Abstract
The assembly of herpesvirus capsids is a complex process involving interactions of multiple proteins in the cytoplasm and in the nucleus. Based on comparative genome analyses, varicella-zoster virus (VZV) open reading frame 23 (ORF23) encodes a conserved capsid protein, referred to as VP26 (UL35) in other alphaherpesviruses. Mutagenesis using a VZV bacterial artificial chromosome system showed that ORF23 was dispensable for replication in vitro. However, the absence of ORF23 disrupted capsid assembly in a melanoma cell line. Expression of ORF23 as a red fluorescent protein (RFP) fusion protein appeared to have a dominant negative effect on replication that was rescued by ORF23 expression from a nonnative site in the VZV genome. In contrast to its VP26 homolog, ORF23 has an intrinsic nuclear localization capacity that was mapped to an SRSRVV motif at residues 229 to 234 in the extreme C terminus of ORF23. In addition, coexpression with ORF23 resulted in nuclear import of the major capsid protein, ORF40. VZV ORF33.5 also translocated ORF40, which may provide a redundant mechanism in vitro but appears insufficient to overcome the dominant negative effect of the monomeric RFP-ORF23 (mRFP23) fusion protein. ORF23 was required for VZV infection of human skin xenografts, indicating that ORF33.5 does not compensate for lack of ORF23 in vivo. These observations suggest a model of VZV capsid assembly in which nuclear transport of the major capsid protein and associated proteins requires ORF23 during VZV replication in the human host. If so, ORF23 expression could be a target for a novel antiviral drug against VZV.
Varicella-zoster virus (VZV) is a member of the subfamily Alphaherpesvirinae of Herpesviridae (4). It is a ubiquitous human pathogen and the causative agent of varicella (chicken pox) and herpes zoster (shingles). Like other alphaherpesviruses, the formation of VZV capsids is predicted to require the products of VZV open reading frames (ORFs) 20, 23, 33, 33.5, 40, and 41 (3, 5, 9, 10). Based on sequence similarities, these proteins are related to the herpes simplex virus type 1 (HSV-1) capsid proteins VP19C (UL38), VP26 (UL35), VP24 and VP21 (UL26), pre-VP22a (UL26.5), VP5 (UL19), and VP23 (UL18), respectively (Table 1). Assuming conservation of functions, ORF20, ORF23, ORF40, and ORF41 comprise the outer capsid shell. However, direct information about the VZV capsid proteins is limited.
TABLE 1.
VZV capsid proteins closely related to HSV capsid proteins
| HSV capsid protein | VZV capsid protein |
|---|---|
| VP19C | ORF20 |
| VP26 | ORF23 |
| VP24/VP21 | ORF33 |
| pre-VP22a | ORF33.5 |
| VP5 | ORF40 |
| VP23 | ORF41 |
ORF23 is the homolog of VP26, which is the smallest HSV-1 capsid protein (1, 13). ORF33.5 has characteristics of a scaffolding protein, like preVP22a, and ORF33 is related to the UL26 protease, which cleaves itself, releasing the minor scaffold proteins VP24 and VP21 (18, 20). ORF40 is related to VP5, the major HSV-1 capsid protein. VP5 makes up 150 hexons and 12 pentons in the mature HSV-1 capsid (15, 24). VP26, which is present in nearly equimolar amounts as VP5, decorates the outer surface of the capsid shell on hexons in cell-free capsids, capsids derived from HSV-1-infected cells, and recombinant capsids (25). VP26 is produced late in the infectious cycle and is present in multiple phosphorylated forms (13). VP26 has been expressed as a green fluorescent protein (GFP) or monomeric red fluorescent protein (mRFP) fusion protein to investigate HSV-1 capsid assembly because of its small size (112 amino acids), position on the capsid, and dispensability for viral replication in cultured cells (5, 8). The ORF23 gene product is predicted to be 235 amino acids, whereas VP26 and the other orthologous alphaherpesvirus UL35 proteins are about 115 amino acids long (6). The first 115 residues of ORF23 retain most of the conserved amino acids of UL35 proteins, while the larger C terminus is unique. ORF23 is the homolog of the small capsid protein (SCP) encoded by UL48/49 in human cytomegalovirus (HCMV) (13). HCMV requires this protein for production of infectious virus, and modifying SCP with GFP was not compatible with HCMV replication. Of interest, the amino acid residues at the extreme C terminus of ORF23 are more similar to the HCMV SCP than to any of its alphaherpesvirus homologs.
Analyses of individual proteins in the HSV-1 capsid assembly pathway using transient-expression systems showed that not all capsid proteins have an inherent nuclear localizing capacity (19). Instead, their nuclear accumulation results from interactions between VP19C and preVP22a, which have nuclear localizing signals, and VP5, VP23, and VP26, which do not. In HSV-1, the VP5 major capsid protein binds to the scaffold protein, VP22a, and is transported to the nucleus (16). In the nucleus, VP5-VP22a units assemble to form the basic framework of capsomeres, to which other capsid proteins can attach (16). Triplex proteins link adjacent capsomeres and appear to be formed from two molecules of VP23 and one molecule of VP19C (22). VP19C, which has a nuclear localizing domain, interacts with VP23 to form triplexes that then move into the nucleus. Triplexes can bind to major protein-scaffold complexes only as preformed complexes (22). While it has an inherent nuclear localizing capacity, VP22a does not influence VP23 transport; however, VP19C can relocate both VP5 and VP23 to the nucleus. VP26 only reaches the nucleus if VP5 is present, along with either VP19C or VP22a (19).
The purpose of these experiments was to investigate the functions of VZV ORF23 in viral replication and its interactions with other putative capsid proteins and to assess its role in the pathogenesis of VZV infection of differentiated human skin cells in vivo, using the SCIDhu mouse model (14). We used the two-step red recombination procedure to mutate ORF23 in an infectious VZV bacterial artificial chromosome (BAC) clone, pOKA-BAC (23). Since ORF23, in contrast to HSV-1 VP26, exhibited independent nuclear localization, we examined the influence of ORF23 on the subcellular localization of the VZV capsid proteins encoded by ORF20, ORF40, and ORF41 when expressed in plasmid vectors. Finally, we analyzed the nuclear localization capacity of ORF33.5 and evaluated ORF23 and ORF33.5 interactions with ORF20, ORF40, and ORF41 in order to investigate the sequence of events that might occur during VZV capsid assembly.
MATERIALS AND METHODS
Generation of recombinant viruses using pOKA-BAC.
The parent OKA (pOKA)-BAC system used in these experiments was developed by N. Osterrieder, Cornell University, Ithaca, NY, from pOka cosmids (17, 23). To prepare electrocompetent cells, an initial starter culture of 5 ml LB broth inoculated with a single colony of Escherichia coli EL250 harboring recombinant pOKA-BAC was grown overnight with chloramphenicol (30 μg/ml) at 32°C. LB broth (500 ml) was inoculated with the starter culture (5 ml) and incubated at 32°C to an optical density at 600 nm of 0.5 to 0.6, chilled for 10 to 15 min, transferred to prechilled centrifuge tubes, and centrifuged at 3,000 × g. The supernatant was removed, and cells were resuspended in an equal volume of ice-cold 10% glycerol; washes were repeated twice, and the bacterial pellet was resuspended in a 0.008× portion of the original culture volume (4 ml) in ice-cold 10% glycerol, mixed, and used immediately or stored at −80°C.
(i) Generation of pOKA-BAC23mRFP.
pOKA-BAC was mutagenized with the two-step red recombination method using the plasmids pEP-mRFP1 and pBAD-I-Sce (a kind gift from Neal Copeland, NIAID) (23). To make the first construct, pOKA-BAC23mRFP (Fig. 1A and B) primers were designed to insert the mRFP gene just before the stop codon of ORF23. The stop codon was inserted after the mRFP gene such that ORF23mRFP was expressed as a single cassette. BAC23mRFP-F and BAC23mRFP-R (Table 2) were designed with a 42-bp homologous region matching the 5′ end of ORF23 corresponding to nucleotides (nt) 42,388 to 42,430 in the forward primer and a 50-bp homologous region match for the 3′ end from nt 42,431 to 42,481 for the reverse primer followed by homology at the 3′ end of primers to pEP-mRFP1. All amplifications were performed with Accuprime Taq DNA polymerase Hi Fidelity (Invitrogen, Carlsbad, CA). Amplification yielded a 1.8-kb cassette, ORF23-mRFP-Kanr-mRFP-ORF23. The product was gel purified (Qiagen gel extraction kit) and sequenced. The PCR product (50 ng) was electroporated into competent E. coli EL250 cells harboring pOKA-BAC. The transformants were plated on LB agar containing 30 μg/ml chloramphenicol and 50 μg/ml kanamycin (positive selection marker for mRFP integration). Positive transformants of pOKA-BAC23mRFP-Kanr-mRFP which appeared 48 h later were screened by colony PCR with primers VBF1 and VBR1 (Table 2) to check for integration of the PCR cassette, visualized as a 2.5-kb amplification product. Two pOKA-BAC23mRFP-Kanr-mRFP clones with the correct insertion, based on sequencing, were selected for further processing. To excise the kanamycin cassette, plasmid pBAD-I-Sce with the ampicillin selection marker was electroporated into electrocompetent pOKA-BAC23mRFP-Kanr-mRFP cells. E. coli EL250 cells harboring both constructs were grown for 1 to 2 h in the presence of chloramphenicol (30 μg/ml), ampicillin (100 μg/ml), and 1% arabinose, to induce Sce endonuclease excision of the kanamycin cassette and incubated for 15 min in a 42°C water bath to induce genes for red recombination and then at 32°C for 1 to 4 h. Negative selection for kanamycin was done by plating transformants on LB agar plates containing chloramphenicol (30 μg/ml) and ampicillin (100 μg/ml). Positive colonies were screened by PCR for the loss of the kanamycin insertion and sequenced to document reconstitution of the intact mRFP fusion gene.
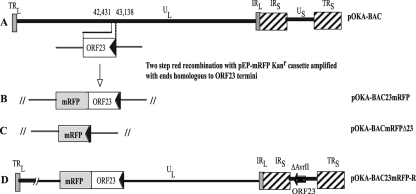
FIG. 1.
Schematic representation of ORF23 mutagenesis in pOKA-BAC. A. Location of ORF23 in the unique long region (UL) segment of the VZV genome; the terminal repeats (TR), unique short region (US), and internal repeats (IR) are indicated. The arrow shows the direction of ORF23 transcription. B. Insertion of mRFP (gray box) just before the ORF23 stop codon. C. Replacement of ORF23 with mRFP. D. ORF23 insertion at the ectopic AvrII site in the US (position 112,851) of pOKA-BAC23mRFP (dark box).
TABLE 2.
Primers used in the study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| BAC23mRFP-F | TCCCACACGGGACAATCTGGAAACGCTTCAAGAAGTCGTAGGGTGATGGCCTCCTCCGAGGACGTCATC |
| BAC23mRFP-R | GCCTATTAAAGAAAAAAAAACAACGATTATTTTCTGTGTATTTTTATTTACAAGGCGCCGGTGGAGTGG |
| 1.4FR2-F | CCCGTAAACGAGCTCACGGCGGCCGCATAAAAATACACAGAAAATAATCGTTGTT |
| 1.4FR2-HincII | ACCGGTCGACGTTATGTCCGAGGGA |
| 1.4FR1-Pci | GCCACGGATGCATACATGTATAAG |
| 1.4FR1-R | TTTTCTGTGTATTTTTATGCGGCCGCCGTGAGCTCGTTTACGGGACAATCGATAA |
| pEP-mRFP1-SacF | ATCCACCGGTCGAGCTCATGGCCT |
| pEP-mRFP1-NotR | TATTTTTATGCGGCCGCTTTACTT |
| pEP-Kan-SacF | AATAAAGAGCTCGGATGACGACGATAAGTAGGG |
| pEP-Kan-NotR | TTAATGCGGCCGCGTTTACGGGACAATCGATAACAGCATACACGTACATCTGCGCACAACCAATTAACCAATTCTGATTAG |
| VerifyΔORF23-F | CCAGGCATGTCGGCGTATGA |
| VerifyΔORF23-R | CGCGTGTCTGGAAACTTGCT |
| ORF23 AvrII-F | GGTGCTTCCTAGGAAATAAATTGCC |
| ORF23 AvrII-R | ATATAAAGTAGCGCCTAGGAAAG |
| VBF1 | CTATCGTACTAAGTCTCGACGAAC |
| VBR1 | ACTGGCCCTGTTAAGCCAGGCAT |
| ORF23-F | CCCGTAAACGAGCTCACGATGACA |
| ORF23-R | CGAATTCTTTTCTGCAGATTTTTATTT |
| ORF23NLS-mut-R | GGTACCGTCGACTGCAGAATTCGCCCTTTTTTTATCTGCAGCCCAGCACCTCCTGAAGCG |
| ORF33.5-F | ACGCAGTTGTGTAGGAACGAAGCTTATATGGCTT |
| ORF33.5-R | TACTTTTATTTTACCGGTCCACACCGCCCCA |
| ORF40-F | GTCGCAGATCTGCTGACAAAATGACAAC |
| ORF40-R | GTATGTGGGCGTGGTAAGCTTATCGCGGAA |
| ORF20-F | AATTTAACCAAGCTTCTATGGGGAGTCAA |
| ORF20-R | GCTATGTTAAATTCGGTACCTTTTACATACAT |
| ORF41-F | CACGCTTCCTAAGCTTACATGGCTATGCCATTT |
| ORF41-R | TTGTGTTTTTATTAACGGTACCTTACACTTGAA |
| ORF23-PGEX2T-F | ACTAAGGATCCATGACACAA |
| ORF23-PGEX2T-R | CTGAATTCTTTTATTTACACCCT |
Portions of sequences shown in bold for BAC23mRFP-F and BAC23mRFP-R indicate homology to 5′ and 3′ ends of ORF23. Italicized portions are homologus to the 5′ and 3′ ends of the RFP cassette in pEP-mRFP. Portions of sequence shown in bold for 1.4FR2-F and 1.4FR1-R indicate the overlap region. Portions of sequence shown in bold for pEP-Kan-SacF and pEP-Kan-NotR indicate introduced restriction enzyme sites. The underlined portion of pEP-Kan-NotR indicates the sequence required for the second red recombination step. Portions of sequence shown in italics for ORF23-F, ORF23-R, ORF33.5-F, ORF33.5-R, ORF40-R, ORF20-F, ORF20-R, ORF41-F, and ORF41-R indicate introduced restriction enzyme sites.
(ii) Generation of pOKA-BACmRFPΔ23.
The red recombination procedure in the first step was modified to achieve higher efficiency in obtaining first-stage recombinants. Briefly, a digested fragment from a construct was used instead of a PCR product for recombination with pOKA- BAC. A 1.4-kb region spanning nt 42,198 to 43,646 of VZV was digested from cosmid pvSpe14 with restriction enzyme AgeI and cloned in pLITMUS28 vector (New England Biolabs). To delete ORF23, two PCR fragments on either side of the ORF23 gene were amplified with primers 1.4FR2-F/1.4FR2-HincII (from nucleotide position 42,198 to 42,430) and 1.4FR1-Pci/1.4FR1-R (from nucleotide position 43,137 to 43,472) (Table 2), such that primers 1.4FR1-R and 1.4FR2-F had an overlap. The two PCR products were quantitated and mixed in equal proportions, and 5 ng of this mixture was used as template for amplification with 1.4FR2-HincII/1.4FR1-Pci. The amplified product resulted in deletion of the ORF23 gene and substitution at that position with Sac and Not restriction sites. This product was cloned into pLITMUS1.4Age at restriction sites HincII/Pci to give pLITMUS1.4AgeΔORF23. To substitute ORF23 with mRFP, primers pEP-mRFP1-SacF/pEP-mRFP1-NotR (Table 2) were used to amplify plasmid pEP-mRFP1 with Sac and Not sites in the forward and reverse primer. The amplified product was cloned in pLITMUS1.4AgeΔORF23 to give the construct pLITMUS1.4Age-mRFPΔORF23, which was selected on LB Amp/Kan plates with ampicillin (100 μg) and kanamycin (30 μg/ml). The kanamycin resistance was introduced by integration of the pep-mRFP1 cassette. This construct was digested with Age, and the digested product was gel purified and used for transfection in the first-stage red recombination with pOKA-BAC. Integration of the mRFP kanamycin cassette in pOKA-BAC was confirmed by PCR and sequencing. The subsequent stages to excise the kanamycin cassette were followed as described previously. The final BAC constructs were electroporated into E. coli Gene Hog cells (Invitrogen, Carlsbad, CA). A negative selection was also done for the pBAD-I-Sce plasmid, to ensure that the Gene Hog cells harbored only the recombinant BAC DNA. BAC DNA was isolated using the Nucleobond BAC maxiprep kit (Clontech, Mountain View, CA).
Transfections were done in MeWo cells using the calcium phosphate mammalian cell transfection kit (Clontech). Cells were maintained in minimal essential medium (Mediatech, Washington, DC) supplemented with 10% fetal bovine serum (Gemini Bio-products, Woodland, CA), nonessential amino acids, and antibiotics (17, 21) and observed for plaques; if no plaques appeared, cells were passaged five to six times to ensure detection of slow-growing viruses. To confirm nucleotide deletions and insertions, DNA was isolated from MeWo cells or skin xenograft tissues with DNAzol reagent (Invitrogen, Carlsbad, CA). PCR was performed with Accuprime Taq DNA polymerase HI Fidelity (Invitrogen, Carlsbad, CA). The resulting PCR products were separated on 0.8% agarose gels and purified for sequencing.
(iii) Generation of pOKA-BAC23mRFP-R virus.
As pOKA-BAC23mRFP transfections did not yield virus, ORF23 was inserted at a nonnative site to create a rescued virus (Fig. 1D). An 851-bp fragment which included ORF23 and the intergenic region between ORF22 (coded on the opposite strand) and ORF24 (nt 42,369 to 43,219) was amplified with primers ORF23AvrII-F and ORF23AvrII-R and cloned into the AvrII restriction site in the pvSpe23ΔAvrII cosmid. Positive transformants were plated on LB agar plates with Kan (50 μg/ml) and Amp (100 μg/ml) and screened with restriction digestion and sequencing to confirm the ORF23 insertion. A cosmid, pvSpe23AvrORF23, that contained ORF23 at the AvrII site and was oriented toward the IRS was identified. To generate the pOKA-BAC23mRFP rescue, pvSpe23AvrORF23 or pvSpe23Avr with no ORF23 insertion (1.5 μg) was digested with AscI in 30 μl total of reaction mix for 3 h. The digestion products were heat inactivated and cotransfected with pOKA-BAC23mRFP DNA (3 μg) using calcium phosphate transfection (12, 21). Primer synthesis and sequencing were done by Elim Biopharmaceuticals, Inc. (Hayward, CA).
(iv) Generation of pOKA-BACΔ23.
A pOKA BAC construct with a deletion of only ORF23 was generated. Two PCR primers were designed, pEP-Kan-SacF, which introduced a SacI site immediately adjacent to the start of the Kanr cassette, and pEP-Kan-Not, which introduced a NotI site adjacent to a 41-nt segment which is repeated upstream of the SacI site (required for the second red recombination step) and is followed by sequences from the Kanr cassette (Table 2). PCRs were carried out using Accuprime Pfx (Invitrogen, Carlsbad, CA) and pEP-Kan as a template. The 1-kb PCR product was gel purified and tailed using Taq polymerase in the presence of dATP. The tailed product was cloned into the TOPO cloning vector pCR4-TOPO (Invitrogen, Carlsbad, CA) per the manufacturer's instructions. Clones were screened by restriction enzyme analysis, and positive clones were sequenced to verify the sequence of the Kanr cassette. The pCR4-TOPO NotKanSac construct and the plasmid pLITMUS1.4AgeΔORF23 were digested with NotI and SacI. The 3,500-nt vector fragment from pLITMUS1.4AgeΔORF23 and the 1,000-nt insert fragment from pCR4-TOPO NotKanSac were isolated and ligated together. Clones were screened by restriction enzyme digest, and positive clones (designated pLitmus 1.4AgeΔORF23Kan) were maxi-prepped using the Invitrogen High Pure plasmid DNA purification kit. pLitmus 1.4AgeΔORF23Kan were digested with AgeI, and the 1,700-nt insert was isolated and purified. The first red recombination was done as follows. The isolated fragment was quantitated spectrophotometrically, and 100 to 200 ng of insert was electroporated into electrocompetent E. coli strain GS1783 containing the wild-type VZV BAC DNA. Cells were placed in SOC medium, rocked at 32°C for 60 min, and then plated on LB-Kan (50 μg/ml)-chloramphenicol (30 μg/ml) plates and placed at 32°C overnight. Colonies were selected and grown overnight in liquid medium containing kanamycin and chloramphenicol, as above, overnight at 32°C. The next day, cultures were lysed by boiling at 100°C for 3 min and subjected to PCR using primers VerifyΔORF23-F and VerifyΔORF23-R (Table 2) and Taq polymerase (Invitrogen). A 1.4-kb PCR product, as opposed to a 1-kb product in the wild-type DNA control, was detected in the majority of cultures checked, indicating that the first red recombination step had occurred. The second recombination step was done as above. Colonies were grown in LB medium with 30 μg/ml chloramphenicol overnight at 32°C, boiled, and subjected to PCR. Positive clones contained a 400-nt product, indicating that the second red recombination event, which removes the Kanr cassette, had occurred. Positive clones were maxi-prepped using the Qiagen large construct purification kit, and deletion of the ORF23 gene was verified by restriction enzyme digest, direct sequencing of the BAC DNA, and PCR of the BAC DNA using the VerifyΔORF23-F/VerifyΔORF23-R primers followed by sequencing of the PCR products. Six independent transfections were done using the CalPhos mammalian transfection system (Clontech) with either two different pOKA-BACΔORF23 clones used for each transfection (three transfections) or four different pOKA-BACΔORF23clones used for each transfection (three transfections), together with wild-type pOKA-BAC (all transfections) or the wild-type pOKA cosmid set (one transfection) as controls.
Growth kinetics and plaque size assays.
The replication kinetics and peak titers of recombinant viruses were assessed by infectious focus assay (14). Viruses recovered from MeWo cell transfections were also propagated in human embryonic lung fibroblasts (HELF). MeWo cells and HELF were seeded in six-well plates and infected with an inoculum of ∼1 × 103 PFU. Cells were trypsinized on days 1 to 5, and titers were determined on 24-well plates seeded with MeWo cells; plaques were visualized by staining with human polyclonal anti-VZV immunoglobulin G (IgG; 1:1,000) in order to identify small plaques. To evaluate plaque size, monolayers were imaged and photographed with a Zeiss Axiovert microscope. The maximum diameter of at least 10 plaques was measured for each test virus on each day of the growth curve. Statistical comparisons were done using the Student t test.
Expression of capsid proteins in plasmid vectors.
ORF23 and ORF23 with targeted mutations were fused to mRFP using the pDS-mRFP-N1 vector (Clontech, Mountain View, CA). ORF23 was amplified from VZV DNA using the primers ORF23-F and ORF23-R (Table 2), containing a SacI site in the forward primer and an EcoRI site in the reverse primer for cloning upstream of the mRFP gene in the multiple cloning site, which yielded ORF23mRFP. ORF23Δ169-228mRFP was constructed by digesting ORF23mRFP with PmlI and XmnI and ligation, which removed amino acids 169 to 228 upstream of the putative nuclear localization signal (NLS). ORF23Δ229-234mRFP was constructed by SalI digestion of ORF23-mRFP and Klenow fill in followed by XmnI digestion and ligation, which removed the putative NLS motif located at amino acids 229 to 234. ORF23Δ169-234mRFP was constructed by SalI digestion of ORF23mRFP and Klenow fill in, followed by PmlI digestion and ligation, which deleted amino acids 169 to 234, including the putative NLS and some upstream sequences. ORF23NLSmRFP had point mutations in the putative NLS motif that were introduced using the PCR primer ORF23NLSmut-R to change the arginine residues to glycine residues (bold) (SRSRRV-SGSGGV) (Table 2). Since pOKA-BAC23mRFP did not yield infectious virus, a construct was made that expressed the ORF23mRFP cassette as it had been inserted into this pOKA-BAC23mRFP construct. This ORF23mRFP cassette lacks the 16-amino-acid linker sequence. It was amplified and cloned using SacI and NotI restriction sites in the forward and reverse primers and expressed under the cytomegalovirus (CMV) immediate-early (IE) promoter in the pDS backbone, generating ORF23*mRFP. A construct, pDS-ORF23, was made that expressed wild-type ORF23 without fusion to mRFP under the CMV IE promoter. HSV-1 VP26 was amplified from HSV DNA and fused to mRFP for use as a positive control for cytoplasmic localization in confocal studies. ORF33.5-mRFP was made by amplifying ORF33.5 from VZV DNA with primers ORF33.5-F and ORF33.5-R, with restriction sites used for cloning in pDS-mRFP-N1 (Table 2). ORF20, ORF40, and ORF41 were amplified with primers ORF20F/R, ORF40F/R, and ORF41F/R and cloned into the multiple cloning site in the pEGFP-C1 vector (Clontech, Mountain View, CA) (Table 2). Plasmids expressing mRFP or GFP fusion proteins were transfected into AD293 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and OptiMEM+Glutamax-I (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and cells were examined at 32 h after transfection.
Confocal microscopy.
Polyclonal anti-ORF23 antiserum was made by expressing ORF23 (nt 43,138 to 42,439) in pGEX-2T vector (Pharmacia Biotech, Milwaukee, WI) using primers ORF23 pGEX2T-F and ORF23 pGEX2T-R (Table 2). Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside. GST-ORF23 protein was purified using glutathione S-transferase (GST) microspin purification columns (Amersham, GE Healthcare, Piscataway, NJ) for rabbit immunization. IgG was isolated from pre- and postimmunization (day 63) sera (ImmunoPure protein a IgG purification column; Pierce, Rockford, IL). Purified IgG was preadsorbed with uninfected MeWo or HELF cell lysates.
For an indirect immunofluorescence microscopy, infected MeWo, HELF, or AD293 cells were fixed at 48 h with 4% paraformaldehyde for 10 min at room temperature and permeabilized in 2% paraformaldehyde-0.2% Triton X-100 for 15 min. Cells were washed five times in phosphate-buffered saline (PBS) for 5 min, blocked with 1% fish gelatin in PBS for 1 h, and incubated 3 to 4 h at room temperature with the polyclonal rabbit anti-ORF23 (1:2,000) or human anti-VZV IgG (1:1,000) in blocking buffer. Cells were washed five times with PBS and incubated for 30 min to 1 h with fluorescein isothiocyanate-labeled goat anti-rabbit IgG or Texas red-labeled goat anti-human IgG (Jackson ImmunoResearch, Inc.) in blocking buffer. After three PBS washes, coverslips were incubated with Hoechst 33258 nuclear stain (1:7,000) in PBS, washed twice, and mounted with Vectashield (Vector Laboratories, Inc., Burlingame, CA). For live cell imaging, AD293 cells were transfected in two-chamber slides (LabTek II chambered coverglass; Nalgene Nunc, Rochester, NY). Nuclei were visualized with Hoechst 33258 (1:7,000). Monolayers were washed twice with PBS, and 0.25 ml of Dulbecco's modified Eagle's medium with 2% fetal calf serum and no phenol red dye (Invitrogen, CA) was added. Imaging was done with a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss, Inc.).
Transmission electron microscopy.
MeWo and HELF cells were infected with ∼1 × 103 PFU of the test virus and grown on glass coverslips for 48 h. The cells were fixed for 35 min with 2% glutaraldehyde in 0.1 M phosphate buffer (PBS), pH 7.0, washed twice in PBS, and postfixed with 1% osmium tetroxide (Polysciences, Inc., Warrington, PA) in PBS for 1 h. After two 10-min washes in double-distilled water, specimens were stained in 0.25% uranyl acetate (Polysciences, Inc.) overnight. The specimens were washed with water and dehydrated through a graded series of alcohol and propylene oxide washes. Cells were embedded by placing gelatin capsules filled with resin on top of the coverslip and placing in a 60°C oven for 24 h. Embedded samples were thin sectioned with a Reichert Ultracut E ultramicrotome (Reichert-Jung, Vienna, Austria), placed on copper grids, stained with uranyl acetate and Reynold's lead citrate, and viewed with a Hitachi H-7000 transmission electron microscope.
Infection of skin xenografts in mice.
Skin xenografts were made in homozygous CB-17scid/scid mice, with human fetal tissues obtained with informed consent according to federal and state regulations. Animal use was in accordance with the Animal Welfare Act and approved by the Stanford University Administrative Panel on Laboratory Animal Care. VZV recombinants, passed three times in HELF cells, were used to inoculate four to five xenografts with each test virus (14); titers were ∼1 × 105 PFU/ml, as determined at the time of inoculation. Skin xenografts were harvested at 10 and 21 days after infection; viral titers were measured by infectious focus assay.
Immunoblotting and immunoprecipitation.
Infected and uninfected MeWo or HELF cells were lysed with radioimmunoprecipitation assay buffer (50 mM Tris [pH 8], 150 mM NaCl, 1% IGEPAL CA-360 [Sigma, St. Louis, MO], 0.1% sodium dodecyl sulfate [SDS; Bio-Rad, Hercules, CA], 0.5% deoxycholic acid Σ) containing a protease inhibitor (Complete Mini tablet; Roche, Inc.) in 1-ml volumes per T75 flask and sonicated. Lysates were boiled in sample buffer and separated by using 10% sodium dodecyl sulfate-polyacrylamide gels. Total protein was quantitated by Bradford assay for normalization. Proteins were transferred to Immobilon-P (polyvinylidene difluoride) membranes (Millipore, Bedford, MA) and analyzed with polyclonal rabbit anti-ORF23 (1:1,000). Immunoblots of the same extracts were analyzed with polyclonal anti-IE4 antibody (1:5,000) as a loading control for viral proteins and with anti-α-tubulin (1:10,000) for cell proteins.
Cytoplasmic and nuclear fractions were prepared from AD293 cells 48 h after transfection by washing twice with cold Dulbecco's phosphate buffered saline (DPBS) containing protease inhibitor in 50 ml DPBS, scraping into 15-ml prechilled conical tubes, and centrifugation at 500 × g at 4°C. Supernatants were discarded, and cell pellets were resuspended in 200 μl hypotonic buffer (20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 10% glycerol). After adding protease inhibitors, 1 mM dithiothreitol, 1 mM sodium orthovanadate, and 50 mM sodium fluoride, the pellet was incubated in hypotonic buffer for 15 min on ice, and 10 μl of hypotonic buffer containing 4% IGEPAL CA-360 (Sigma, St. Louis, MO) was added and vortexed for 10 s. The lysate was centrifuged for 1 min at 10,000 × g at 4°C. The supernatant was the cytoplasmic fraction, and the pellet was used for nuclear fraction collection. The pellet was washed, resuspended, and treated as described above. The supernatant was collected as the soluble nuclear fraction. Total protein was verified by Bradford assay (Bio-Rad) and normalized. Proteins were transferred to Immobilon-P (polyvinylidene difluoride) membranes (Millipore, Bedford, MA) and analyzed with DsRed polyclonal antibody (1:3,000) or GFP (JL-8) monoclonal antibody (1:16,000) (Clontech, Mountain View, CA). The extracts were also analyzed for anti-α-tubulin to assess separation of the cytoplasmic and nuclear cell fractions. Anti-DsRed antibody was used because nonspecific bands were present in blots of transfected AD293 cell lysates probed with rabbit polyclonal anti-ORF23 antibody.
For coimmunoprecipitation, protein A slurry was washed three times with wash buffer (0.5% bovine serum albumin and 0.5% IGEPAL CA-360 in DPBS) and once with water; the beads were collected in DPBS with 1% bovine serum albumin. The cell lysate was separated into cytoplasmic and nuclear fractions, and samples were diluted with 2 volumes of PBS wash buffer containing protease inhibitor cocktail (Roche, CA). The lysate was precleared by incubation of the sample with 30 μl protein A slurry for 1 h at 4°C on an orbital mixer. The slurry was spun down and the supernatant collected; the beads were washed in wash buffer three times, resuspended in 30 μl of 1× sample buffer, and boiled for 10 min. This preparation was used as a negative control. The precleared lysate was incubated with 1 μl of polyclonal rabbit anti-DsRed at 4°C on an orbital shaker for 1 h. Protein A slurry (50 μl per immunoprecipitation mixture) was added to the immune complex and incubated at 4°C overnight. The beads were spun down, the supernatant was discarded, and the pellet was washed twice in sucrose buffer (1 M sucrose, 0.5 M sodium chloride, 0.5% IGEPAL CA-630), four times in wash buffer, and twice in PBS. The pellet was resuspended in 25 μl of PBS and 5 μl of SDS-polyacrylamide gel electrophoresis sample buffer, boiled for 10 min, and centrifuged at high speed for 3 min; the supernatant was tested by immunoblotting with appropriate antibodies.
RESULTS
ORF23 mutagenesis in pOKA-BAC and generation of recombinant viruses.
Transfections of MeWo cells with pOKA-BAC yielded virus consistently. However, no virus was recovered from six independent transfections of pOKA-BAC23mRFP, in which mRFP was inserted in frame with ORF23 at position 42,431 (Fig. 1B) or from six independent transfections with four independently derived OKA-BACΔORF23 clones. No infectious virus was recovered from any of the transfections of pOKA-BAC23mRFP or pOKA-BACΔORF23; the transfected cells were passaged for 4 weeks. Virus was present in cells transfected with the pOKA-BAC and pOKA cosmids within 5 to 8 days posttransfection. Bioinformatics analysis of the ORF23mRFP cassette using Protean Lasergene software (DNAstar Inc., Madison, WI) suggested that the hydrophilicity index of the ORF23 C terminus was changed when mRFP was directly fused just before the ORF23 stop codon. To determine whether this mutation could be rescued, ORF23 was inserted into the cosmid pvSpe23ΔAvr at the AvrII cloning site. Infectious virus was recovered from cotransfections of pOKA-BAC23mRFP and pvSpe23AvrORF23 but not from cotransfections with pvSpe23ΔAvr without the ORF23 insertion. The rescued virus was designated pOKA-BAC23mRFP-R (Fig. 1D). The ORF23 insertion at nt 112,851 was confirmed by PCR, sequencing, and Southern hybridization (data not shown). Whereas recombinant virus was not recovered when ORF23 was fused to mRFP or when the ORF23 sequence was deleted without any insertion, deleting ORF23 and replacing the ORF23 coding sequence with the mRFP cassette yielded infectious virus (Fig. 1C). This virus, designated pOKA-BACmRFPΔ23, had the expected mutations by PCR, sequencing, and Southern hybridization. Thus, the expression of an aberrant form of ORF23 as an mRFP fusion protein or removing the ORF23 sequence appeared to block viral replication, whereas deleting the gene by inserting the foreign mRFP DNA sequence was not lethal.
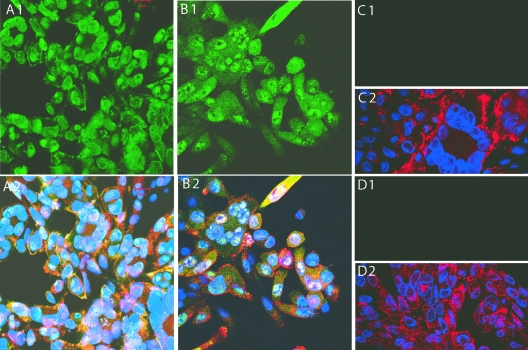
The evaluation of ORF23 expression in Mewo and HELF cells infected with pOKA-BAC showed that the most ORF23 protein was detected in the nucleus (Fig. 2A); ORF23 expression in HELFs had the same pattern (data not shown). ORF23 expression was also observed on plasma membranes, but cytoplasmic staining was faint. In contrast, more cytoplasmic expression of ORF23 was observed in cells infected with pOKA-BAC23mRFP-R, which has the ORF23mRFP fusion at the native site and ORF23 at the ectopic site in the US region (Fig. 2B). As expected, cells infected with pOKA-BACmRFPΔ23 showed no ORF23 expression (Fig. 2C) and pOKA-BAC infected cells showed no reactivity with preimmune rabbit IgG (Fig. 2D).
FIG. 2.
ORF23 protein expression in MeWo cells infected with pOKA-BAC, pOKA-BAC23mRFP-R, and pOKA-BACmRFPΔORF23. MeWo cells infected with pOKA-BAC (A1 and A2), pOKA-BAC23mRFP-R (B1 and B2), or pOKA-BACmRFPΔ23 (C1 and C2) were stained with rabbit anti-ORF23 IgG (1:2,000) and detected with fluorescein isothiocyanate-labeled goat anti-rabbit IgG (top panel) and merged with staining with human polyclonal anti-VZV IgG (1:1,000) detected with Texas red-labeled goat anti-human IgG and Hoechst nuclear stain (bottom panel). The preimmune rabbit IgG showed no reactivity with MeWo cells infected with OKA-BAC (D1 and D2).
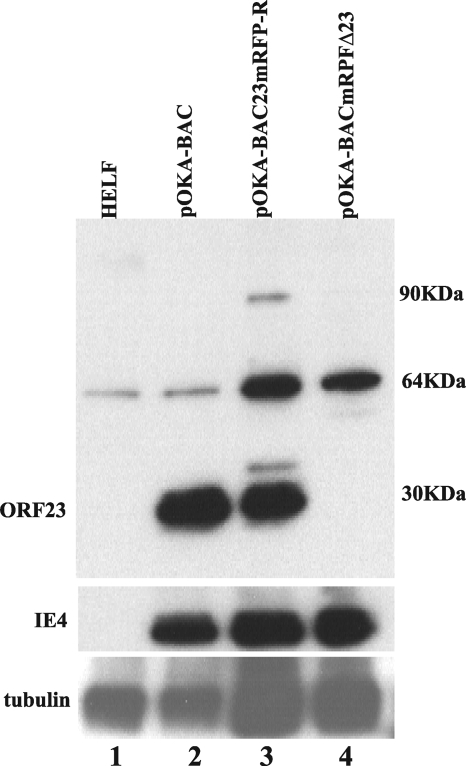
By immunoblot analysis, ORF23 was detected as a 30-kDa band in lysates of HELF infected with pOKA-BAC (Fig. 3, lane 2); results in MeWo cells were similar (data not shown). This band was slightly higher than the predicted 26 kDa, which may have been due to phosphorylation, since ORF23 has 13 (6 serine and 7 threonine) predicted phosphorylation sites. ORF23 was also expressed as a 30-kDa band in cells infected with pOKA-BAC23mRFP-R, which is consistent with its expression from the ectopic site (Fig. 3, lane 3). Expression of ORF23mRFP from this virus was also predicted to yield a 52-kDa band, which was not detected; ORF23 epitopes may have been disrupted or masked by mRFP. Two additional 90- and 35-kDa bands were observed, which may have represented modified or degraded forms of the fusion protein. As expected, ORF23 was not expressed in cells infected with pOKA-BACmRFPΔ23. A nonspecific band was observed at 64 kDa in uninfected as well as in infected HELF lysates. The prominence of this band was greatest in lanes testing ORF23 mutants. This was due to increased protein loading, which was required to normalize to IE4 levels in pOKA-ORF23mRFP-R-infected and pOKA-BACΔ23-infected cell lysates.
FIG. 3.
ORF23 expression in HELF cells infected with pOKA-BAC, pOKA-BAC23mRFP-R, or pOKA-BACmRFPΔ23. Uninfected and infected HELF lysates were run on a 10% SDS-polyacrylamide gel and probed with anti-ORF23 (1:2,000). Lane 1, uninfected HELF; lane 2, pOKA-BAC; lane 3, pOKA-BAC23mRFP-R; lane 4, pOKA-BACmRFPΔ23. The blots were probed with anti-IE4 to show similar viral protein loading, which required adding more infected cell lysate for the ORF23 mutants (center panel). Anti-α-tubulin (1:16,000) was used to assess cell protein loading, which was higher, as expected, in lanes with lysates of ORF23 mutant-infected HELF (lower panel).
Plaque morphologies and growth kinetics of pOKA-BAC and ORF23 mutants.
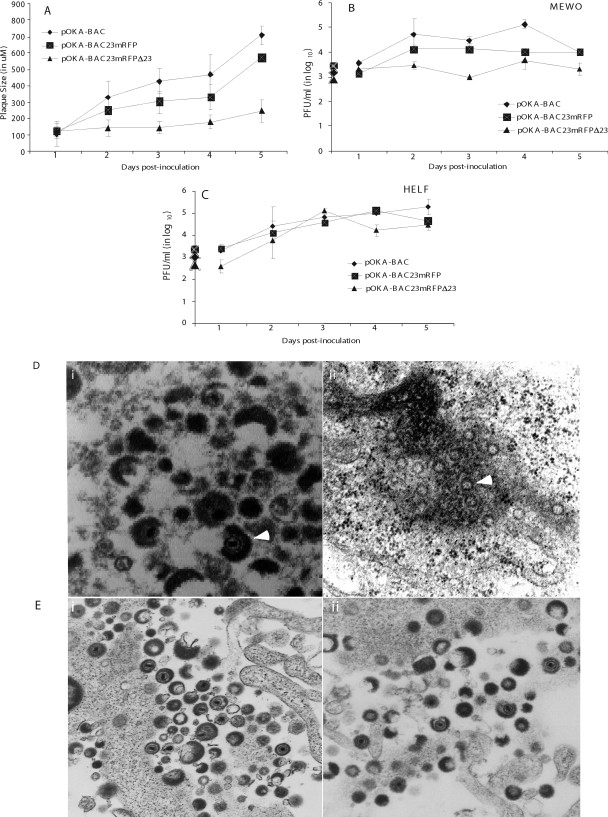
The plaque morphologies of pOKA-BAC, pOKA-BAC23mRFP-R, and pOKA-BACmRFPΔ23 were evaluated over 5 days in MeWo cells (Fig. 4A). When compared to pOKA-BAC, the pOKA-BAC23mRFP-R rescue virus showed a modest but statistically significant reduction in plaque size (mean, 550 μm versus 700 μm at 5 days; P < 0.01). In contrast, pOKA-BACmRFPΔ23, from which ORF23 was deleted, had a very small plaque phenotype in MeWo cells (mean, 180 μm at 5 days; P < 0.001 compared to pOKA-BAC). Evaluation of the growth kinetics of pOKA-BAC, pOKA-BAC23mRFP-R, and pOKA-BACmRFPΔ23 in MeWo cells showed that titers of pOKA-BAC increased to a peak of 1.3 × 105 PFU/ml on day 4 (Fig. 4B). pOKA-BAC23mRFP-R displayed a moderate reduction compared to pOKA-BAC, reaching a peak titer of 1.3 × 104 PFU/ml on day 2. The pOKA-BACmRFPΔ23 mutant showed only a slight increase from the initial inoculum of 1 × 103 PFU, reaching a maximum of 4.6 × 103 PFU/ml on day 4. In contrast, all three viruses displayed similar growth characteristics in HELF (Fig. 4C). The peak titers in HELF cells were 2 × 105 for pOKA-BAC, 1.33 × 105 for pOKA-BAC23mRFP-R, and 1.3 × 105 for pOKA-BACmRFPΔ23.
FIG. 4.
Plaque morphologies and growth kinetics of pOKA-BAC and ORF23 mutants in MeWo and HELF cells. (A) Plaque diameters were measured in MeWo cells infected with ∼1 × 103 PFU of pOKA-BAC, pOKA-BAC23mRFP-R, and pOKA-BACmRFPΔ23 from days 1 through 5. The x axis indicates the days after inoculation when infected cell monolayers were stained with anti-VZV IgG, and the y axis indicates the size of the plaques in micrometers. (B and C) MeWo cells (B) and HELF (C) cells were inoculated with pOKA-BAC23mRFP-R and pOKA-BACmRFPΔ23 at ∼1 × 103 PFU. Infectious virus yields were determined from days 1 to 5 after infection. Titers are expressed as means for triplicate wells at each time point. The x axes indicate the days after inoculation when infected cell monolayers were harvested, and the y axes indicate PFU determined by infectious focus assay. (D) Analysis of virion formation of ORF23 mutants and pOKA BAC in Mewo cells by TEM. Left panel: accumulation of typical capsids in the nucleus of pOKA-BAC-infected MeWo cells 48 h after inoculation. Magnification, ×35,000. Right panel: accumulation of empty spherical particles in the nucleus of pOKA-BACmRFPΔ23-infected MeWo cells; white arrows point to the empty spherical capsids. Magnification, ×35,000. (E) Analysis of virion formation of ORF23 mutants and pOKA BAC in HELF cells by TEM. Left panel: accumulation of typical capsids in the nucleus of pOKA-BAC-infected HELF cells 48 h after inoculation. Magnification, ×10,000. Right panel: accumulation of capsids in the nuclei of pOKA-BACmRFPΔ23-infected HELF cells. Magnification, ×10,000.
Effects of deleting ORF23 on virion formation in vitro.
When virion morphogenesis was evaluated in MeWo cells infected with pOKA-BAC and pOKA-BACmRFPΔ23 by transmission electron microscopy (TEM), cells infected with pOKA-BAC showed the characteristic accumulation of capsids and cytoplasmic enveloped virions (Fig. 4D, left panel). However, 48 h after infection of MeWo cells with pOKA-BACmRFPΔ23, empty spherical structures accumulated near the periphery of the nuclei (Fig. 4D, right panel). This pattern was similar in 20 infected nuclei; no complete capsids or enveloped cytoplasmic virions were identified in cells infected with the ORF23 deletion virus for as long as 5 days after infection. In contrast to MeWo cells, virion morphology was indistinguishable in HELF infected with pOKA-BAC (Fig. 4E, left panel) and pOKA-BACmRFPΔ23 (Fig. 4E, right panel), and the characteristic accumulation of capsids and cytoplasmic enveloped virions was observed with both viruses.
Replication of pOka-BAC and ORF23 mutants in skin xenografts in SCIDhu mice in vivo.
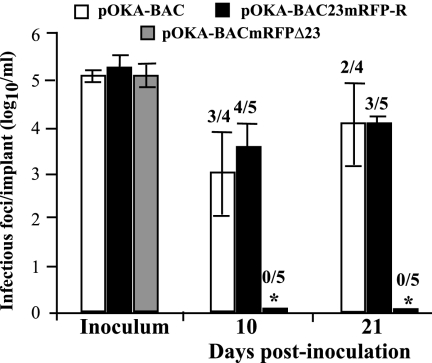
The replication of pOKA-BAC and pOKA-BAC23mRFP-R was similar in skin xenografts on days 10 and 21 after inoculation (Fig. 5). Three of four pOKA-BAC infected xenografts and four of five xenografts infected with pOKA-BAC23mRFP-R yielded virus on day 10; the mean titers were 5.3 × 103 PFU/ml and 9.3 × 103 PFU/ml, respectively. On day 21, virus was recovered from two of four pOKA-BAC-infected xenografts and three of five xenografts infected with pOKA-BAC23mRFP-R; the mean titers were 4 × 104 PFU/ml and 2.2 × 104 PFU/ml, respectively. Persistence of mRFP expression was observed in monolayers infected with these viruses after skin replication. In contrast, pOKA-BACmRFPΔ23 was not recovered from any of five xenografts infected with this ORF23 null mutant at day 10 or day 21 after inoculation.
FIG. 5.
Replication of pOKA-BAC and ORF23 mutants in skin xenografts in SCIDhu mice. Skin xenografts were inoculated with pOKA-BAC, pOKA-BAC23mRFP-R, and pOKA-BACmRFPΔ23. Each bar represents the mean titer of infectious virus (error bars show standard deviations) recovered from four to five xenografts harvested at 10 and 21 days after inoculation. Inoculum titers are shown at the left. The asterisk indicates no viral growth in any implants. The number of xenografts per number inoculated from which infectious virus was recovered is shown above each bar. Mean titers were based on data from xenografts that yielded virus.
Analysis of intracellular localization of ORF23 and ORF23 mutant proteins in transient-expression assays.
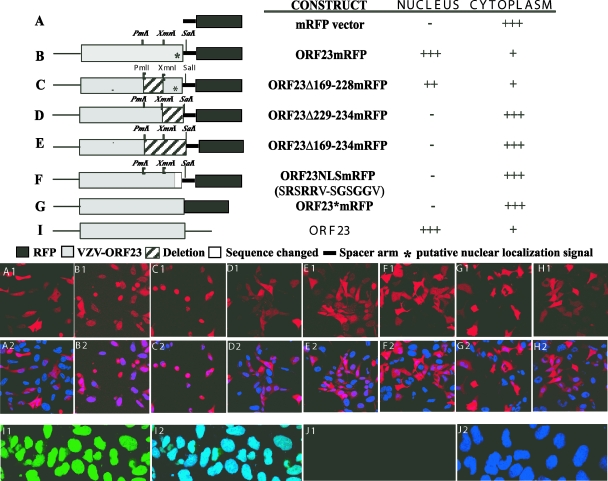
Transfections of AD293 cells were done using plasmids expressing intact ORF23 or mutant ORF23 fused in frame to the N terminus of mRFP in the pDS monomer vector (Fig. 6). The cloning procedure used to generate these plasmids left a 16-amino-acid spacer before the mRFP start codon (Fig. 6, lines B to F). When expressed in AD293 cells, ORF23mRFP was predominantly nuclear at 32 h (Fig. 6B). This observation indicated that ORF23 differs from its UL35 homologs, which do not exhibit independent nuclear localization (15). A bioinformatics analysis of the unique C-terminal residues of ORF23 did not reveal motifs matching the conventional NLS (http://cubic.bioc.columbia.edu/papers/2000_nls/paper.html). However, a motif that had abundant positively charged residues, SRSRRV, consistent with a possible NLS, was found in the ORF23 C terminus between amino acids 229 and 234. This region was evaluated for NLS function using ORF23 mutant constructs (Fig. 6C to F). ORF23 protein was primarily nuclear when the putative NLS was retained, as it was in ORF23mRFP and ORF23Δ169-228mRFP (Fig. 6B and C). However, ORF23 was predominantly cytoplasmic when the putative NLS was deleted from ORF23Δ229-234mRFP or ORF23Δ169-234mRFP, or when it was disrupted in ORF23NLSmRFP (Fig. 6D to F). The pDS mRFP vector alone showed predominantly cytoplasmic mRFP expression (Fig. 6A). The ORF23mRFP cassette was also amplified from the pOKA-BAC23mRFP construct that had failed to yield infectious virus. This ORF23mRFP cassette, which does not have the 16-amino-acid spacer between ORF23 and mRFP, was expressed under the CMV IE promoter. This construct, designated ORF23*mRFP, showed only cytoplasmic expression (Fig. 6G). Thus, fusion of mRFP just before the ORF23 stop codon appears to have introduced structural changes that disrupted the putative NLS. HSV-1 VP26 expressed as an mRFP fusion protein had the expected cytoplasmic localization (Fig. 6H). ORF23 expressed without an mRFP tag in pDS-ORF23 localized to the nucleus, as detected using anti-ORF23 (Fig. 6I), indicating that the presence of the mRFP did not alter the intracellular localization of ORF23 in the pDS backbone; anti-ORF23 showed minimal background fluorescence in untransfected AD293 cells (Fig. 6J).
FIG. 6.
Analysis of ORF23 localization using transient-expression constructs. (Top) The schematic illustrates the mRFP vector (A), the ORF23mRFP fusion protein (B), truncations (C, D, and E), and point mutations (F) in the sequence encoding ORF23 fused to mRFP or ORF23 directly fused to mRFP without any linker (G), and ORF23 not fused to mRFP (I). The solid black box indicates the mRFP vector, and the gray shaded box indicates the ORF23 sequence. The * marks the location of the putative nuclear localizing signal, the hatched box indicates regions that were deleted, and the dark box indicates amino acid changes from the intact ORF23 protein. The ORF23 cassette is joined to the mRFP fusion protein with a 16-amino-acid spacer arm, indicated by the solid black line, which is part of the vector backbone. The names of the constructs and their subcellular localizations when expressed in AD293 cells are indicated to the right of each schematic. (Bottom panel) Detection of cellular localization of mRFP in AD293 cells transfected with ORF23 wild-type and mutant constructs and controls by direct fluorescence confocal microscopy at 32 h after transfection. The alphabetical notation in the panels (i.e., A1, B1, C1, D1, E1, F1, G1, and I1) correspond with each construct mentioned at the top, and the corresponding lower panel (A2, B2, C2, D2, E2, F2, G2, and I2) shows merged images of the same construct with nuclear Hoechst stain. (H1) Direct fluorescence images from HSV-1 VP26mRFP; (H2) same construct merged with Hoechst nuclear stain. (I1) AD293 cells transfected with pDS-ORF23, stained with rabbit anti-ORF23 IgG (1:2,000), was detected with fluorescein isothiocyanate-labeled goat anti-rabbit IgG; (I2) merged image with Hoechst nuclear stain. Control panels J1 and J2 depict background fluorescence in AD293 cells with anti-ORF23 IgG.
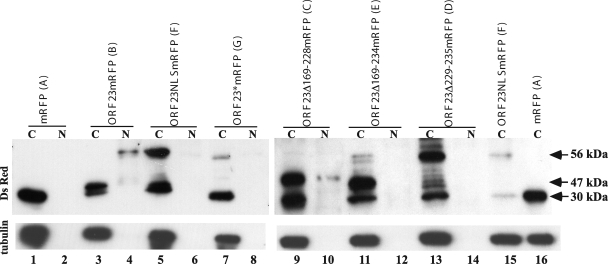
The subcellular localization of ORF23mRFP and ORF23mRFP mutants was confirmed by immunoblotting of nuclear and cytoplasmic fractions of transfected AD293 cells using rabbit polyclonal anti-DsRed antibody (Clontech, Mountain View, CA). When blots were probed with anti-ORF23, many nonspecific bands were observed in mock samples from transfected AD293 cells; hence, experiments were done only with DSred antibody. When expressed alone, mRFP was detected as a 30-kDa band in the cytoplasmic fraction only (Fig. 7, lanes 1 and 2), whereas ORF23mRFP was detected as a 56-kDa band that was predominantly nuclear (Fig. 7, lanes 3 and 4). Although ORF23-mRFP is predicted to be 52 kDa, it is likely to have isoforms due to phosphorylation. ORF23NLSmRFP was completely cytoplasmic, which was consistent with disrupting the putative NLS (Fig. 7, lanes 5 and 6). ORF23*mRFP was also cytoplasmic and slightly smaller than 56 kDa because it lacked the 16-amino-acid linker (Fig. 7, lanes 7 and 8). Lysates of cells transfected with ORF23Δ169-228mRFP had a 47-kDa band in both fractions, which was consistent with the predicted size after the 59-amino-acid deletion (Fig. 7, lanes 9 and 10). The retention of the NLS motif appears to allow translocation of the mRFP fusion protein to the nucleus, although cytoplasmic expression was prominent, in contrast to intact ORF23, which was completely translocated to the nucleus. This observation suggested that removing the upstream 169 to 228 amino acids compromised the efficiency of nuclear trafficking. Lysates of cells transfected with ORF23Δ169-234mRFP showed a 45-kDa band in the cytoplasm, which was consistent with the 66-amino-acid deletion and the disruption of the putative NLS (Fig. 7, lanes 11 and 12). Lysates of cells transfected with ORF23Δ229-234mRFP, which has only a 6-amino-acid deletion, had a prominent 56-kDa band in the cytoplasmic fraction, indicating that deleting the NLS motif alone was sufficient to block the nuclear translocation of the mutant ORF23 fusion protein (Fig. 7, lanes 13 and 14). The band at approximately 30 kDa in all cytoplasmic fraction lanes is likely to represent some level of mRFP translation from the vector and was consistently present.
FIG. 7.
Immunoblot analysis of the intracellular localization of ORF23 and ORF23 mutant proteins. Control vector, ORF23, and ORF23 mutant constructs were transfected into AD293 cells, and the cell lysate was separated into cytoplasmic (C) and nuclear (N) fractions. Blots were probed with polyclonal dsRED antibody. ORF23mRFP expression yielded a 56-kDa band; mRFP protein expressed alone was 30 kDa. Blots were probed with anti-α-tubulin as a control for separation of cytoplasmic and nuclear fractions. Letters A to G in parentheses following the construct names correspond to the letters in the confocal images in Fig. 6. Lanes: 1 and 2, vector control; 3 and 4, ORF23-RFP; 5 and 6, ORF23NLSmRFP; 7 and 8, ORF23*RFP; 9 and 10, ORF23Δ169-228mRFP; 11 and 12, ORF23Δ169-234mRFP; 13 and 14, Δ229-234mRFP. Lanes 15 and 16, ORF23NLSmRFP and mRFP vector, were reloaded to have a size standard for 56 kDa and 30 kDa in the second blot.
Analysis of ORF20, ORF33.5, ORF40, and ORF41 in transient-expression assays.
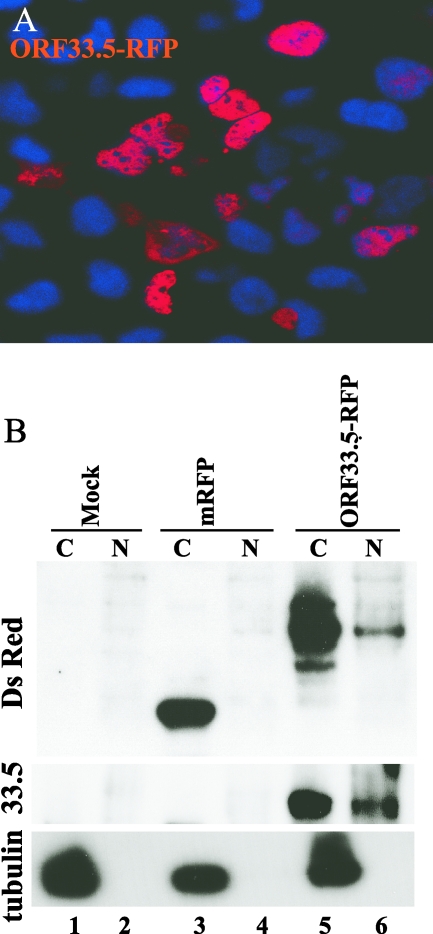
Since the nuclear translocation of ORF23 differed from the cytoplasmic localization of its alphaherpesvirus homologs, we examined subcellular localization patterns of other putative VZV capsid proteins using mRFP and GFP fusion constructs. Because ORF33.5, the putative VZV scaffold protein, can replace HSV-1 preVP22a in capsid assembly in the baculovirus expression system, we predicted that it would also have an inherent nuclear localization capacity (18). Nuclear ORF33.5mRFP expression was evident at 32 h after transfection (Fig. 8A), which was confirmed by immunoblotting using rabbit polyclonal anti-DsRed antibody (Fig. 8B). The specificity of the band and the nuclear localization were also confirmed by probing the blot with anti-ORF33.5 (kindly provided by V. G. Preston, MRC Virology Unit, Glasgow, United Kingdom) (18) (Fig. 8B).
FIG. 8.
Analysis of ORF33.5 intracellular localization. A. AD293 cells were transfected with the ORF33.5mRFP construct and visualized by direct fluorescence confocal microscopy at 32 h after transfection. In this merged image, the red signal is fluorescence from the fusion protein and the blue signal is nuclear Hoechst stain. B. Control vector and ORF33.5 constructs were transfected into AD293 cells, and the cell lysates were separated into cytoplasmic (C) and nuclear (N) fractions. The blot was probed with polyclonal anti-dsRED antibody and with anti-α tubulin as a control for separation of cytoplasmic and nuclear fractions. The blot was reprobed with anti-ORF33.5 antibody (kindly provided by V. G. Preston). Lanes: 1 and 2, mock; 3 and 4, vector control (30 kDa); 5 and 6, ORF33.5mRFP (60 kDa).
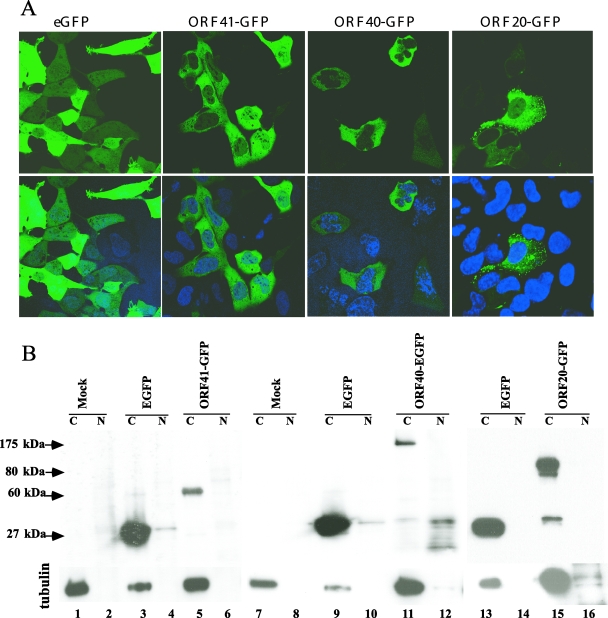
ORF40, the homolog of HSV-1 VP5, and two homologs of the HSV-1 triplex protein complex including ORF41, which is related to HSV-1 VP23 and ORF20, the homolog of HSV-1 VP19C, were also expressed as fusion proteins. In HSV-1, VP5 and VP23 are cytoplasmic; VP5 transport to the nucleus requires preVP22a (19). ORF40 and ORF41 were made as GFP fusion constructs; ORF20 was expressed as an mRFP fusion construct and as a GFP fusion construct. Expression of ORF40-GFP, ORF41-GFP, and ORF20-GFP was localized to the cytoplasm, as was GFP alone (Fig. 9, upper panel). The cytoplasmic restriction was confirmed by immunoblotting of transfected cell lysates using mouse monoclonal anti-GFP (Fig. 9, lower panel). ORF41-GFP appeared as a 60-kDa band in the cytoplasm (Fig. 9, lanes 5 and 6). ORF40-GFP was detected as a 175-kDa band in the cytoplasmic fraction (Fig. 9, lanes 11 and 12), and ORF20-GFP was detected as a 80-kDa band in the cytoplasm (Fig. 9, lanes 15 and 16). All fusion protein constructs also had some cytoplasmic GFP expression, detected as a 27-kDa band. Thus, of the five proteins that were tested which are predicted to constitute VZV capsids, only ORF23 and ORF33.5 had intrinsic nuclear localization capacity when expressed as fusion proteins, while ORF40, ORF41, and ORF20 remained in the cytoplasm.
FIG. 9.
Analysis of ORF20, ORF40, and ORF41 intracellular localization. A. AD293 cells were transfected with the ORF20-GFP, ORF40-GFP, and ORF41-GFP constructs and visualized by direct fluorescence confocal microscopy at 48 h after transfection. The lower panel shows the merged images of the same constructs with nuclear Hoechst stain. B. Control vector and ORF20-GFP, ORF40-GFP, and ORF41-GFP constructs were transfected into AD293 cells, and the cell lysates were separated into cytoplasmic (C) and nuclear (N) fractions. The blot was probed with monoclonal GFP antibody. Lanes: 1 and 2, mock; 3 and 4, vector enhanced GFP (EGFP) control; 5 and 6, ORF41-GFP (60 kDa); 7 and 8, mock; 9 and 10, vector EGFP control; 11 and 12, ORF 40-GFP (175 kDa); 13 and 14, vector EGFP control; 15 and 16, ORF 20-GFP (80 kDa). The same blot was probed with anti-α tubulin as a control for separation of cytoplasmic and nuclear fractions. Less of the cytoplasmic fraction from the EGFP vector control was added, as indicated by reduced α-tubulin, because expression of EGFP alone was high compared to the EGFP fusion protein constructs.
Influence of ORF23 on intracellular localization of ORF20, ORF40, and ORF41.
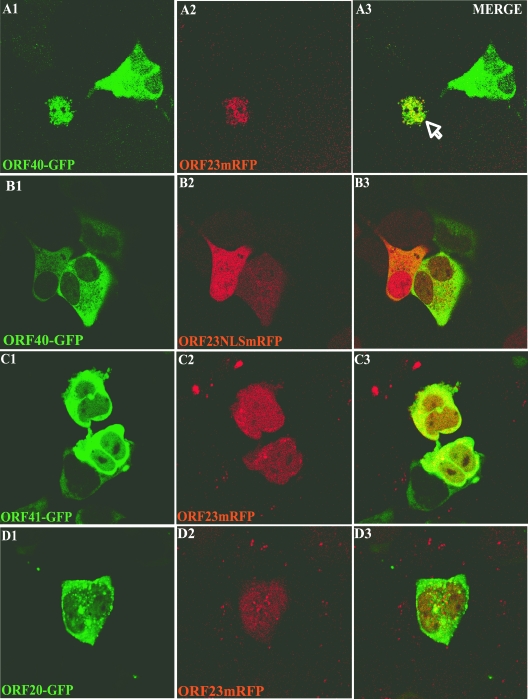
Whereas ORF33.5 had a cellular localization pattern that was similar to its HSV homolog, ORF23 and ORF20 seemed to have switched their roles with regard to inherent nuclear localization capacity. Therefore, we examined whether ORF23 was able to mediate nuclear import of the capsid proteins that were cytoplasmic when expressed alone. ORF23mRFP was cotransfected with ORF40-GFP, ORF41-GFP, or ORF20-GFP; cotransfection with ORF23NLSmRFP, which has disrupted nuclear localization, was used as a control. Coexpression of ORF23 caused the nuclear translocation of ORF40, the major capsid protein (Fig. 10A), whereas ORF23NLSmRFP did not alter the cytoplasmic restriction of ORF40 (Fig. 10B). ORF23 did not appear to induce the nuclear transport of ORF41 (Fig. 10C) or ORF20 (Fig. 10D), which are predicted to be part of the triplex protein complex.
FIG. 10.
Influence of ORF23 on intracellular localization of ORF20, ORF40, and ORF41. AD293 cells were cotransfected with pairs of constructs, including ORF23mRFP and ORF40-GFP (A), ORF23NLSmRFP and ORF40-GFP (B), ORF23mRFP and ORF41-GFP (C), and ORF23mRFP and ORF20-GFP (D), and examined 32 h after transfection by direct fluorescence confocal microscopy. The left, center, and right panels show visualization of GFP expression, RFP expression, and merged images, respectively.
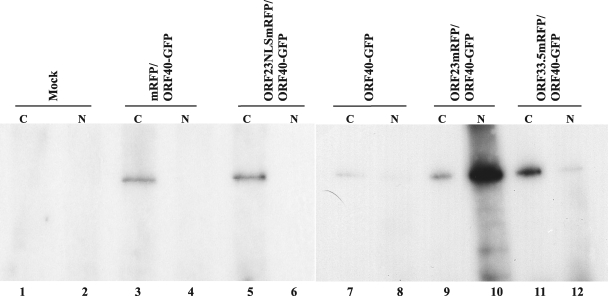
Coimmunoprecipitation experiments were done to assess the capacity of ORF23 to bind to and translocate ORF40 into the nucleus. Cotransfections were done with ORF23mRFP and ORF40-GFP, ORF23NLSmRFP and ORF40-GFP, mRFP/ORF40-GFP, and ORF40-GFP alone. Immunoprecipitation after cotransfection of ORF40-GFP with the mRFP vector or ORF23NLSmRFP did not result in ORF40-GFP transfer into the nuclear fractions (Fig. 11, lanes 4 and 6). ORF40-GFP alone was not detected in the nuclear fraction, although a very faint band was detected in the cytoplasmic fraction (Fig. 11, lane 7), probably due to some ORF40-GFP binding to the Sepharose beads. In contrast, coexpression of ORF40-GFP with ORF23mRFP resulted in high levels of OFR40-GFP in the nuclear fraction (Fig. 11, lane 10).
FIG. 11.
Interactions of ORF23 and ORF33.5 with ORF40. AD293 cells were cotransfected with ORF23 and ORF33.5 RFP fusion constructs and ORF40-GFP or controls. Cell lysates were separated into cytoplasmic (C) and nuclear (N) fractions. Polyclonal anti-DsRed antibody was used to immunoprecipitate the protein complexes, and blots were probed with monoclonal anti-GFP (JL-8). ORF40-GFP was detected as a 175-kDa band. Lanes: 1 and 2, mock; 3 and 4, vector mRFP control and ORF40-GFP; 5 and 6, ORF23NLSmRFP and ORF40-GFP; 7 and 8, ORF40GFP alone; 9 and 10, ORF23mRFP and ORF40-EGF; 11 and 12, ORF33.5-RFP and ORF40-GFP.
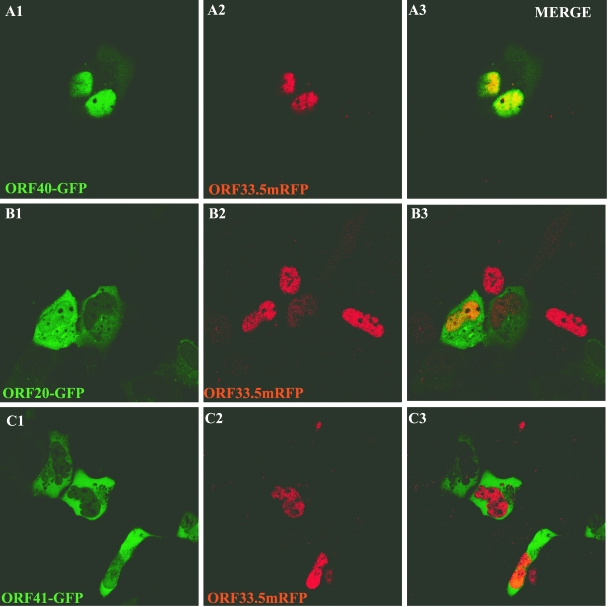
Influence of ORF33.5 on intracellular localization of ORF20, ORF40, and ORF41.
Since ORF33.5 also had intrinsic nuclear localization capacity, its effects on the distribution of ORF20, ORF40, and ORF41 were examined in cotransfection experiments. ORF33.5mRFP expression was associated with nuclear translocation of ORF40-GFP (Fig. 12A). ORF20-GFP and ORF41-GFP remained cytoplasmic when coexpressed with ORF33.5mRFP (Fig. 12B and C). When coimmunoprecipitation experiments were done to assess the capacity of ORF33.5 to translocate ORF40 into the nucleus, some ORF40 nuclear translocation was also observed (Fig. 11, lane 12). The results were reproducible when coimmunoprecipitation experiments were done five times, confirming that the ORF33.5-ORF40 interaction was weaker than the ORF23-ORF40 interaction.
FIG. 12.
Influence of VZV ORF33.5 on intracellular localization of ORF20, ORF40, and ORF41. Cells were cotransfected with pairs of constructs, including ORF33.5-RFP and ORF40-GFP (A), ORF33.5-RFP and ORF41-GFP (B), and ORF33.5-RFP and ORF20-GFP (C), and examined 48 h after transfection by direct fluorescence confocal microscopy. The left, center, and right panels show visualization of GFP expression, RFP expression, and merged images, respectively.
DISCUSSION
The assembly of herpesvirus capsids is a complex process involving interactions between multiple proteins in the cytoplasm and in the nucleus. Based on comparative genome analyses, VZV ORF23 encodes a conserved capsid protein, referred to as VP26 (UL35) in other alphaherpesviruses (3, 5, 10) Our analyses of ORF23 demonstrated that while it is dispensable for VZV replication in vitro, ORF23 is an essential protein for VZV infection and lesion formation in human skin xenografts in the SCIDhu mouse model of VZV pathogenesis in vivo. Using a rabbit polyclonal anti-ORF23 antibody, we showed that ORF23 was expressed as a nuclear protein and at plasma membranes in VZV-infected cells in vitro. Replacing ORF23 with mRFP in a VZV BAC construct was compatible with VZV replication in vitro. In contrast, pOKA-BACΔORF23 constructs did not yield virus. Since the ORF23 and mRFP genes are approximately the same size (700 bp), it is possible that there is some constraint related to the structure of the VZV genomic DNA at this locus such that replacement with the foreign DNA sequence is compatible with viral replication.
Of interest, the growth kinetics, plaque morphology, and virion morphology of pOKA-BACmRFPΔ23 were indistinguishable from the intact pOKA-BAC recombinant in human fibroblasts, whereas pOKA-BACmRFPΔ23 replication in a melanoma cell line was characterized by a very small plaque phenotype and viral titers that were only fourfold above the inoculum. This cell-type-specific deficiency of VZV replication in the absence of ORF23 was associated with intranuclear accumulation of many empty spherical particles, consistent with a requirement for ORF23 functions in VZV capsid assembly. Although deleting ORF23 did not affect growth in cultured fibroblasts and virion morphology was not altered, the failure of pOKA-BACmRFPΔ23 to infect skin xenografts indicates that ORF23 is a virulence determinant and is required when VZV infects differentiated human skin cells within their intact tissue microenvironment in vivo.
Expressing HSV-1 VP26 (UL35) as a GFP fusion protein does not block replication (8, 25). In contrast, recombinant VZV was not produced when mRFP was inserted in frame with ORF23 in the pOKA-BAC23mRFP construct, suggesting that ORF23 expression as an mRFP fusion protein at the C terminus was incompatible with capsid formation. Fusion of mRFP at the N terminus of ORF23 or as an internal fusion within a flexible amino acid loop was not done; fusion at either of these sites could be compatible with VZV replication.
Since deleting ORF23 did not block VZV replication, it appears that expression of the mutant form of ORF23 with RFP at the C terminus interfered with other steps necessary for capsid assembly. The long C-terminal region of ORF23 is unique among the herpesvirus homologs. However, ORF23 has a C-terminal motif, SRSRRV, which resembles the C-terminal SRTRR motif of the HCMV small capsid protein, although HCMV SCP is only 75 amino acids long. Similar to our observations with ORF23, expression of SCP as a GFP fusion protein was incompatible with viral replication; however, SCP deletion also prevented HCMV replication (2). Introducing ORF23 into pOKA-BAC23mRFP at a nonnative site in the US region permitted replication, indicating that ORF23 expression from the ectopic site was sufficient to compensate for the apparent dominant negative effects of the ORF23mRFP protein expressed in the same recombinant. Replication of pOKA-BAC23mRFP-R was equivalent to pOKA-BAC in fibroblasts, but ORF23 expression from the ectopic location was less than optimal for replication in melanoma cells, which were also less permissive for VZV replication when ORF23 was deleted. Nevertheless, ectopic ORF23 expression in the presence of the ORF23mRFP fusion protein did not impair VZV virulence in skin xenografts, indicating that the mutant ORF23 protein did not have adverse effects in vivo.
Analyses of the subcellular localization of ORF23 by transient-expression methods revealed that the nuclear localization of ORF23 observed in VZV-infected cells was an intrinsic characteristic of ORF23 when expressed independently of other VZV proteins and demonstrated that the SRSRRV motif was a nonconventional nuclear localization signal. These experiments showed that ORF23 differs from its VP26 homolog, which localizes to the nucleus only when VP5, the major capsid protein, is nuclear, as a result of coexpression of VP5 with VP19C or VP22a (1). The importance of the unique ORF23 C terminus for its nuclear localization was suggested by the cytoplasmic retention of ORF23 when it was expressed as a direct RFP fusion protein, which should disrupt the structural conformation of the ORF23 C terminus. ORF23 nuclear localization was preserved when a linker that was predicted to maintain the structural integrity of the C terminus was inserted between ORF23 and RFP sequences. The nuclear localization function of ORF23 was mapped to residues SRSRVV (positions 229 to 234) in the extreme C terminus. These residues comprise a motif compatible with general criteria for nuclear localization signals, which have abundant positively charged residues and a cluster of basic residues preceded by a helix-breaking residue, and are typically hexapeptides with at least four basic residues and neither acidic nor bulky residues (http://cubic.bioc.columbia.edu/papers/2000_nls/paper.html).
Considered together, the transient-expression and pOKA-BAC mutagenesis experiments indicate that the nonconserved ORF23 C terminus confers a novel nuclear localizing capacity and that this function is compensated for in cultured cells to a variable extent depending on the cell type but is required for the pathogenesis of VZV skin infection. To investigate possible mechanisms for this essential role of ORF23 in vivo, we examined the intracellular trafficking of the ORF20, ORF33.5, ORF40, and ORF41 putative VZV capsid proteins using GFP fusion constructs. ORF20 did not undergo nuclear translocation, whereas the homolog, HSV-1 VP19C (UL38), does (19). Thus, VZV ORF23 has nuclear localization capacity not shared by its homologs and ORF20 lacks this function, whereas its homologs have it. However, the ORF33.5 scaffold protein exhibited the nuclear localization phenotype of its homolog, preVP22a (UL26.5) (15). ORF40 and ORF41 were exclusively cytoplasmic when expressed in the absence of other VZV proteins, which is similar to the cytoplasmic retention of the related proteins, VP5 (UL19) and VP23 (UL18) (19).
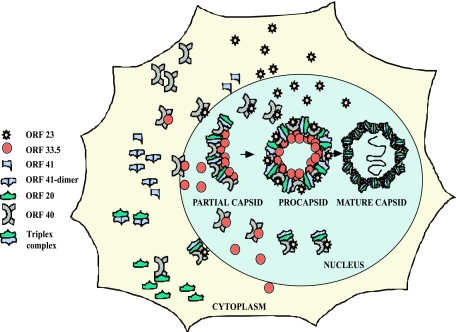
As the major VZV capsid protein, ORF40 must be transported into the nuclei of VZV-infected cells. We found that both ORF23 and ORF33.5 have the capacity to mediate ORF40 translocation. HSV-1 VP26 (UL35) does not translocate the major capsid protein, VP5; in HCMV, SCP and the large capsid protein also remain cytoplasmic when coexpressed (11, 19). However, preVP22a (UL26.5), like its VZV homolog, ORF33.5, has this capacity (15). Nevertheless, ORF23 expression appeared to result in more efficient translocation of ORF40 than ORF33.5 expression, based on coimmunoprecipitation experiments. As predicted, ORF40 nuclear translocation by ORF23 required the intact SRSRRV nuclear localization signal at residues 229 to 234, and the expression of ORF23 as an RFP fusion protein resulted in the cytoplasmic retention of most ORF40 protein. Failure of the ORF23-RFP fusion protein to translocate ORF40 to the nucleus in the transient-expression system suggests that the inability to recover virus from transfections using the pOKA-BAC23mRFP construct was due to lack of ORF40 nuclear transfer in the presence of the aberrant OR23 fusion protein, whereas the pOKA-BACmRFPΔ23 construct yielded infectious virus. We speculate that ORF33.5 can transport sufficient ORF40 into the nucleus to allow VZV replication in the absence of ORF23 in vitro. Thus, ORF33.5 binding to ORF40, which would be necessary for its probable scaffold protein function, may provide a redundant mechanism that moves the ORF40 major capsid protein into the nuclei in cultured cells. This mechanism appears to be more or less robust depending upon the cell type, since the consequences of deleting ORF23 were pronounced in melanoma cells but not discernible in human fibroblasts. However, we propose that this alternative ORF33.5 mechanism for ORF40 nuclear import is not sufficient for VZV pathogenesis in differentiated human skin, since pOKA-BACmRFPΔ23 did not replicate in vivo. The intracellular localization patterns of the VZV capsid proteins that we evaluated and parallels drawn from steps in capsid assembly that have been established for other herpesviruses provide some information relevant for modeling VZV capsid assembly (Fig. 13). In VZV, ORF23 and ORF33.5 are the two proteins among the putative capsid proteins that have nuclear localizing capacity. Both of these proteins can translocate the ORF40 major capsid protein into the nucleus. However, neither ORF23 nor ORF33.5 mediated the transport of ORF20 or ORF41 from the cytoplasm into the nucleus. In HSV-1, the ORF20 and ORF41 homologs, which are VP19C (UL38) and VP23 (UL18), form a trimeric complex that is transported into the nucleus only when bound to VP5, the major capsid protein. VZV assembly may occur by formation of a similar trimeric complex, consisting of ORF20 and ORF41 bound to ORF40 in the cytoplasm. According to the model proposed, this larger complex may then be transferred into the nucleus for the final assembly of capsomeres through ORF40 binding to either ORF23 or ORF33.5. The fact that deleting ORF23 is lethal for skin infection suggests a model in which nuclear transport of the larger complex requires ORF23 to support VZV pathogenesis in the human host in vivo. If so, the inhibition of ORF23 expression could be a target for a novel antiviral drug against VZV.
FIG. 13.
Model of events in VZV capsid assembly. ORF23 and ORF33.5 are the two capsid proteins that have nuclear localization signals. Both of these proteins can translocate ORF40, the major capsid protein, into the nucleus. Neither ORF23 nor ORF 33.5 can translocate the ORF20 and ORF41 capsid proteins into the nucleus. Events in the cytoplasm depict single representative interactions. Since the HSV homologs form a trimeric complex, this model proposes that capsid assembly occurs by interaction of this trimeric complex with ORF40, which is transferred into the nucleus by either ORF23 or ORF33.5.
Acknowledgments
We thank Nikolaus Osterrieder (Department of Microbiology and Immunology, Cornell University, Ithaca, NY) for kindly providing us the necessary reagents for red recombination mutagenesis, Jens Von Einem for his help with technical queries related to BAC mutagenesis, Valerie G. Preston (MRC, Virology Unit, Glasgow, United Kingdom) for providing us with rabbit anti-ORF33.5 antibody, and Mike Reichelt, Stefan Oliver, and Nafisa Ghori for their technical assistance and advice.
This work was supported by NIH grants AI20459 and AI053846.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Booy, F. P., B. L. Trus, W. W. Newcomb, J. C. Brown, J. F. Conway, and A. C. Steven. 1994. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc. Natl. Acad. Sci. USA 915652-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borst, E. M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 751450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, J. I., S. E. Straus, and A. M. Arvin. 2007. Varicella-zoster virus, p. 2773-2818. In D. H. Knipe, D. E. Griffin, R. A. Lamb, S. E. Straus, P. M. Howley, M. A. Martin, and B. Roizman (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 5.del Rio, T., T. H. Ch'ng, E. A. Flood, S. P. Gross, and L. W. Enquist. 2005. Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J. Virol. 793903-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, P., J. C. Akpa, and S. Person. 2003. Residues of VP26 of herpes simplex virus type 1 that are required for its interaction with capsids. J. Virol. 77391-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247115-124. [DOI] [PubMed] [Google Scholar]
- 8.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 727563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 101044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kut, E., and D. Rasschaert. 2004. Assembly of Marek's disease virus (MDV) capsids using recombinant baculoviruses expressing MDV capsid proteins. J. Gen. Virol. 85769-774. [DOI] [PubMed] [Google Scholar]
- 11.Lai, L., and W. J. Britt. 2003. The interaction between the major capsid protein and the smallest capsid protein of human cytomegalovirus is dependent on two linear sequences in the smallest capsid protein. J. Virol. 772730-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallory, S., M. Sommer, and A. M. Arvin. 1998. Analysis of the glycoproteins I and E of varicella-zoster virus (VZV) using deletional mutations of VZV cosmids. J. Infect. Dis. 178(Suppl. 1)S22-S26. [DOI] [PubMed] [Google Scholar]
- 13.McNabb, D. S., and R. J. Courtney. 1992. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J. Virol. 662653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffat, J. F., M. D. Stein, H. Kaneshima, and A. M. Arvin. 1995. Tropism of varicella-zoster virus for human CD4t+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J. Virol. 695236-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232499-511. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson, P., C. Addison, A. M. Cross, J. Kennard, V. G. Preston, and F. J. Rixon. 1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J. Gen. Virol. 751091-1099. [DOI] [PubMed] [Google Scholar]
- 17.Niizuma, T., L. Zerboni, M. H. Sommer, H. Ito, S. Hinchliffe, and A. M. Arvin. 2003. Construction of varicella-zoster virus recombinants from parent Oka cosmids and demonstration that ORF65 protein is dispensable for infection of human skin and T cells in the SCID-hu mouse model. J. Virol. 776062-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preston, V. G., J. Kennard, F. J. Rixon, A. J. Logan, R. W. Mansfield, and I. M. McDougall. 1997. Efficient herpes simplex virus type 1 (HSV-1) capsid formation directed by the varicella-zoster virus scaffolding protein requires the carboxy-terminal sequences from the HSV-1 homologue. J. Gen. Virol. 781633-1646. [DOI] [PubMed] [Google Scholar]
- 19.Rixon, F. J., C. Addison, A. McGregor, S. J. Macnab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 772251-2260. [DOI] [PubMed] [Google Scholar]
- 20.Sheaffer, A. K., W. W. Newcomb, J. C. Brown, M. Gao, S. K. Weller, and D. J. Tenney. 2000. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 746838-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommer, M. H., E. Zagha, O. K. Serrano, C. C. Ku, L. Zerboni, A. Baiker, R. Santos, M. Spengler, J. Lynch, C. Grose, W. Ruyechan, J. Hay, and A. M. Arvin. 2001. Mutational analysis of the repeated open reading frames, ORFs 63 and 70 and ORFs 64 and 69, of varicella-zoster virus. J. Virol. 758224-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer, J. V., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 1998. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J. Virol. 723944-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 24.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. USA 8911508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 21026-1030. [DOI] [PubMed] [Google Scholar]