Google Scholar]
- 9.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 768989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252324-330. [DOI] [PubMed] [Google Scholar]
- 11.Garoufalis, E., I. Kwan, R. Lin, A. Mustafa, N. Pepin, A. Roulston, J. Lacoste, and J. Hiscott. 1994. Viral induction of the human beta interferon promoter: modulation of transcription by NF-κB/rel proteins and interferon regulatory factors. J. Virol. 684707-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagen, M., T. D. Chung, J. A. Butcher, and M. Krystal. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 681509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale, B. G., D. Jackson, Y. H. Chen, R. A. Lamb, and R. E. Randall. 2006. Influenza A virus NS1 protein binds p85β and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. USA 10314194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay, A. J., G. Abraham, J. J. Skehel, J. C. Smith, and P. Fellner. 1977. Influenza virus messenger RNAs are incomplete transcripts of the genome RNAs. Nucleic Acids Res. 44197-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayakawa, M., H. Miyashita, I. Sakamoto, M. Kitagawa, H. Tanaka, H. Yasuda, M. Karin, and K. Kikugawa. 2003. Evidence that reactive oxygen species do not mediate NF-κB activation. EMBO J. 223356-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-κB. Genes Dev. 182195-2224. [DOI] [PubMed] [Google Scholar]
- 17.Hayden, M. S., A. P. West, and S. Ghosh. 2006. NF-κB and the immune response. Oncogene 256758-6780. [DOI] [PubMed] [Google Scholar]
- 18.Hiscott, J., T. L. Nguyen, M. Arguello, P. Nakhaei, and S. Paz. 2006. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene 256844-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 976108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18621-663. [DOI] [PubMed] [Google Scholar]
- 21.Krug, R. M., F. V. Alonso-Caplen, I. Julkunen, and M. G. Katze. 1989. Expression and replication of the influenza virus genome, p. 89-152. In R. M. Krug (ed.), The influenza viruses. Plenum, New York, NY.
- 22.Lamb, R. A., and R. M. Krug. 2003. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 23.Leahy, M. B., D. C. Pritlove, L. L. Poon, and G. G. Brownlee. 2001. Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J. Virol. 75134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leahy, M. B., G. Zecchin, and G. G. Brownlee. 2002. Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters. J. Virol. 762019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 161977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, Y., Y. Hong, and T. G. Parslow. 2005. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 7910348-10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63609-620. [DOI] [PubMed] [Google Scholar]
- 28.Nimmerjahn, F., D. Dudziak, U. Dirmeier, G. Hobom, A. Riedel, M. Schlee, L. M. Staudt, A. Rosenwald, U. Behrends, G. W. Bornkamm, and J. Mautner. 2004. Active NF-κB signalling is a prerequisite for influenza virus infection. J. Gen. Virol. 852347-2356. [DOI] [PubMed] [Google Scholar]
- 29.Pahl, H. L., and P. A. Baeuerle. 1995. Expression of influenza virus hemagglutinin activates transcription factor NF-κB. J. Virol. 691480-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierce, J. W., R. Schoenleber, G. Jesmok, J. Best, S. A. Moore, T. Collins, and M. E. Gerritsen. 1997. Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J. Biol. Chem. 27221096-21103. [DOI] [PubMed] [Google Scholar]
- 31.Pleschka, S., T. Wolff, C. Ehrhardt, G. Hobom, O. Planz, U. R. Rapp, and S. Ludwig. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3301-305. [DOI] [PubMed] [Google Scholar]
- 32.Pons, M. W. 1971. Isolation of influenza virus ribonucleoprotein from infected cells. Demonstration of the presence of negative-stranded RNA in viral RNP. Virology 46149-160. [DOI] [PubMed] [Google Scholar]
- 33.Poon, L. L., D. C. Pritlove, J. Sharps, and G. G. Brownlee. 1998. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J. Virol. 728214-8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regan, J. F., Y. Liang, and T. G. Parslow. 2006. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol. 80252-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronni, T., S. Matikainen, T. Sareneva, K. Melen, J. Pirhonen, P. Keskinen, and I. Julkunen. 1997. Regulation of IFN-alpha/beta, MxA, 2′,5′-oligoadenylate synthetase, and HLA gene expression in influenza A-infected human lung epithelial cells. J. Immunol. 1582363-2374. [PubMed] [Google Scholar]
- 36.Scholtissek, C. 1976. Inhibition of influenza RNA synthesis by virazole (ribavirin). Arch. Virol. 50349-352. [DOI] [PubMed] [Google Scholar]
- 37.Stack, J., I. R. Haga, M. Schroder, N. W. Bartlett, G. Maloney, P. C. Reading, K. A. Fitzgerald, G. L. Smith, and A. G. Bowie. 2005. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 2011007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, S. C., and S. Yamaoka. 2005. Activation of NF-κB by HTLV-I and implications for cell transformation. Oncogene 245952-5964. [DOI] [PubMed] [Google Scholar]
- 39.Tchatalbachev, S., R. Flick, and G. Hobom. 2001. The packaging signal of influenza viral RNA molecules. RNA 7979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wurzer, W. J., C. Ehrhardt, S. Pleschka, F. Berberich-Siebelt, T. Wolff, H. Walczak, O. Planz, and S. Ludwig. 2004. NF-κB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J. Biol. Chem. 27930931-30937. [DOI] [PubMed] [Google Scholar]
- 41.Wurzer, W. J., O. Planz, C. Ehrhardt, M. Giner, T. Silberzahn, S. Pleschka, and S. Ludwig. 2003. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 222717-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao, G., A. Fong, and S. C. Sun. 2004. Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J. Biol. Chem. 27930099-30105. [DOI] [PubMed] [Google Scholar]
- 43.Xiao, G., E. W. Harhaj, and S. C. Sun. 2001. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7401-409. [DOI] [PubMed] [Google Scholar]