Abstract
The human germ cell tumor line Tera-1 produces retroviral particles which are encoded by the human endogenous retrovirus family HERV-K(HML-2). We show here, by quantitative reverse transcriptase PCR, that HML-2 gag and env RNA transcripts are selectively packaged into Tera-1 retroviral particles, whereas RNAs from cellular housekeeping genes and from other HERV families (HERV-H and HERV-W) are nonselectively copackaged. Assignment of cloned HML-2 gag and env cDNAs from Tera-1 retroviral particles to individual HML-2 loci in the human genome demonstrated that HML-2 RNA transcripts packaged into Tera-1 retroviral particles originate almost exclusively from an HML-2 provirus on chromosome 22q11.21. Based on relative cloning frequencies, this provirus was the most active among a total of eight transcribed HML-2 loci identified in Tera-1 cells. These data suggest that at least one HML-2 element, that is, the HML-2 provirus on 22q11.21, has retained the capacity for packaging RNA into HML-2-encoded retroviral particles. Given its elevated transcriptional activity and the presence of a full-length Gag open reading frame, the 22q11.21 HML-2 provirus may also significantly contribute to Gag protein and thus particle production in Tera-1 cells. Our findings provide important clues to the generation and biological properties of HML-2-encoded particles. In addition, copackaging of non-HML-2 HERV transcripts in HML-2-encoded particles should inform the debate about endogenous retroviral particles putatively encoded by non-HML-2 HERV families that have previously been described for other human diseases, such as multiple sclerosis.
The human genome contains a relatively large fraction (∼8%) of sequences that resemble retrovirus sequences. Such elements, named human endogenous retroviruses (HERVs), represent traces of ancestral germ line infections by active retroviruses, which have thereafter been transmitted in a Mendelian manner (1, 14). Due to the acquisition of numerous mutations, deletions, or truncations, most HERVs are nonfunctional. However, some HERVs have retained the capacity to code for functional proteins or even retrovirus-like particles (RVLP) (6, 15, 34, 36). The HERV-K(HML-2) family, in short HML-2, appears to play an exceptional role among the approximately 30 HERV families present in the human genome (26). HML-2 is the most “active” HERV family, as suggested by HML-2 proviral insertions which formed after the divergence of humans and chimpanzees and by some HML-2 proviruses still being polymorphic within the human population (2, 4, 24, 39, 51). Furthermore, several proviral HML-2 loci are almost or completely intact, that is, they contain full-length open reading frames (ORFs) for Gag, Pro, Pol, and Env proteins (3, 38, 50, 51). However, although engineered HML-2 consensus sequences or a chimeric construct of three recombined HML-2 proviruses have been shown to be infectious and to form new proviruses, a fully replication-competent “natural” HML-2 allele could not be identified (16, 30). HERVs have been suspected to play a role in human pathologies (e.g., cancer or autoimmunity), and evidence suggests that HML-2 may possibly be involved in the development of germ cell tumors (8, 21, 46).
HML-2 is currently the only HERV family that has conclusively been shown to be capable of producing RVLP (10, 49). Nevertheless, an ability for particle production has also been proposed for the HERV-H and HERV-W families, which have previously been implicated in the chronic inflammatory and demyelinating neurological disease multiple sclerosis (12, 28). Since HML-2-encoded RVLP have especially been observed in cell lines derived from human germ cell tumors (11, 29), they were originally designated human teratocarcinoma-derived virus particles (HTDV) but were renamed HTDV/HERV-K after they were found to be encoded by HERV-K(HML-2) (9, 34). HTDV/HERV-K RVLP are recognized by anti-HML-2 Gag sera in immuno-electron microscopy (5, 9, 32), harbor reverse transcriptase (RT) activity (31, 33), and were shown to contain HML-2 pol RNA when expressed in a recombinant baculovirus expression system (49). The RNA content of HTDV/HERV-K RVLP and the packaging of HML-2 RNA into those RVLP have not been investigated to a greater extent so far. In addition, because HML-2 is a multicopy family that comprises about 50 full-length proviral HML-2 loci in the human genome (14, 44), the question arises as to which HML-2 proviruses are transcriptionally active in germ cell tumor cell lines and may thus be responsible for RVLP production or represent the origin of HML-2 RNA in HTDV/HERV-K RVLP.
In this work, we show that there is a selective packaging of HML-2 gag and env RNA transcripts into HML-2-encoded RVLP produced by the human germ cell tumor cell line Tera-1. In contrast, RNAs from cellular housekeeping genes and RNAs from other HERV families are nonselectively copackaged. We furthermore identify the proviral origin of HML-2 transcripts packaged into Tera-1 RVLP, as well as transcriptionally active HML-2 proviruses in Tera-1 cells. Our findings provide important clues to the generation and biological properties of HML-2-encoded RVLP in a human germ cell tumor cell line. Additionally, they may also have implications for endogenous RVLP supposedly encoded by non-HML-2 HERV families that have been described for other human diseases, such as multiple sclerosis.
MATERIALS AND METHODS
Cells.
Tera-1 cells (ATCC HTB-105), derived from a lung metastasis of a human embryonal testicular carcinoma (20), were cultured in McCoy's 5A modified medium (Sigma) containing 10% fetal calf serum (FCS; PAA), penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were maintained at 37°C in a 5% CO2 atmosphere.
Antibodies, IP, immunoblotting, and immunocytochemistry.
Generation of polyclonal anti-HML-2 Gag rabbit sera by immunization of rabbits with Escherichia coli anthranilate synthetase (TrpE) HML-2 Gag fusion proteins was previously described (42, 47). For immunoprecipitation (IP) of HML-2 Gag, confluent Tera-1 cells were scraped from two 175-cm2 tissue culture flasks and washed in phosphate-buffered saline (PBS). As an input control, 10% of the cells were lysed in sample buffer (125 mM Tris-HCl [pH 6.8], 6% [wt/vol] sodium dodecyl sulfate, 10% [vol/vol] mercaptoproprandiol, 10% [vol/vol] glycerol), sonicated, and stored at −70°C. The remaining cells were resuspended in IP lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1% [vol/vol] Triton X-100, 1 mM dithiothreitol, aprotinin [1 μg/ml], phenylmethylsulfonyl fluoride [0.8 mM]) and lysed for 1 h on ice. Following centrifugation, IP buffer lysates (1 ml) were incubated with 50 μl of either anti-HML-2 Gag serum or the corresponding preimmune serum for 1 h at 4°C. After the addition of protein G Sepharose beads (GE Healthcare) and incubation on a rotator for 2 h at 4°C, beads were washed three times with IP lysis buffer and once with PBS and then resuspended in sample buffer. Samples were boiled for 5 min and centrifuged, and supernatants were stored at −70°C until further processing. For immunoblot analyses, protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were probed with anti-HML-2 Gag rabbit serum and developed with secondary peroxidase-labeled immunoglobulin G antibodies (Sigma) and enhanced chemiluminescence. In competition experiments, the anti-HML-2 Gag serum was preadsorbed overnight with either bacterially expressed TrpE-HML-2 Gag fusion protein or TrpE alone (22). For immunocytochemistry, Tera-1 cells were plated at 4 × 105 cells per well (growth area, 0.9 cm2 per well) on microscope slides, using flexible eight-well tissue culture chambers attached to the slides (flexiPerm; Greiner Bio-One). The following day, cells were fixed with 4% (vol/vol) paraformaldehyde in PBS, permeabilized with 0.2% (vol/vol) Triton X-100 in PBS, blocked with 10% (vol/vol) FCS in PBS, and stained with anti-HML-2 Gag rabbit serum or the corresponding preimmune serum diluted 1:200 in PBS with 10% FCS. Following incubation with secondary antibody (goat anti-rabbit immunoglobulin G-Alexa Fluor 568 diluted 1:1,000; Molecular Probes), cells were dehydrated in 70%, 80%, and 100% (vol/vol) ethanol and mounted with Vectashield (Vector) medium containing 4′,6′-diamidino-2-phenylindole (DAPI). Slides were inspected with a Leitz Aristoplan fluorescence microscope, and images were acquired using AxioVision 3.0 software. Digital acquisition parameters and further processing (Corel Photo-Paint 12) were identical for all images.
Isolation of RNA and protein from Tera-1 cells and retroviral particles.
Total RNA was prepared from 5 × 106 Tera-1 cells by use of an RNeasy Mini kit (Qiagen) and was eluted in 60 μl of double-distilled H2O. Tera-1 cell RNAs were quantitated by measuring the A260. For isolation of Tera-1 RVLP, supernatants from two 175-cm2 tissue culture flasks in which Tera-1 cells had been grown to confluence for 4 to 6 days were collected and cleared of cellular debris by three subsequent rounds of low-speed centrifugation (1,200 × g, 10 min) and filtration through 0.45-μm-pore-size filters. Filtered supernatants were layered over a cushion of 5 ml of 30% glycerol in PBS (vol/vol) in Ultra-Clear centrifuge tubes (Beckman) and ultracentrifuged using a Beckman SW28 rotor (100,000 × g for 3 h at 4°C) in a Centrikon T-2060 (Kontron) ultracentrifuge. Throughout this work, we refer to the material obtained by this procedure as pelletable RVLP (pRVLP). Protein extracts from pRVLP were generated by resuspending pellets in sample buffer and were stored at −70°C. For purification of encapsidated RNA from pRVLP, pellets were digested with 30 U of RNase One (Promega) for 45 min at 37°C in order to remove potentially contaminating nonencapsidated RNAs (48). RNAs were then extracted from virions with a QIAamp viral RNA mini kit (Qiagen), eluted in 50 μl of double-distilled H2O, and stored at −70°C until further processing. Note that the presence of excess amounts of carrier RNA required for the purification procedure did not permit us to measure the amount of RNA extracted from Tera-1 pRVLP. To verify the absence of reagent contaminations, mock RNA purifications were carried out in parallel from ultracentrifuged Tera-1 cell culture medium that had not been in contact with cells.
Conventional RT-PCR.
Contaminating DNA was removed from Tera-1 cell and pRVLP RNAs by use of a Turbo DNA-free kit (Ambion Inc.) by following the protocol for rigorous DNase treatment. In brief, 2 units of Turbo DNase was added to a 50-μl reaction mix containing 10 μg of cellular RNA (or 40 μl of Tera-1 pRVLP RNA) and incubated for 30 min at 37°C, when another 2 units of Turbo DNase was added and incubation was continued for 30 min at 37°C. After removal of DNase by use of 10 μl of the provided DNase inactivation reagent, 1 μg of DNase-digested cellular RNA (or 11 μl of Tera-1 pRVLP RNA) was reverse transcribed in a 20-μl reaction mix with Superscript II (Invitrogen) and 25 μM random hexamer primers (MWG-Biotech AG). Negative controls were generated in parallel for each sample by omitting Superscript II from the reaction mix. Genomic DNA from Tera-1 cells was extracted with a QIAamp DNA blood mini kit (Qiagen). Conventional RT-PCR was performed in a 50-μl reaction mix containing 1 μl of cDNA or ∼50 ng of genomic DNA, 0.5 μM of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, reaction buffer (10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2), and 0.05 units/μl of Taq DNA polymerase (D1806; Sigma). Cycling parameters were as follows: 5 min at 94°C; 40 cycles of 45 s at 94°C, 45 s at 57°C, and 1 min at 72°C; and 10 min at 72°C. Amplification of the chromosome 22q11.21 HML-2 locus from genomic Tera-1 DNA was carried out using the Expand long-template system (Roche) with buffer 1 according to the instructions of the manufacturer. A list of PCR primers used in this study is provided in Table S1 in the supplemental material.
qRT-PCR.
Quantitative real-time RT-PCR (qRT-PCR) was carried out on a LightCycler 1.5 instrument (Roche). For qRT-PCR analysis, cDNAs and control RNAs not submitted to RT were diluted (1:20 for Tera-1 cells and 1:10 for Tera-1 pRVLP) in nuclease-free water (Roche), aliquoted, and stored at −20°C. A new aliquot was used for each qRT-PCR run. Tera-1 cell and pRVLP cDNAs were analyzed in duplicate in 20-μl reaction mixtures containing 1 μM of each primer and reagents provided in a LightCycler FastStart DNA MasterPLUS SYBR green I kit (Roche) according to the manufacturer's protocol. An aliquot of a calibrator sample containing a fixed amount of cDNA was included in each qRT-PCR run to control for interassay variability. Cycling parameters were as follows: 10 min at 95°C and 45 cycles of 10 s at 95°C, 3 s at 60°C, and extension at 72°C for various times depending on amplicon length. A melting curve analysis was performed after each qRT-PCR run. The correct sizes of the amplicons and the absence of contaminations in RT-negative controls were also verified by agarose gel electrophoresis. Extensive setup experiments were performed for each qRT-PCR assay; in case these revealed the presence of primer dimers, SYBR green fluorescence was acquired during an additional step following the extension step at a temperature above the melting peak of the primer dimers but below that of the specific PCR product.
Calculation of normalized relative encapsidation efficiencies.
Relative expression levels of different targets were quantitated by determining the threshold cycle (CT) for each target by qRT-PCR with cDNA from either Tera-1 cells or pRVLP. Raw CT values were further processed by the formula 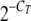 × 1012 (where CT is the threshold cycle of the analyzed gene and values were multiplied by 1012 to obtain more convenient numbers). Relative expression of each target was measured in the cDNA equivalent of 12.5 ng total RNA from Tera-1 cells. Since we could not measure the amount of RNA extracted from Tera-1 pRVLP (see above), we did not know the absolute amount of total RNA from Tera-1 pRVLP, for whose cDNA equivalent relative expression of each target was determined. As an index of the tendencies of individual RNAs to become packaged into Tera-1 pRVLP (“encapsidation efficiency”), independent of the absolute amounts of RNA analyzed, we therefore calculated “normalized relative encapsidation efficiencies.” To this end, we first calculated the means of the expression levels of each analyzed target gene in Tera-1 cell RNAs from three to five independent experiments. Those values were considered to represent the constitutive expression of each gene in Tera-1 cells. We then divided the expression level of a given target gene in RNAs extracted from Tera-1 pRVLP by the corresponding constitutive expression level of that gene in Tera-1 cell RNA. The obtained ratios were subsequently normalized to the ratio of HERV-K env, which was arbitrarily set at 1. Note that the normalized relative encapsidation efficiency depends only on the relative abundances of the different individual RNAs in the total RNA samples from Tera-1 cells and Tera-1 pRVLP and can thus be calculated without knowing the absolute amounts of RNA analyzed (as was the case for Tera-1 pRVLP RNA).
× 1012 (where CT is the threshold cycle of the analyzed gene and values were multiplied by 1012 to obtain more convenient numbers). Relative expression of each target was measured in the cDNA equivalent of 12.5 ng total RNA from Tera-1 cells. Since we could not measure the amount of RNA extracted from Tera-1 pRVLP (see above), we did not know the absolute amount of total RNA from Tera-1 pRVLP, for whose cDNA equivalent relative expression of each target was determined. As an index of the tendencies of individual RNAs to become packaged into Tera-1 pRVLP (“encapsidation efficiency”), independent of the absolute amounts of RNA analyzed, we therefore calculated “normalized relative encapsidation efficiencies.” To this end, we first calculated the means of the expression levels of each analyzed target gene in Tera-1 cell RNAs from three to five independent experiments. Those values were considered to represent the constitutive expression of each gene in Tera-1 cells. We then divided the expression level of a given target gene in RNAs extracted from Tera-1 pRVLP by the corresponding constitutive expression level of that gene in Tera-1 cell RNA. The obtained ratios were subsequently normalized to the ratio of HERV-K env, which was arbitrarily set at 1. Note that the normalized relative encapsidation efficiency depends only on the relative abundances of the different individual RNAs in the total RNA samples from Tera-1 cells and Tera-1 pRVLP and can thus be calculated without knowing the absolute amounts of RNA analyzed (as was the case for Tera-1 pRVLP RNA).
Cloning of HERV-K(HML-2) gag and env cDNAs and assignment to distinct proviral loci.
Primer pairs HERV-K gag+/gag− and HERV-K env 8146/env 8665 were used to amplify cDNAs from Tera-1 cells and pRVLP. PCR products were excised from agarose gels, purified (NucleoSpin Extract II; Macherey-Nagel), and ligated into the pGEM-T vector (Promega). Plasmid DNAs from randomly selected insert-containing clones were purified with a QIAprep miniprep kit (Qiagen) and sequenced on an Applied Biosystems 3730x capillary sequencer, using vector-specific primers (Institut für Immunologie und Genetik, Kaiserlautern, Germany). Sequences were analyzed using either BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat; March 2006 human genome assembly) or the in-house Bio-Python script Locus-Assigner, with the latter basically performing pairwise sequence comparisons between cDNA and reference sequences and cataloging numbers of differences between cDNA and reference sequences. Assignment of cDNA sequences to corresponding HML-2 proviruses is based on characteristic nucleotide differences between the various HML-2 proviruses. The proviral HML-2 locus with no or very few nucleotide mismatches to an HML-2 cDNA sequence can be assumed to represent the origin of this cDNA if all alternative loci display more nucleotide differences. As reference sequences for the assignment of HML-2 gag and env cDNA sequences, HML-2 proviral loci were collected from the human genome sequence (March 2006 assembly) at the Human Genome Browser, using the HERV-K(HML-2.HOM) sequence as a probe for BLAT searches. Retrieved sequences were aligned using DiAlign (40) and MAFFT (25), and the alignment was subsequently manually optimized. Subregions of the alignment were used for further analyses. The sequence assignment strategy and the Locus-Assigner software were previously discussed in more detail (19). cDNA sequences with more than 17 mismatches to the best-matching reference proviral sequence were excluded from further analysis because they very likely represent recombined cDNAs from different proviral transcripts that arose ex vivo during cDNA generation (18).
RESULTS
Expression of HML-2 Gag protein and particles by Tera-1 cells.
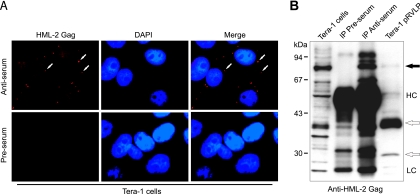
Before analyzing the RNA content of Tera-1 pRVLP, we aimed to reconfirm the production of HML-2 Gag protein and particles by the human germ cell tumor cell line Tera-1, which was used in this investigation. Immunocytochemistry with a polyclonal anti-HML-2 Gag rabbit serum revealed a dot-like staining pattern in Tera-1 cells (Fig. 1A), possibly representing groups of RVLP, as described before (9). Such immunoreactivity was not seen when Tera-1 cells were stained with the corresponding preimmune serum. Additionally, immunoblots of Tera-1 cell lysates demonstrated expression of an 80-kDa HML-2 Gag precursor protein which could be immunoprecipitated with the HML-2 Gag antiserum but not the corresponding preimmune serum (Fig. 1B). In protein lysates from ultracentrifuged Tera-1 cell supernatants, we observed a prominent band of about 39 kDa and a faint band of approximately 30 kDa, both corresponding to processed HML-2 Gag proteins. Preadsorption of the HML-2 Gag antibody with bacterially expressed HML-2 Gag protein abolished the 80-, 39-, and 30-kDa Gag protein bands, further confirming their identity (data not shown). Taken together with former observations from others and ourselves (5, 9, 22, 47), these data suggest that Tera-1 cells constantly produce HML-2 Gag protein and that the pelletable supernatant material from Tera-1 cells contains HML-2-encoded RVLP.
FIG. 1.
Analysis of HML-2 Gag protein expression in Tera-1 cells and pRVLP. (A) Immunocytochemistry was performed on fixed and permeabilized Tera-1 cells, using an HML-2 Gag-specific polyclonal rabbit serum (Anti-serum) or the corresponding preimmune serum (Pre-serum) as a control. Nuclei were stained with DAPI. Arrows indicate immunoreactivity for HML-2 Gag. Magnification, ×1,000. (B) Immunoblot analysis of HML-2 Gag protein in Tera-1 cells and pRVLP. The blot was probed with anti-HML-2 Gag polyclonal rabbit serum. Tera-1 cells, protein lysate of Tera-1 cells; IP Pre-serum, protein lysate of Tera-1 cells immunoprecipitated with HML-2 Gag preimmune serum; IP Anti-serum, protein lysate of Tera-1 cells immunoprecipitated with anti-HML-2 Gag antiserum; Tera-1 pRVLP, protein lysate of the pellet from an ultracentrifuged supernatant of Tera-1 cells. Filled arrow, position of HML-2 Gag precursor protein; open arrows, positions of processed HML-2 Gag proteins; HC and LC, positions of immunoglobulin heavy and light chains in the IP lanes.
Detection of HML-2 transcripts in Tera-1 particle-associated RNA.
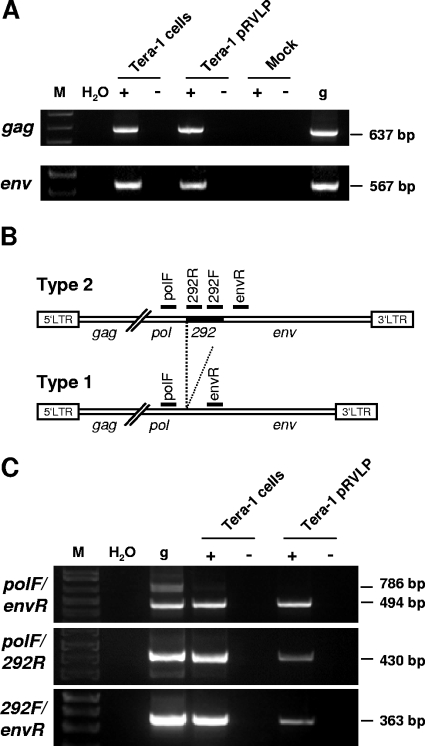
The presence of HML-2 gag and env RNA transcripts in Tera-1 pRVLP was first studied by conventional RT-PCR, using cDNAs generated from the pelleted material obtained by ultracentrifugation of Tera-1 cell supernatants. As shown in Fig. 2A, HML-2 gag and env transcripts were readily detectable in those cDNAs, whereas no amplification products could be observed in controls lacking RT or in mock purifications.
FIG. 2.
Analysis of HML-2 RNA transcripts in Tera-1 cells and pRVLP. (A) RT-PCR for HML-2 gag (primers HERV-K gag+ and gag−) and env (primers HERV-K env 7036 and env 7602) was carried out on total RNAs isolated from Tera-1 cells and pRVLP, which were subjected (+) or not subjected (−) to reverse transcription. Mock RNA purifications were performed as described in Materials and Methods. Amplicon sizes are indicated on the right. g, Tera-1 cell genomic DNA; M, DNA size marker; H2O, PCR negative control. (B) There are two types of HERV-K(HML-2) proviruses in the human genome, characterized by the presence (type 2) or absence (type 1) of a stretch of 292 bp (filled area) at the pol-env junction. Positions and names of the primers used for detection of type 1 and 2 transcripts are indicated. (C) Analysis of HML-2 type 1 and type 2 RNA transcripts in Tera-1 cells and pRVLP. Note that the primer pair polF/envR amplifies a 786-bp fragment from type 2 and a 494-bp fragment from type 1 proviruses.
HML-2 proviruses in the human genome have been classified into two subtypes, termed type 1 and type 2 proviruses. Type 2 proviruses contain a characteristic 292-bp sequence at the pol-env junction, which is absent in type 1 proviruses (Fig. 2B) (32). Using primers that span the 292-bp sequence, transcripts from type 2 proviruses were weakly detectable in Tera-1 cell but not Tera-1 pRVLP RNA (Fig. 2C). While this seemed compatible with an enrichment of type 1 proviral sequences in Tera-1 pRVLP, failure to detect type 2 sequences could also be related to preferential amplification of the shorter type 1 proviral sequences. Indeed, employing two primer pairs, with one primer located within the 292-bp sequence each, type 2 transcripts became detectable in Tera-1 pRVLP RNA, suggesting that HML-2 type 2 transcripts are present in Tera-1 pRVLP (Fig. 2C).
Active packaging of HML-2 RNAs into Tera-1 retroviral particles.
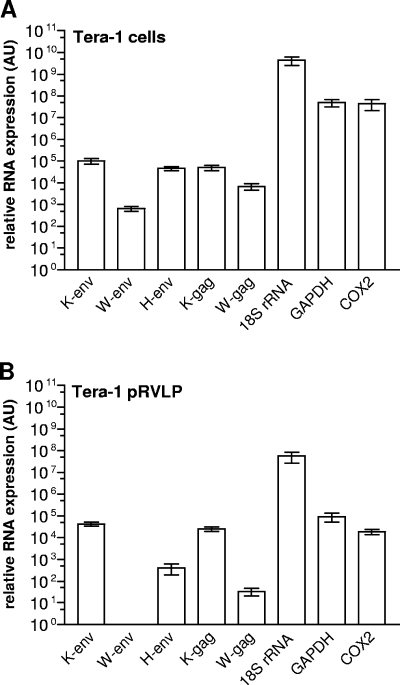
Having established that Tera-1 pRVLP contain HML-2 gag and env RNAs, we asked whether HML-2 transcripts are selectively enriched in Tera-1 pRVLP RNA, as would be expected if HML-2 RNAs were actively packaged in Tera-1 pRVLP (17, 45). To this end, we determined, by qRT-PCR, relative expression levels of HML-2 gag and env in comparison to two cellular housekeeping genes (18S rRNA and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]), a mitochondrially carried gene (cytochrome c oxidase II [COX2]), and transcripts from two other HERV families (HERV-W gag and env and HERV-H env) in total RNAs from Tera-1 cells and Tera-1 pRVLP. Figure 3A represents the relative expression levels (arbitrary units) of these different target genes in total RNA from Tera-1 cells. The much lower expression levels of HERV RNAs than those of 18S rRNA, GAPDH, and COX2 in Tera-1 cells demonstrate that HERV RNAs comprise only a very small proportion of the total RNA pool in Tera-1 cells. RNA expression levels of a given HERV family, as measured in this study, depend on the cumulative transcriptional activity of the different proviruses from each HERV family that are recognized by the PCR primer pairs used for amplification. The exact number of those proviruses is difficult to estimate, in particular because proviruses with minor mismatches to the primers are likely amplified as well but it is hard to predict their precise number. Taking this into account, HML-2 env and gag appeared to be the most abundant among the different HERV RNAs investigated in Tera-1 cells, but expression levels of other endogenous retroviral transcripts (HERV-H env and HERV-W gag) were significant in these cells as well.
FIG. 3.
Relative RNA expression levels of HML-2 env (K-env), HERV-W env (W-env), HERV-H env (H-env), HML-2 gag (K-gag), HERV-W gag (W-gag), 18S rRNA (18S rRNA), GAPDH, and the mitochondrially encoded protein COX2 were analyzed by qRT-PCR in total RNAs extracted from Tera-1 cells (A) and Tera-1 pRVLP (B). The bar charts show the relative abundances (expressed in arbitrary units [AU]) of individual RNAs and represent the means ± standard errors of the means for three to five independent experiments.
Although we could not quantitate the absolute amount of total RNA extracted from Tera-1 pRVLP, RNA yields from Tera-1 pRVLP were obviously lower than those from Tera-1 cells. The expression levels of the different targets in total RNA from Tera-1 pRVLP were therefore lower than those obtained for total RNA from Tera-1 cells (Fig. 3B). However, interestingly, expression levels of individual RNAs in Tera-1 pRVLP appeared to be roughly proportional to their expression levels in Tera-1 cells. There were two remarkable exceptions, though. Levels of HERV-K gag and env in Tera-1 pRVLP were almost as high as those in Tera-1 cells, consistent with active packaging of those RNAs into Tera-1 pRVLP. To better assess the relative tendency of each of the investigated RNA targets to become packaged into Tera-1 pRVLP, we calculated their normalized relative encapsidation efficiencies, which provide an index of the encapsidation efficiencies of individual RNAs relative to the encapsidation efficiency of HERV-K env (for details, see Materials and Methods). As summarized in Table 1, the normalized relative encapsidation efficiencies of HML-2 gag and env RNAs were strikingly similar to each other and substantially higher than those of all other analyzed RNA species. Normalized relative encapsidation efficiencies of HML-2 gag and env RNAs were elevated at least ∼35-fold (compared to 18S rRNA) and maximally ∼620-fold (compared to COX2). While no normalized relative encapsidation efficiency could be calculated for HERV-W env, as it was not detectable in Tera-1 pRVLP, normalized relative encapsidation efficiencies of HERV-H env and HERV-W gag were largely in the same range as those of the cellular 18S rRNA and GAPDH RNAs. The very low normalized relative encapsidation efficiency of COX2 RNA may indicate that this mitochondrially encoded RNA tends to be excluded from Tera-1 pRVLP. In summary, these data strongly suggest that HML-2 RNAs are selectively packaged into Tera-1 pRVLP. In contrast, RNAs from cellular housekeeping genes as well as from non-HML-2 HERV families seem to become nonselectively copackaged into those particles.
TABLE 1.
Normalized relative encapsidation efficiencies of individual RNAs analyzed in this studya
| RNA | Normalized relative encapsidation efficiency (mean ± SEM) | Mean fold change relative to HERV-K env efficiency |
|---|---|---|
| HERV-K env | 1 | 1 |
| HERV-K gag | 1.1 ± 0.19 | 0.9 |
| HERV-H env | 1.99 × 10−2 ± 8.05 × 10−3 | −50.4 |
| HERV-W env | NA | NA |
| HERV-W gag | 1.2 × 10−2 ± 4.04 × 10−3 | −83.6 |
| 18S rRNA | 2.9 × 10−2 ± 1.31 × 10−2 | −34.5 |
| GAPDH | 4.41 × 10−3 ± 7.53 × 10−4 | −227 |
| COX2 | 1.61 × 10−3 ± 6.78 × 10−4 | −622.1 |
Normalized relative encapsidation efficiencies were calculated by dividing expression levels of individual RNAs in Tera-1 pRVLP by their expression levels in Tera-1 cells. The obtained ratios were subsequently normalized to the ratio of HERV-K env, which was arbitrarily set at 1 (for details, see Materials and Methods). Values are presented as means ± standard errors of the means for three to five independent experiments. NA, due to the absence of HERV-W env RNA from Tera-1 pRVLP, calculation of an encapsidation efficiency was not applicable for that target.
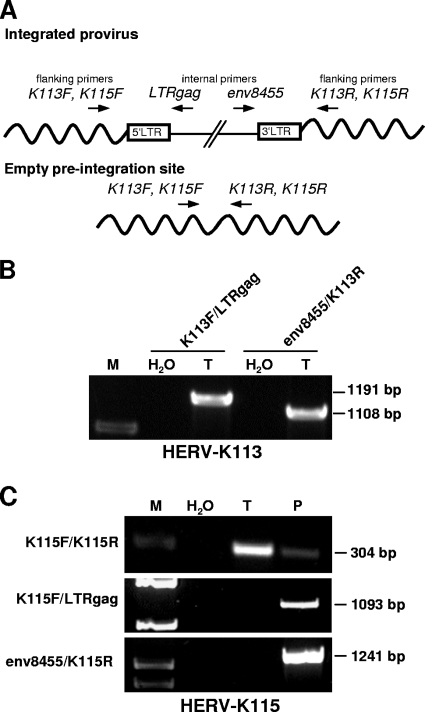
Tera-1 cell genomes harbor the HERV-K113 but not the HERV-K115 provirus.
Several HML-2 insertions are known to be polymorphic in the human population, that is, those HML-2 proviruses are not present in all human individuals (24, 41, 51). The polymorphic HERV-K113 provirus cloned into a baculovirus expression vector is capable of producing retroviral particles similar to those observed in germ cell tumor cell lines. It has therefore been proposed as a candidate provirus for particle production in germ cell tumor cell lines (10). Besides HERV-K113, which harbors full-length ORFs for all retroviral proteins, the polymorphic HERV-K115 provirus also carries full-length ORFs for Gag and Env (51). We therefore determined whether the genomes of Tera-1 cells harbor HERV-K113 and/or HERV-K115, using PCR primer pairs located within the proviruses and their flanking genomic DNAs (Fig. 4A). As shown in Fig. 4B and C, Tera-1 cells were found to harbor HERV-K113 but not HERV-K115. Thus, for the identification of transcriptionally active HML-2 proviruses in Tera-1 cells (see below), the HERV-K113 sequence (GenBank accession no. AY037928.1), which is absent in the March 2006 human genome assembly (27), was included in the list of HML-2 reference proviruses.
FIG. 4.
(A) PCR strategy for detection of HERV-K113 and HERV-K115 proviruses or empty preintegration sites in genomic DNA from Tera-1 cells. Locations, orientations, and names of flanking and internal primers used for detection of HERV-K113 and HERV-K115 are indicated. (B) PCR was performed on Tera-1 cell genomic DNA (T), using the indicated primer pairs, for detection of HERV-K113. Amplicon sizes are given on the right. M, DNA size marker; H2O, PCR negative control. (C) The presence of HERV-K115 was analyzed in genomic DNA from Tera-1 cells (T) and a positive control genomic DNA (P) from peripheral blood mononuclear cells of a HERV-K115-positive human individual. Detection of the HERV-K115 empty preintegration site in the positive control DNA indicates heterozygosity.
Assignment of HML-2 gag and env transcripts from Tera-1 cells and retroviral particles to HML-2 proviral loci.
The strategy for the detection of transcriptionally active HML-2 proviruses in Tera-1 cells consisted of the amplification of HML-2 gag and env transcripts from Tera-1 cells by RT-PCR, using primer pairs located within the gag and env genes, and subsequent assignment of a number of cloned and sequenced HML-2 gag and env cDNAs to distinct HML-2 proviral loci, taking advantage of characteristic nucleotide differences between different HML-2 proviruses. By this approach, a total of eight transcriptionally active HML-2 proviruses were identified in Tera-1 cells (Table 2). Three of these proviruses (located on 5q33.3, 7p22.1, and 22q11.21) were detected by both HML-2 gag and env transcripts, whereas the other five proviruses were detected by only one type of transcript. The fact that we could not detect both gag and env transcripts from some HML-2 proviruses might be related to a somewhat different spectrum of HML-2 proviruses recognized by the employed HML-2 gag and env PCR primers.
TABLE 2.
Proviral origins and relative cloning frequencies of HML-2 gag and env cDNAs from Tera-1 cells and pRVLPa
| HML-2 provirus chromosomal location (biblographic name) | Provirus ID | Relative cloning frequency (%)b
|
Type | Presence of full-length ORFs
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| HML-2 gag
|
HML-2 env
|
||||||||
| Tera-1 cells | Tera-1 pRVLP | Tera-1 cells | Tera-1 pRVLP | Gag | Env | Rec | |||
| 1q22 (HERV-K102) | c1_B | 16.3 | 1 | (+) | |||||
| 3q13.2 | c3_B | 4.2 | 1 | + | |||||
| 5q33.3 (HERV-K10, HERV-K107) | c5_A | 4.2 | 5 | 4.7 | 1 | + | |||
| 6q14.1 (HERV-K109) | c6_A | 2.3 | 2 | + | + | + | |||
| 7p22.1 (HML-2.HOM, HERV-K108, HERV-K[C7]) | c7_A | 16.7 | 5 | 20.9 | 2 | + | + | + | |
| 11q22.1 | c11_A | 2.3 | 2 | + | + | ||||
| 11q23.3 | c11_B | 25 | 1 | ||||||
| 22q11.21 (HERV-K101) | c22_A | 50 | 90 | 53.5 | 100 | 1 | + | ||
The chromosomal locations as well as bibliographic names of transcribed HML-2 proviruses identified in this study in Tera-1 cells and pRVLP are given in the first column. The provirus identifiers indicate the designations of the different HML-2 loci, as used in reference 19. Relative cloning frequencies of HML-2 proviruses (%) in cDNAs from Tera-1 cells and pRVLP are listed separately for gag and env transcripts. Type 1/2 indicates the presence (type 2) or absence (type 1) of a 292-bp sequence at the pol-env boundary. Full-length ORFs for HML-2 Gag and Env proteins were identified in HML-2 proviral sequences given in the March 2006 version of the human genome sequence at the Human Genome Browser. (+), the provirus on chromosome 1q22 displays a premature stop codon in gag according to the human genome reference sequence, but an allele with a full-length Gag ORF appears to exist in Caucasians as well (J. Mayer, unpublished observation). Proviruses with an ORF for the Rec protein are from reference 37.
The total numbers of clones analyzed were 24, 20, 43, and 21 for HML-2 gag in Tera-1 cells and Tera-1 pRVLP and for HML-2 env in Tera-1 cells and Tera-1 pRVLP, respectively.
As listed in Table 2, based on the March 2006 human genome assembly at the Human Genome Browser, several HML-2 proviruses identified as transcriptionally active in Tera-1 cells carry full-length ORFs for HML-2 Gag and/or Env protein. HML-2 type 2 proviruses can also encode the accessory protein Rec, which, similar to human immunodeficiency virus type 1 (HIV-1) Rev and human T-cell leukemia virus type 1 Rex, acts as an RNA-binding nuclear export protein that stabilizes unspliced HML-2 transcripts and enhances their transport out of the nucleus (35). Previous studies have also suggested that Rec may possibly play a role in the development of germ cell tumors (8, 21). It should therefore be noted that according to a former investigation (37), the three HML-2 type 2 proviruses transcribed in Tera-1 cells harbor an ORF for Rec protein (Table 2).
Assuming that the relative cloning frequencies of transcripts from individual HML-2 proviruses correlate with their transcriptional activities, the most active HML-2 locus in Tera-1 cells was a provirus on chromosome 22q11.21, with at least 50% of HML-2 gag and env cDNAs each having originated from this HML-2 locus (Table 2). Notably, the 22q11.21 provirus, previously also designated HERV-K101 (2), harbors an ORF for a full-length Gag protein. Given its strong transcriptional activity, the provirus on 22q11.21 seems to be a particularly attractive candidate to explain Gag production in Tera-1 cells. Moreover, and quite strikingly, in cDNAs from Tera-1 pRVLP, the 22q11.21 HML-2 provirus accounted for 90% of cloned HML-2 gag and 100% of cloned HML-2 env sequences, indicating that HML-2 transcripts packaged into Tera-1 pRVLP originate almost exclusively from that particular locus. Indeed, those results were verified with a different pair of PCR primers (HERV-K env 7036/7602) located in a different region of HML-2 env. All of the 16 additional HML-2 env cDNA sequences generated with that primer pair from Tera-1 pRVLP cDNA were unambiguously derived from the HML-2 locus on 22q11.21 as well (data not shown).
Although the HML-2 locus on 22q11.21 clearly dominated HML-2 transcripts in Tera-1 pRVLP, a minority of HML-2 gag sequences in Tera-1 pRVLP originated from other proviruses, that is, HML-2 loci on chromosomes 5q33.3 and 7p22.1; the latter locus was previously described as HERV-K(HML-2.HOM), HERV-K108, and HERV-K(C7) (2, 38, 50). In contrast to the proviruses on 5q33.3 and 22q11.21, the HML-2 provirus on 7p22.1 is a type 2 HML-2 provirus (Table 2). The identification of transcripts from the 7p22.1 HML-2 locus is therefore consistent with the detection of HML-2 type 2 RNAs in Tera-1 pRVLP by conventional RT-PCR (Fig. 2C). Finally, in spite of the fact that Tera-1 cells harbor HERV-K113 (Fig. 4B), none of the gag and env transcripts from Tera-1 cells or pRVLP were assignable to HERV-K113, suggesting that HERV-K113 is either transcriptionally silent or transcribed only at very low levels in Tera-1 cells. As far as the Tera-1 cell line is concerned, our data therefore do not support the suggestion that HML-2 RVLP produced by germ cell tumor cell lines might be encoded by HERV-K113 (10).
Two nonsynonymous mutations in HML-2 22q11.21 gag and env in Tera-1 cells.
Closer inspection of cDNA sequences from the 22q11.21 locus revealed two G-to-A exchanges, one in gag and the other in env, compared to the human genome March 2006 assembly. The G-to-A exchange in gag was located at nucleotide position +1946 of the HML-2 provirus on 22q11.21 (first nucleotide of the 5′ long terminal repeat [5′LTR] = +1), corresponding to chromosome 22 nucleotide position 17308132. This mutation was present in all of the HML-2 gag cDNA sequences from Tera-1 cells and pRVLP assigned to the 22q11.21 locus. Similarly, all of the 16 HML-2 env cDNA sequences amplified by the primer pair HERV-K env 7036/7602 from Tera-1 pRVLP cDNA, which were assigned to the 22q11.21 provirus, displayed a G-to-A exchange at nucleotide position +7155 of the HML-2 provirus on 22q11.21, corresponding to chromosome 22 nucleotide position 17313341.
Sequencing of the respective regions of the HML-2 provirus on 22q11.21 in genomic DNA from Tera-1 cells revealed the presence of the G-to-A exchange in gag in six of six sequenced clones and of the G-to-A exchange in env in two of two sequenced clones. It is noteworthy that these two hitherto undescribed polymorphisms in the transcriptionally most active HML-2 provirus in Tera-1 cells are nonsilent: the exchange in gag results in an Ala279Thr exchange with respect to the HERV-K101 Gag protein (GenBank accession number P63145), and the exchange in env results in a Gly222Asp exchange in HERV-K101 Env (P61566). The functional consequences, if any, of these polymorphisms are currently unknown. Nevertheless, the presence of these peculiar polymorphisms in both genomic DNA from the HML-2 22q11.21 provirus and cDNA transcripts assigned to this locus further corroborates that the 22q11.21 HML-2 locus represents the origin of those cDNAs.
DISCUSSION
In the present investigation, we show that there is a selective packaging of HML-2 RNA into pRVLP produced by the human germ cell tumor line Tera-1. In contrast, cellular RNAs or RNAs from non-HML-2 HERV families are nonselectively copackaged into those particles.
The selective packaging of retroviral RNA into particles is mediated by the interaction of specific segments of the retroviral RNA (called packaging signal or ψ sites) with RNA binding structures within the nucleocapsid (NC) domain of the assembling Gag polyproteins (17). The packaging signal of the HML-2 family has not yet been identified. However, since the vast majority of HML-2 transcripts in Tera-1 pRVLP originated from a single HML-2 provirus on chromosome 22q11.21, this locus likely has retained a functional packaging signal. It may thus prove useful for further characterization of the molecular mechanisms involved in HML-2 RNA packaging. Although we have no direct information on whether there are full-length transcripts from the 22q11.21 provirus in Tera-1 pRVLP, this seems quite likely. Indeed, our findings that the normalized relative encapsidation efficiencies of HML-2 gag and env transcripts were practically identical (Table 1) and that both types of transcripts originated almost exclusively from the same provirus on 22q11.21 appear consistent with the presence of full-length RNA from that provirus in Tera-1 pRVLP.
HML-2-encoded RVLP from all different germ cell tumor cell lines (including Tera-1 cells) analyzed so far seem to be defective (5, 10). Accordingly, attempts to prove their infectivity have failed up to now (33, 34). The biological significance/consequences of HML-2 RNA packaging into Tera-1 pRVLP are therefore unclear. It seems possible that the capacity for RNA packaging simply represents an intact relict, which has not yet become nonfunctional, of an otherwise degenerating HERV family. Nevertheless, the data obtained with Tera-1 RVLP may shed light on aspects of HML-2 which might have been relevant when it was active. For instance, copackaging of transcripts from different HML-2 proviruses in Tera-1 pRVLP (Table 2) opens the possibility for recombinations between those proviral transcripts.
Copackaging of cellular RNAs into RVLP is a well-documented phenomenon (43, 45). According to a study on murine leukemia virus (MLV) and HIV-1 Gag particles, copackaging of cellular RNAs is largely nonselective, and the distribution of cellular RNA transcripts in RVLP in most cases simply reflects the expression levels of individual cellular RNAs in the total cellular RNA pool (45). Our results from Tera-1 pRVLP overall are in accordance with these previous findings, with levels of cellular RNAs copackaged into Tera-1 particles being roughly proportional to their levels in Tera-1 cells. Interestingly, normalized relative encapsidation efficiencies of RNA transcripts from HERV families other than HML-2 (HERV-H and HERV-W) were not substantially different from those of cellular housekeeping genes, indicating that non-HML-2 HERV RNAs are nonselectively copackaged into Tera-1 pRVLP, just like other cellular RNAs. Moreover, these data suggest that the signals involved in packaging of HML-2 RNAs are specific for the HML-2 family and do not extend to HERV-H or HERV-W.
The low normalized relative encapsidation efficiency of the mitochondrial COX2 RNA is likely due to the fact that assembling Gag proteins do not have access to mitochondrial RNAs as easily as to cellular RNAs (45). However, exclusion of COX2 RNA from RVLP appears to be even more pronounced in MLV and HIV-1 than in the Tera-1 system (45). The strategy for isolation of Tera-1 RVLP employed in this study was specifically designed to avoid contaminations with cellular debris and nonencapsidated RNAs. We still cannot completely exclude that in addition to RVLP, some cellular debris, microvesicles, or exosomes, possibly containing mitochondrial or other RNAs, were present in the ultracentrifuged pellets from Tera-1 cell supernatants. Nevertheless, even if this had been the case, it would be very unlikely to have affected the key finding of our study, that is, the selective packaging of HML-2 RNA into Tera-1 pRVLP. Alternatively, the less strong exclusion of COX2 RNA from Tera-1 than from MLV and HIV-1 RVLP could also be due to intrinsic cell type-specific differences between Tera-1 cells and 293T cells (which were used in the MLV and HIV-1 study) (45).
The transcriptional activity of HML-2 is known to be elevated in germ cell tumors (23) and in germ cell tumor cell lines (32, 47), but the individual HML-2 proviruses responsible for increased HML-2 RNA expression in Tera-1 cells were unknown. We identified eight transcriptionally active HML-2 proviruses in Tera-1 cells. It is conceivable that more than those eight HML-2 proviruses are transcribed in Tera-1 cells. For instance, HML-2 proviruses that are transcribed at very low rates could be missed in the cloning procedure unless much larger numbers of clones are generated. Some HML-2 proviruses also show mismatches with the primers used in this study and may therefore escape detection. Based on the March 2006 assembly at the Human Genome Browser, we detected seven HML-2 loci in the human genome harboring full-length ORFs for HML-2 Gag proteins. Because all of those loci could, in principle, be amplified by the HML-2 gag primers employed in this work, it is still unlikely that we missed one of the HML-2 proviruses of potential functional relevance for particle production (which requires only a Gag protein) just for technical reasons.
Remarkably, transcripts from the HML-2 provirus on 22q11.21 were most prevalent within the pool of HML-2 gag and env transcripts in Tera-1 cells. This suggests that, for reasons currently not understood, transcription of this provirus is upregulated in Tera-1 cells. The HML-2 provirus on 22q11.21 therefore appears to be of particular interest among the five transcribed HML-2 proviruses in Tera-1 cells that carry full-length Gag ORFs (Table 2) and may contribute significantly to Gag protein and thus RVLP production in Tera-1 cells. Future studies should clarify whether HML-2 transcription patterns similar to those observed in Tera-1 cells can also be found in other germ cell tumor cell lines known to produce HML-2-encoded proteins and RVLP. The transcription patterns of HML-2 proviruses in primary germ cell tumor (n = 10), orchitic (n = 1), atrophic (n = 1), and normal testis (n = 3) samples were recently studied (19). In that analysis, transcripts from the 22q11.21 HML-2 provirus were found in all germ cell tumor samples as well as in orchitic and atrophic testis samples, but not in normal testis samples. Together with its strong expression in Tera-1 cells, these data suggest that the HML-2 locus on 22q11.21 may be activated in diseased testicular tissue.
The data obtained in this investigation, using Tera-1 pRVLP as a model system, might possibly also prove useful for further studies on endogenous RVLP supposedly encoded by other HERV families. In particular, it will be interesting to apply our experimental approach to endogenous RVLP that have been described for cell culture supernatants from patients with multiple sclerosis and which have been proposed to be encoded by HERV-H and HERV-W (12, 13, 28). The origin and identity of these particles are still not entirely resolved (7, 52). In both instances, the existence of HERV-H- and HERV-W-encoded RVLP has essentially been deduced from the detection of HERV-H- or HERV-W-related sequences in RVLP-associated RNA. However, the nonselective copackaging of HERV-H and HERV-W transcripts in HML-2-encoded particles observed in the present study clearly indicates that the mere detection of RNA from a specific HERV family in RVLP does not allow one to conclude that the respective RVLP are also encoded by this HERV family. Our findings may even provoke speculations that the putative HERV-H- and HERV-W-encoded RVLP could in fact consist of HML-2 proteins. An analysis of selective packaging of HERV-H or HERV-W RNAs into RVLP previously observed in cell cultures from patients with multiple sclerosis could therefore significantly contribute to their further characterization.
In summary, we have shown that at least one HML-2 element has retained the capacity for RNA packaging into HML-2-encoded particles. The functional significance of HML-2 RNA packaging into Tera-1 RVLP, which are generally considered noninfectious, currently remains unknown. Identification of a distinct number of transcriptionally active HML-2 loci in a human germ cell tumor cell line may aid future investigations on the suspected role of HML-2 in germ cell tumorigenesis to focus on relevant HML-2 loci. In this respect, the HML-2 provirus on 22q11.21 appears to be of particular interest. Since that provirus apparently harbors a functional packaging signal, it may also prove useful for further elucidation of the molecular mechanisms involved in HML-2 RNA packaging into HML-2-encoded RVLP. Finally, copackaging of non-HML-2 HERV RNAs into HML-2-encoded RVLP may have implications for non-HML-2 RVLP previously described in other human diseases, such as multiple sclerosis.
Supplementary Material
Acknowledgments
This study was supported by grants from DFG and HOMFOR.
Footnotes
Published ahead of print on 6 August 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. USA 101(Suppl. 2)14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9861-868. [DOI] [PubMed] [Google Scholar]
- 3.Beimforde, N., K. Hanke, I. Ammar, R. Kurth, and N. Bannert. 2008. Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology 371216-225. [DOI] [PubMed] [Google Scholar]
- 4.Belshaw, R., A. L. Dawson, J. Woolven-Allen, J. Redding, A. Burt, and M. Tristem. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 7912507-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieda, K., A. Hoffmann, and K. Boller. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82591-596. [DOI] [PubMed] [Google Scholar]
- 6.Blaise, S., N. de Parseval, L. Benit, and T. Heidmann. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 10013013-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomberg, J., D. Ushameckis, and P. Jern. 2005. Evolutionary aspects of human endogenous retroviral sequences (HERVs) and disease, p. 204-238. In E. D. Sverdlov (ed.), Retroviruses and primate genome evolution. Landes Bioscience, Austin, TX.
- 8.Boese, A., M. Sauter, U. Galli, B. Best, H. Herbst, J. Mayer, E. Kremmer, K. Roemer, and N. Mueller-Lantzsch. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 194328-4336. [DOI] [PubMed] [Google Scholar]
- 9.Boller, K., H. Konig, M. Sauter, N. Mueller-Lantzsch, R. Lower, J. Lower, and R. Kurth. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196349-353. [DOI] [PubMed] [Google Scholar]
- 10.Boller, K., K. Schonfeld, S. Lischer, N. Fischer, A. Hoffmann, R. Kurth, and R. R. Tonjes. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89567-572. [DOI] [PubMed] [Google Scholar]
- 11.Bronson, D. L., E. E. Fraley, J. Fogh, and S. S. Kalter. 1979. Induction of retrovirus particles in human testicular tumor (Tera-1) cell cultures: an electron microscopic study. J. Natl. Cancer Inst. 63337-339. [PubMed] [Google Scholar]
- 12.Christensen, T. 2005. Association of human endogenous retroviruses with multiple sclerosis and possible interactions with herpes viruses. Rev. Med. Virol. 15179-211. [DOI] [PubMed] [Google Scholar]
- 13.Christensen, T., P. Dissing Sorensen, H. Riemann, H. J. Hansen, and A. Moller-Larsen. 1998. Expression of sequence variants of endogenous retrovirus RGH in particle form in multiple sclerosis. Lancet 3521033. [DOI] [PubMed] [Google Scholar]
- 14.de Parseval, N., and T. Heidmann. 2005. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet. Genome Res. 110318-332. [DOI] [PubMed] [Google Scholar]
- 15.Dewannieux, M., S. Blaise, and T. Heidmann. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 7915573-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet, G. Pierron, and T. Heidmann. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 161548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3643-655. [DOI] [PubMed] [Google Scholar]
- 18.Flockerzi, A., J. Maydt, O. Frank, A. Ruggieri, E. Maldener, W. Seifarth, P. Medstrand, T. Lengauer, A. Meyerhans, C. Leib-Mosch, E. Meese, and J. Mayer. 2007. Expression pattern analysis of transcribed HERV sequences is complicated by ex vivo recombination. Retrovirology 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flockerzi, A., A. Ruggieri, O. Frank, M. Sauter, E. Maldener, B. Kopper, B. Wullich, W. Seifarth, N. Müller-Lantzsch, C. Leib-Mösch, E. Meese, and J. Mayer. 2008. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV transcriptome project. BMC Genomics 9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogh, J., and G. Trempe. 1975. New human tumor cell lines, p. 115-138. In J. Fogh (ed.), Human tumor cells in vitro. Plenum Press, New York, NY.
- 21.Galli, U. M., M. Sauter, B. Lecher, S. Maurer, H. Herbst, K. Roemer, and N. Mueller-Lantzsch. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 243223-3228. [DOI] [PubMed] [Google Scholar]
- 22.Götzinger, N., M. Sauter, K. Roemer, and N. Mueller-Lantzsch. 1996. Regulation of human endogenous retrovirus-K gag expression in teratocarcinoma cell lines and human tumours. J. Gen. Virol. 772983-2990. [DOI] [PubMed] [Google Scholar]
- 23.Herbst, H., M. Sauter, and N. Mueller-Lantzsch. 1996. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. Am. J. Pathol. 1491727-1735. [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 1011668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh, K., K. Kuma, T. Miyata, and H. Toh. 2005. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 1622-33. [PubMed] [Google Scholar]
- 26.Katzourakis, A., and M. Tristem. 2005. Phylogeny of human endogenous and exogenous retroviruses, p. 186-203. In E. D. Sverdlov (ed.), Retroviruses and primate genome evolution. Landes Bioscience, Austin, TX.
- 27.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komurian-Pradel, F., G. Paranhos-Baccala, F. Bedin, A. Ounanian-Paraz, M. Sodoyer, C. Ott, A. Rajoharison, E. Garcia, F. Mallet, B. Mandrand, and H. Perron. 1999. Molecular cloning and characterization of MSRV-related sequences associated with retrovirus-like particles. Virology 2601-9. [DOI] [PubMed] [Google Scholar]
- 29.Kurth, R., R. Löwer, J. Löwer, R. Harzmann, R. Pfeiffer, C. G. Schmidt, J. Fogh, and H. Frank. 1980. Oncovirus synthesis in human teratocarcinoma cultures and an increased anti-viral immune reactivity in corresponding patients, p. 835-846. In M. Essex, G. J. Todaro, and H. zur Hausen (ed.), Viruses in naturally occurring cancers. Cold Spring Harbor Laboratory, New York, NY.
- 30.Lee, Y. N., and P. D. Bieniasz. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lower, J., E. M. Wondrak, and R. Kurth. 1987. Genome analysis and reverse transcriptase activity of human teratocarcinoma-derived retroviruses. J. Gen. Virol. 682807-2815. [DOI] [PubMed] [Google Scholar]
- 32.Lower, R., K. Boller, B. Hasenmaier, C. Korbmacher, N. Muller-Lantzsch, J. Lower, and R. Kurth. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. USA 904480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lower, R., J. Lower, H. Frank, R. Harzmann, and R. Kurth. 1984. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J. Gen. Virol. 65887-898. [DOI] [PubMed] [Google Scholar]
- 34.Löwer, R., J. Löwer, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 935177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magin, C., R. Lower, and J. Lower. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 739496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallet, F., O. Bouton, S. Prudhomme, V. Cheynet, G. Oriol, B. Bonnaud, G. Lucotte, L. Duret, and B. Mandrand. 2004. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl. Acad. Sci. USA 1011731-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer, J., S. Ehlhardt, M. Seifert, M. Sauter, N. Muller-Lantzsch, Y. Mehraein, K. D. Zang, and E. Meese. 2004. Human endogenous retrovirus HERV-K(HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology 322190-198. [DOI] [PubMed] [Google Scholar]
- 38.Mayer, J., M. Sauter, A. Racz, D. Scherer, N. Mueller-Lantzsch, and E. Meese. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21257-258. [DOI] [PubMed] [Google Scholar]
- 39.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 729782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgenstern, B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15211-218. [DOI] [PubMed] [Google Scholar]
- 41.Moyes, D., D. J. Griffiths, and P. J. Venables. 2007. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 23326-333. [DOI] [PubMed] [Google Scholar]
- 42.Mueller-Lantzsch, N., M. Sauter, A. Weiskircher, K. Kramer, B. Best, M. Buck, and F. Grasser. 1993. Human endogenous retroviral element K10 (HERV-K10) encodes a full-length gag homologous 73-kDa protein and a functional protease. AIDS Res. Hum. Retrovir. 9343-350. [DOI] [PubMed] [Google Scholar]
- 43.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 985246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono, M. 1986. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to type A and B retrovirus genes. J. Virol. 58937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rulli, S. J., C. S. Hibbert, J. Mirro, T. Pederson, S. Biswal, and A. Rein. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 816623-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruprecht, K., J. Mayer, M. Sauter, K. Roemer, and N. Mueller-Lantzsch. Endogenous retroviruses and cancer. Cell. Mol. Life Sci., in press. [DOI] [PMC free article] [PubMed]
- 47.Sauter, M., S. Schommer, E. Kremmer, K. Remberger, G. Dolken, I. Lemm, M. Buck, B. Best, D. Neumann-Haefelin, and N. Mueller-Lantzsch. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seifarth, W., H. Skladny, F. Krieg-Schneider, A. Reichert, R. Hehlmann, and C. Leib-Mosch. 1995. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J. Virol. 696408-6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tonjes, R. R., K. Boller, C. Limbach, R. Lugert, and R. Kurth. 1997. Characterization of human endogenous retrovirus type K virus-like particles generated from recombinant baculoviruses. Virology 233280-291. [DOI] [PubMed] [Google Scholar]
- 50.Tonjes, R. R., F. Czauderna, and R. Kurth. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 739187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 111531-1535. [DOI] [PubMed] [Google Scholar]
- 52.Voisset, C., R. A. Weiss, and D. J. Griffiths. 2008. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol. Mol. Biol. Rev. 72157-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.