Abstract
Xenotropic murine leukemia virus-related virus (XMRV) is a new human gammaretrovirus identified in prostate cancer tissue from patients homozygous for a reduced-activity variant of the antiviral enzyme RNase L. Neither a casual relationship between XMRV infection and prostate cancer nor a mechanism of tumorigenesis has been established. To determine the integration site preferences of XMRV and the potential risk of proviral insertional mutagenesis, we carried out a genome-wide analysis of viral integration sites in the prostate cell line DU145 after an acute XMRV infection and compared the integration site pattern of XMRV with those found for murine leukemia virus and two human retroviruses, human immunodeficiency virus type 1 and human T-cell leukemia virus type 1. Among all retroviruses analyzed, XMRV has the strongest preference for transcription start sites, CpG islands, DNase-hypersensitive sites, and gene-dense regions; all are features frequently associated with structurally open transcription regulatory regions of a chromosome. Analyses of XMRV integration sites in tissues from prostate cancer patients found a similar preference for the aforementioned chromosomal features. Additionally, XMRV integration sites in cancer tissues were associated with cancer breakpoints, common fragile sites, microRNA, and cancer-related genes, suggesting a selection process that favors certain chromosomal integration sites. In both acutely infected cells and cancer tissues, no common integration site was detected within or near proto-oncogenes or tumor suppressor genes. These results are consistent with a model in which XMRV may contribute to tumorigenicity via a paracrine mechanism.
Prostate cancer is the most common noncutaneous cancer diagnosed in men in developed countries and is responsible for the deaths of approximately 30,000 men per year in the United States (43). Despite its impact on male health, the molecular mechanisms involved in the pathogenesis of prostate cancer, particularly the events contributing to initiation and progression, remain relatively unknown in comparison with those for other common cancers. Epidemiological studies of kindreds with hereditary prostate cancer, who often display early-onset disease and account for 9% of all cases (16), identified HPC1 as a susceptibility locus for prostate cancer (94). HPC1 is linked to RNASEL, which encodes a regulated endoribonuclease for single-stranded RNA and functions in the antiviral action of interferon (IFN) (15, 17). In response to stimulation by viral double-stranded RNA, IFN treatment of cells induces a family of 2′-5′ oligoadenylate synthetases that produce 2′-5′-linked oligoadenylates, which then activate the latent and ubiquitous protein RNase L, resulting in degradation of viral and cellular RNA and apoptosis induction (112). Several germ line variants of HPC1 and RNASEL have been observed in hereditary prostate cancer (91), including a common (35% allelic frequency) missense variant of RNase L in which a G-to-A transition at nucleotide position 1385 results in a Gln instead of an Arg at amino acid position 462 (R462Q). The R462Q RNase L variant has a threefold decrease in catalytic activity compared with the wild-type enzyme (17, 109), and individuals who are homozygous for the R462Q mutation (QQ) have a twofold-increased risk of prostate cancer (17). However, additional genetic and epidemiological studies examining the role of RNASEL as a prostate cancer susceptibility gene have provided mixed evidence, some confirmatory (80, 82, 89) and others not (61, 74, 106), suggesting that either population differences or environmental factors may modulate the impact of RNASEL on prostate cancer formation.
The association of RNASEL mutations with prostate cancer suggests that inherited defects of RNase L may enhance susceptibility to infectious agents, leading to tumorigenesis. Testing this hypothesis led to the identification of a new human retrovirus, xenotropic murine leukemia virus (MLV)-related virus (XMRV), in prostate cancer patients with the QQ variant of RNASEL (101). XMRV was detected in 40% (8 of 20) of the prostate cancers from QQ patients, compared with 1.5% among heterozygous (RQ) and wild-type (RR) patients (1 of 66). XMRV is 8,185 nucleotides in length, harbors no host-derived oncogenes, and shares up to 95% overall nucleotide sequence identity with known MLVs (101). A molecular clone of XMRV capable of infecting human prostate and nonprostate cell lines has been constructed (28). Replication of the cloned virus is sensitive to IFN-β inhibition, and RNase L is required for a complete IFN antiviral response, both findings consistent with the observation that XMRV is associated with patients having the QQ genotype (28). Expression of the human cell surface receptor XPR1 (xenotropic and polytropic retrovirus receptor 1) is required for XMRV infection, implicating XPR1 as an XMRV receptor.
Retroviruses that do not carry oncogenes, such as avian leukosis virus and Moloney MLV, usually induce tumors in their susceptible host animals by proviral insertional mutagenesis, in which proto-oncogenes are activated via promoter or enhancer insertion as a consequence of integrating the viral DNA genome into the host cell chromosome (65). Previous studies showed that most of the host genome is accessible for retroviral integration but that target site selection is not random (67, 86, 108). Furthermore, the viruses studied thus far for their positions of integrated provirus in the human genome show different patterns of target site preference and can be divided into three groups (27, 67). Human immunodeficiency virus type 1 (HIV-1), simian immunodeficiency virus, feline immunodeficiency virus, and equine infectious anemia virus form one group and share a common feature of integrating predominantly within transcription units (25, 38, 45, 48, 86). The second group comprises MLV, porcine endogenous retrovirus, and foamy virus, and their integration favors transcription start sites or CpG islands (67, 69, 73, 100, 108). The third group consists of human T-cell leukemia virus type 1 (HTLV-1), avian sarcoma-leukosis virus, and mouse mammary tumor virus. They have the most random distribution of integration sites and show only a slight preference for transcription units, transcription start sites, or CpG islands (5, 27, 34, 67, 71). The preference for transcription start sites may contribute to the observation that MLV-based vectors are more prone to activate proto-oncogenes via insertional mutagenesis than HIV-based vectors (3, 70, 88). Therefore, integration site preference may have important significance for the potential impact of a retrovirus on its host and the safety of retrovirus-based vectors in gene therapy approaches. This concern is poignantly demonstrated by the subsequent development of leukemia in three children with X-linked severe combined immunodeficiency after an otherwise successful gene therapy trial by use of an MLV-derived vector (36, 37). Analysis of leukemic cells from two patients found integration of the vector in the 5′ region of the LMO2 oncogene.
The high frequency of XMRV detection in prostate cancer from QQ patients and the validation of XMRV as a bona fide human retrovirus (28, 101) raised the possibility that XMRV might be involved in prostate cancer formation. If the initiation and progression of prostate cancer are affected by the ability of RNase L to suppress XMRV replication, the virus could contribute to the geographical prevalence of the disease in developed countries. Involvement of a viral mechanism may also partly explain the morphological and multifocal heterogeneities that distinguish prostate cancer from other common cancers. To determine the integration site preference of XMRV and the potential risk of proviral insertional mutagenesis, we carried out a genome-wide analysis of viral integration sites in a prostate cell line after an acute XMRV infection. In comparison with that of MLV and two human retroviruses, HIV-1 and HTLV-1, integration of XMRV shows a strong preference for transcription start sites, CpG islands, gene-dense regions, and DNase-hypersensitive sites. In prostate cancer tissues, in addition to the aforementioned chromosomal features, XMRV integration sites are associated with frequent cancer breakpoints, common fragile sites, microRNA (miRNA), and cancer-related genes. These associations in prostate cancer tissues may represent a selection event for particular XMRV integration sites and suggest that XMRV may play a role in prostate cancer development.
MATERIALS AND METHODS
XMRV infection.
To isolate virus, plasmid VP62/pcDNA3.1(−) containing the molecular clone of XMRV (28) was transfected into LNCaP cells with Lipofectamine 2000 (Invitrogen), and the cell culture supernatants were harvested at 13 days after transfection and passed through a 0.2-μm filter. Virus infection was performed with six-well plates by using DU145 cells plated 1 day before infection, and the complete RPMI 1640 medium was replaced with medium containing 100 μl of XMRV stock and 8 μg/ml polybrene and incubated for 3 h to allow virus adsorption. The multiplicity of infection was estimated to be 0.1. Cells were then washed once with phosphate-buffered saline, and fresh medium containing 10% fetal bovine serum was added to the cells. After 3 days of infection, the cells were harvested and genomic DNA was isolated with a QIAamp DNA mini kit (Qiagen), following the manufacturer's instructions.
XMRV RNA and DNA determination by PCR.
Peripheral zones from tumor-bearing prostate tissues were frozen at the time of surgery and stored at −80°C. RNA or DNA was isolated from frozen tissue by using Trizol (Invitrogen) or a QIAamp DNA mini kit (Qiagen), respectively, following manufacturer's instructions. XMRV gag sequences were detected using nested reverse transcriptase PCR (RT-PCR), nested PCR, or quantitative RT-PCR. For nested RT-PCR, the first-strand cDNA was synthesized with 1.5 μg of total RNA by using an iScript Select cDNA synthesis kit (Bio-Rad) containing random hexamer oligonucleotides as a primer. Nested PCR on cDNA (1/10 of the total) or on genomic DNA (∼0.5 μg) was performed with duplicates for the detection of gag sequences as described previously, with modifications (the first round of PCR was at 52°C for 35 cycles; the second round of PCR was at 54°C for 35 cycles) (101). Platinum Taq polymerase (Invitrogen) was used for nested PCR. Two-step quantitative RT-PCR was performed with total RNA (1.5 μg) and the Q528R primer (28), using the iScript Select cDNA synthesis kit. Applied Biosystems TaqMan universal PCR master mix was used for the quantitative PCR mixture, containing 900 nM each of Q445T and Q528R and 250 nM of TaqMan probe, and PCR was performed with an Applied Biosystems 7500 instrument (28). Reaction mixtures were incubated at 50°C for 2 min (for optimal AmpErase UNG activity), followed by incubation at 95°C for 10 min (for deactivation of AmpErase UNG and activation of AmpliTaq Gold). The cycling conditions were 50 cycles at 95°C for 15 s and 60°C for 1 min. Known copy numbers of XMRV RNA were used as standards.
Cloning XMRV integration sites.
The assay for determining XMRV integration sites in DU145 cells was similar to that described previously for HIV-1 (48). Briefly, genomic DNA from XMRV-infected DU145 cells was digested with PstI, which cuts once in the XMRV genome at nucleotide position 7150 and produces on average 4-kbp DNA fragments. After digestion, DNA was denatured and annealed with a biotinylated primer, bXMRV7550 (5′-biotin-ATCCTACTCTTCGGACCCTGT), which is complementary to nucleotide positions 7550 to 7571 within the env gene, about 140 bp upstream of the right long terminal repeat (LTR). The annealed primer was then extended using the PicoMaxx high-fidelity PCR system (Stratagene) to produce biotinylated double-stranded DNA containing the virus-human DNA junction region (Int-DNA). The Int-DNA was isolated by binding to streptavidin-agarose Dynabeads (Dynal) and digested with TaqαI (5′-T↓CGA), a 4-bp cutter that does not cleave the vial DNA portion of the Int-DNA and produces on average 250-bp DNA fragments. After digestion, the Int-DNA was ligated with TaqLinker, which was prepared by annealing BHLinkAl (5′-CGGATCCCGCATCATATCTCCAGGTGTGACAGTTT) with TaqLinkS (5′-CACCTGGAGATATGATGCGGGATC). The TaqLinker contains a 2-nucleotide 5′ overhang (in bold type) complementary with the TaqαI-digested Int-DNA. The linker-ligated Int-DNA was amplified by a two-step PCR. The first PCR was carried out using primers XMRV8027F (5′-AACCAATCAGCTCGCTTCTC) and Linker1 (5′-TAACTGTCACACCTGGAGATA) in a final volume of 300 μl with 0.5 μM of each primer, 0.2 mM deoxynucleoside triphosphates, and 12 U Pfu DNA polymerase (Stratagene) under the following conditions: 2 min of preincubation at 94°C, followed by 29 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 4 min. The PCR product was purified using a PCR purification kit (Qiagen) and was used as the template for the second PCR, which used two nested primers, XMRV8147F (5′-CGGGTACCCGTGTTCCCAATA) and Linker2 (5′-TAGATATGATGCGGGATCCG), which anneal downstream of the XMRV8027F and Linker1 binding sites, respectively. The conditions for the second PCR were identical to those for the first PCR, except the second PCR was conducted with only 18 cycles. The second PCR product was electrophoresed on a 1.5% agarose gel, and diffused bands between 200 bp and 2 kbp were extracted using a gel extraction kit (Qiagen). Extracted DNA was cloned into a pCR-Blunt vector by using a Zero Blunt PCR cloning kit (Invitrogen).
Cloning of integration sites from human prostate cancer tissues was similar to the procedure described earlier for DU145 cells, except that the virus-host DNA junction was first subjected to linear amplification. This was necessary because only ∼1% of prostate cells from homozygous QQ patients showed XMRV infection (101). Ten micrograms of genomic DNA isolated from prostate peripheral zones (containing tumor cells) as described previously (28) was mixed with 50 pM bXMRV7550, 0.2 mM deoxynucleoside triphosphates, and PicoMaxx polymerase (Stratagene) under the following conditions: 3 min of preincubation at 94°C, followed by 80 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 4 min. Two units of fresh PicoMaxx enzyme was added to the reaction mixture after 40 cycles. Biotinylated DNA was isolated from the PCR product by binding to 200 mg of streptavidin-agarose Dynabeads, and the remaining procedure was identical to that described earlier for XMRV-infected DU145 cells.
Sequence analysis and mapping integration sites.
The sequence of the cloned DNA was determined by dideoxy sequencing, and sequencing ambiguities were resolved by repeated sequencing on both strands. The authenticity of the integration site sequence was verified by the following criteria: (i) the sequence contained both the XMRV LTR and the linker sequence, (ii) a match to the human genome started after the end of the LTR (5′-…CA-3′) and ended with the linker sequence, and (iii) the host DNA region (containing 20 or more nucleotides) from the putative integration site sequence showed 96% or greater identity to the human genomic sequence. The authenticated integration site sequences were then mapped to the human genome hg18 (UCSC March 2006 freeze; NCBI build 36.1) by using the BLASTN program (http://www.ensembl.org/index.html) or BLAT (UCSC; http://genome.ucsc.edu/). Transcription units in the vicinity of the integration sites were identified using the RefSeq gene database (NCBI Reference Sequence Project; http://www.ncbi.nih.gov/RefSeq/). Similarities to repetitive sequences were analyzed as described previously (48). All the genomic feature data sets were downloaded from the UCSC genome database (http://genome.ucsc.edu/cgi-bin/hgTracks).
Statistical analysis of integration site sequences.
To determine integration site selection bias, a comparative set of 10,000 random positions in the human genome were generated in silico by choosing random numbers between 1 and 3,093,120,360, which represents the total length of the 22 autosomal chromosomes plus the X and Y sex chromosomes. To test for differences in proportions, we used r × c contingency table analysis (by Fisher's exact test when individual cell counts were small [<10] or by chi-square approximation). To test for equality of distribution, we used the two-sample Kolmogorov-Smirnov test.
Nucleotide sequence accession numbers.
The GenBank accession numbers for integration site sequences from DU145 cells and prostate cancer tissues are EU981292 to EU981799 and EU981800 to EU981813, respectively.
RESULTS
Mapping of XMRV integration sites in DU145 cells.
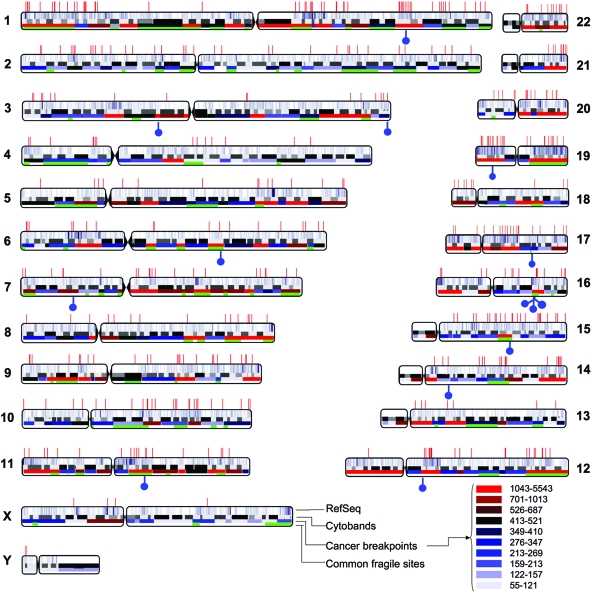
Due to the limited availability of XMRV-positive prostate cancer tissues and the low number of XMRV-infected cells (101), a genome-wide analysis of XMRV integration sites was first carried out using a human prostate cancer cell line, DU145. The cellular DNA of infected cells was isolated, and a linker ligation-mediated PCR assay was used to specifically amplify host-virus DNA junction sequences (48). We have used this assay successfully to analyze XMRV integration sites in human tumor tissue samples with a sensitivity of ∼1 copy/100 cells (28). The amplified junction sequence was cloned and sequenced. The authenticity of the sequence was then verified and the location mapped to the human genome. We sequenced a total of 508 authentic XMRV integration sites from DU145 cells, and 472 of these sites were mapped to unique locations in the human genome. Integration events were found in all 24 human chromosomes (22 autosomes and the sex chromosomes X and Y) (Fig. 1). The frequencies of integration of XMRV were generally proportional to chromosome size, but the overall frequency of XMRV integration into human chromosomes was different from that of uniformly random integration (P < 0.0001). Notably, chromosomes 1, 17, and 19 were significantly overrepresented (P values of 0.0015, 0.0021, and <0.0001, respectively), while chromosomes 5, 13, and X were significantly underrepresented (P = 0.0081, 0.0099, and 0.0002, respectively). Different integration frequencies among the different human chromosomes have also been observed for other retroviruses (38, 55, 67, 71, 73, 86). Additionally, using the criteria previously defined for integration hot spot (86), which is three or more integrations within a 100-kbp region, we identified four integration hot spots for XMRV (Table 1).
FIG. 1.
Positions of XMRV integration sites in the human genome. The human chromosomes are shown numbered. Centromere locations are denoted by chromosomal indentations. Sites of XMRV integration in DU145 cells are indicated as red vertical lines along the top, and XMRV integration sites in prostate cancer tissues are indicated as blue “lollipops” on the bottom. Within each chromosome, the top bar shows the relative densities of RefSeq genes, with higher gene-dense regions shown as a more intense cyan. The second bar shows the chromosome cytobands. The third bar shows the cancer breakpoints, and the frequencies of breakpoints in different chromosomal regions are denoted by different colors (see the key at the bottom right-hand corner). The green shading in the bottom bar denotes the locations of common fragile sites.
TABLE 1.
Integration hotspots of XMRV
| Chromosomal region | Integration site positions | ||
|---|---|---|---|
| 7q36.1 | 151018217 | 151071783 | 151075248 |
| 8q11.21 | 49590648 | 49665683 | 49668524 |
| 19p13.2 | 7363696 | 7366399 | 7366635 |
| 20q11.22 | 33358459 | 33364040 | 33370664 |
Association of integration sites with transcription units and repetitive elements.
Genome-wide analysis of integration sites of several retroviruses showed that integration is not random, and the association of integration sites with certain chromosomal features varies among different retroviruses. The distribution of XMRV integration sites and their association with chromosomal features were compared to levels for other retroviruses as well as a random control that comprises 10,000 chromosomal sites randomly generated in silico. Since the DU145 cell has not been used previously for analyzing integration site preference, the association of XMRV integration sites with chromosomal features was compared to that for other retroviruses by using published data sets generated from primary cells or other cell lines (Table 2). We first determined whether each XMRV integration site was associated with a transcription unit and repetitive elements by using the National Center for Biotechnology Information (NCBI) human Reference Sequence (RefSeq) gene database and Repeat Masker, respectively. The 24,837 RefSeq genes are curated on the basis of known mRNA transcripts, which should avoid computational bias associated with methods that rely on gene prediction programs (79). Consistent with the published data (27, 48, 67, 86, 108), our analysis showed that HIV-1 integration favored transcription units, while MLV and HTLV-1 showed modest preferences for genes (Table 3). Integration of XMRV also favored transcription units, and the degree of preference was significantly less than that for HIV-1 (P < 0.0001) but indistinguishable from those for MLV and HTLV-1 (P = 0.0789 and 0.327, respectively). We also repeated the analysis by using Known Genes from the University of California, Santa Cruz (UCSC), genome database as another human gene annotation table, and a similar pattern was observed for each virus (data not shown).
TABLE 2.
Integration site datasets used in this study
| Data set (no. of integration sites) | Cell type(s) | Source or reference | GenBank accession no. |
|---|---|---|---|
| XMRV (472) | DU145 | This report | EU981292 to EU981799 |
| MLV (1,026) | HeLa | 108 | AY515855 to AY516880 |
| HIV-1 (3,716) | IMR-90, PBMC | 67 | CL528773 to CL529239 and CL529240 to CL529767 |
| CEM | 48 | EF035624 to EF035928 | |
| SupT1 | 86 | BH609398 to BH610086 | |
| H9, HeLa | 108 | AY516881 to AY517469 | |
| CD34+ cell | 100 | DU799519 to DU800849 | |
| HTLV-1 (541) | HeLa | 27 | EF580177 to EF580913 |
TABLE 3.
Genomic features associated with retroviral integration sitesa
| Genomic featureb | % of integration sites near indicated genomic feature (P)c
|
||||
|---|---|---|---|---|---|
| Random control | XMRV | MLV | HIV-1 | HTLV-1 | |
| RefSeq genes | 34.8 | 50.4 (<0.0001) | 45.4 (<0.0001) | 72.1 (<0.0001) | 47.5 (<0.0001) |
| Repeat elements | |||||
| L1 (LINE) | 16.3 | 7.0 (<0.0001) | 7.1 (<0.0001) | 14.3 (0.0107) | 11.1 (0.0004) |
| Alu (SINE) | 10.3 | 14.8 (0.0015) | 6.2 (0.0001) | 15.2 (<0.0001) | 7.9 (0.0492) |
| LTR | 8.1 | 3.8 (0.0008) | 5.6 (0.0110) | 4.0 (<0.0001) | 6.2 (0.0848) |
The gene densities, or numbers of RefSeq genes within ±500 kbp of the integration site, are as follows: for the random control, 4.1; for XMRV, 9.2; for MLV, 7.2; for HIV-1, 8.8; and for HTLV-1, 5.6. The densities for XMRV, MLV, and HIV-1 were significantly different from that for the random control by Student's t test (P < 0.01).
LINE, long interspersed nuclear element; SINE, short interspersed nuclear element.
The percentages are relative to all sites in the data set. The P values (chi-square) are for comparison to the random control.
Besides transcription units, integration of many retroviruses, including MLV, HIV-1, and HTLV-1, is preferred in regions enriched with genes (27). For each integration site, we determined the number of RefSeq genes within ±500 kbp of the site (Table 3). The integration sites of all retroviruses analyzed had an average gene density significantly higher than that of the random control (4.1 genes). The highest gene density was seen with XMRV integration sites, with an average of 9.2 genes. MLV and HIV-1 also showed strong preferences for gene-dense regions, with averages of 7.2 and 8.8 genes, respectively. HTLV-1 integration sites had the lowest gene density, with an average of 5.6 genes.
Integration of MLV also showed a weak tendency for favoring active genes (67, 108). To examine the effect of transcriptional activity on XMRV integration, the expression levels of genes whose transcription start sites are closest to and within 10 kbp of an integration site were analyzed using transcriptional profiling of DU145 cells (GSM133589; NCBI Gene Expression Omnibus). The results showed that the percentage of XMRV integration increased proportionally with increasing gene activity (P = 0.013) (Fig. 2).
FIG. 2.
Frequency of XMRV integration as a function of gene activity. All the genes in the transcription profile data of DU145 cells (GSM133589; NCBI Gene Expression Omnibus) were ranked by their relative levels of expression and distributed into five “bins” according to their levels of expression (x axis). The leftmost bin contains genes with the lowest expression levels, and the rightmost bin contains those with the highest. Genes whose transcription start sites were closest to and within 10 kbp of the XMRV integration site (solid line) or the random control (dotted line) were then distributed into the same bins based on their expression levels and summed and the values expressed as the percentages of all genes in the indicated bin (y axis). The chi-square test was used to compare the trend to the null hypothesis of no bias due to expression level (67).
For repetitive elements, previous reports indicate that HIV-1 favors the Alu repeats of the short interspersed nuclear element family and disfavors L1 of the long interspersed nuclear element and LTR elements (48, 86). Our analysis showed the same integration preference for HIV-1 (Table 3). Also, we found that MLV disfavored L1, Alu repeats, and LTR elements, whereas HTLV-1 had no preference for Alu repeats and LTR elements and disfavored L1. Like that of HIV-1, integration of XMRV favored the Alu repeats and disfavored L1 and LTR elements (Table 3). The different preferences of retroviruses for repetitive elements may be partly related to the differential distribution of the repeat sequences. In gene-rich regions of the human genome, Alu repeats are enriched, while L1 and LTR elements are scarce (52).
Integration frequency near and within transcription units.
Although integrations of MLV, HIV-1, and HTLV-1 all favor transcription units, MLV and HTLV-1 integrate preferentially near the start of transcriptional units, whereas HIV-1 integrates throughout the entire length of the transcriptional region (27, 67, 86, 108). To examine the preference of XMRV integration sites within and near transcription units, we normalized the lengths of all RefSeq genes by dividing the genes into 10 bins and dividing 40-kbp regions upstream and downstream of the genes into 5-kbp windows. The relative integration frequency was calculated by dividing the number of integration sites in each bin or window by that in the random control (Fig. 3A). The preferences for integration of MLV, HIV-1, and HTLV-1 into transcription units and intergenic regions were consistent with previous reports (27, 67, 86, 108). In comparison to those of the other retroviruses examined, the integration pattern of XMRV was most similar to that of MLV. The distribution curve for frequency of integration site was bell shaped, with the peak centered near the transcription start site (Fig. 3A).
FIG. 3.
Integration site distribution of XMRV and other retroviruses. (A) Integration intensity within and near transcription units. All RefSeq genes, demarcated by transcription start site (TSS) and transcription termination (TT), were normalized to a common length and then divided into 10 bins (shaded area) to allow comparison. Chromosomal regions up to a distance of 40 kbp upstream and downstream of the transcription unit were divided into 5-kbp windows. The number of integration sites in each bin or 5-kbp window was divided by the number of random control sites in the same bin or window and the value plotted. A value of 1 indicates no difference between the experimental sites and the random control. (B) Integration frequency near transcription start sites. Chromosomal regions within ±12 kbp of the RefSeq transcription start site were divided into 2-kbp windows. The distance upstream of the TSS is denoted by the minus sign. The numbers of integration sites of various retroviruses or random sites in each 2-kbp window were determined and expressed as percentages of the total integration sites.
To examine more closely the integration preferences of XMRV and other retroviruses for transcription start sites, we divided 12-kbp regions upstream and downstream of the transcription sites of all RefSeq genes into 2-kbp windows and determined the percentage of integration events in each 2-kbp window (Fig. 3B). Of the retroviruses previously studied, MLV had the highest preference for transcription start sites, with 19.9% of total integration sites found within ±2 kbp of the transcription start sites (versus 3.0% in the random control; P < 0.0001). Compared to the random control, HTLV-1 showed a modest preference (6.2%; P < 0.0001) and HIV-1 had no preference (2.5%; P = 0.774) for transcription start sites. Notably, 28.2% of the total integration sites of XMRV were located within ±2 kbp of the transcription start sites, and the frequency was significantly higher than that of MLV (P = 0.0005).
Integration frequency near genomic features associated with gene regulatory regions.
The strong preference of XMRV integration near transcription start sites prompted us to examine other genomic features that are frequently associated with gene regulatory regions, such as CpG islands, DNase-hypersensitive sites, and transcription factor-binding sites. CpG islands are regions (at least 200 bp) rich in the CpG dinucleotide and thought to be involved in transcription regulation of nearby genes (9, 54). Previous results have shown that MLV integration favors CpG islands, whereas HIV-1 disfavors CpG islands, and HTLV-1 has no preference (27, 67, 86, 108). In our analysis, it was evident that integration of both XMRV and MLV strongly favored CpG islands (Fig. 4A). Within the ±2-kbp window of the CpG island midpoint, the percentages of XMRV and MLV integration sites were 27.8% and 17.9%, respectively. The preference of XMRV for CpG islands was significantly stronger than that of MLV (P < 0.0001). Compared with the random control (3.2%), HIV-1 (2.5%; P = 0.0440) weakly disfavored CpG islands, whereas HTLV-1 (4.9%; P = 0.0177) showed a moderate preference.
FIG. 4.

Integration frequencies of XMRV and other retroviruses near CpG islands, DNase-hypersensitive sites, and transcription factor-binding sites. (A) CpG islands. Chromosomal regions within ±12 kbp of the CpG island were divided into 2-kbp windows. The distance upstream of the CpG island is denoted by the minus sign. The numbers of integration sites of various retroviruses or random sites in each 2-kbp window were determined and expressed as percentages of the total integration sites. (B) DNase-hypersensitive sites. Random sites or integration sites of the indicated retroviruses within ±1 kbp of DNase-hypersensitive sites were determined and the values expressed as percentages of the total integration sites. (C) Transcription factor-binding sites. Random sites or integration sites of the indicated retroviruses within ±1 kbp of known transcription factor-binding sites (NCBI) were determined and the values expressed as percentages of the total integration sites.
DNase-hypersensitive sites are believed to be a structurally open region of chromatin associated with regulatory elements and are enriched upstream of genes and near CpG islands (23, 24, 31). Previous studies found that, at short intervals (1 to 2 kbp), DNase-hypersensitive sites are preferred integration targets for MLV but not HIV-1 or HTLV-1 (6, 27, 58, 81, 102). To examine the association between DNase-hypersensitive sites and XMRV integration, we determined the percentages of integration sites within ±1 kbp of DNase-hypersensitive sites (Fig. 4B). Similar to those for CpG islands, XMRV and MLV integrations (15.5% and 8.5%, respectively) were significantly higher (P < 0.0001) near the DNase-hypersensitive site than that of the random control (0.9%), while HIV-1 (0.8%; P = 0.583) and HTLV-1 (1.2%; P = 0.424) integrations were not significantly different from that of the random control. Between XMRV and MLV, the preference of XMRV for DNase-hypersensitive sites was significantly stronger (P < 0.0001).
Retroviral integration targeting has also been linked to transcription factors. For instance, binding sites for the AP-1 and Bach1 transcription factors are enriched near MLV integration sites (58), and HIV-1 integration site pattern is altered by the absence or presence of the LEDGF/p75 transcription factor (21, 90). We analyzed the frequencies of integration sites of different retroviruses within ±1 kbp of known transcription factor-binding sites (Fig. 4C), and the percentages of integration sites for XMRV, MLV, HIV-1, and HTLV-1 were 62.1%, 68.0%, 51.1%, and 55.5%, respectively. All viruses showed a significant preference for transcription factor-binding sites compared to the random control (41.3%; P < 0.0001). Statistical analysis linking the preference of retroviral integration to individual transcription factors was not possible, due to the limited number of integration sites against a large array of transcription factors. However, we did notice that the percentages of XMRV integration sites within ±1 kbp of the binding sites for transcription factors Bach2, MZF-1, and NF-E2 p45 were five- to sixfold higher than those of the random control.
To better understand the preferences of XMRV integration for transcription start sites, CpG islands, and DNase-hypersensitive sites, we generated three data sets and constructed Venn diagrams to examine their relationships (Fig. 5). One data set contained integration sites within ±2 kbp of transcription start sites, one contained integration sites within 2 kbp of CpG islands, and one contained integration sites within 1 kbp of DNase-hypersensitive sites. For each data set representing one genomic feature, we determined the extents of the presence of the other two genomic features. As expected from the earlier results (Fig. 4 and 5), the percentages of integration sites in the three data sets for XMRV and MLV were high and accounted for 23.7% and 16.4%, respectively, of the total integration sites. These values were significantly higher (P < 0.0001) than those for the random control (4.5%), HIV-1 (3.9%), and HTLV-1 (6.5%). The sums of overlap areas involving any two or all three data sets for XMRV and MLV were 89.3% and 90.3%, respectively, of the total area, and were significantly higher (P < 0.0001) than those for the random control (34.9%), HIV-1 (27.6%), and HTLV-1 (61.9%). The results suggested that XMRV and MLV integrate preferentially into chromosomal regions containing two or more of the following genomic features: transcription start sites, CpG islands, and DNase-hypersensitive sites.
FIG. 5.

Venn diagrams of the relationship among transcription start sites, CpG islands, and DNase-hypersensitive sites in affecting retroviral integration site preference. Red circles represent integration sites within ±2 kbp of transcription start sites (TSS), green circles represent integration sites within ±2 kbp of CpG islands (CpG), and purple circles represent integration sites within ±1 kbp of DNase-hypersensitive sites (DH). The combined numbers of integration sites in the three data sets for the random control, XMRV, MLV, HIV-1, and HTLV-1 were 4.5%, 3.9%, 6.5%, 23.7%, and 16.4%, respectively, of the total integration sites. Yellow, pink, and blue denote integration sites that contain TSS and CpG, TSS and DH, and CpG and DH, respectively. White denotes integration sites that contain TSS, CpG, and DH. The percentage of overlap areas involving any two or all three data sets is indicated at the top right corner of each panel.
Integration frequency within cancer breakpoints, within common fragile sites, and near miRNAs.
Since XMRV has a strong link with RNASEL variant QQ in prostate cancer, we also examined the associations of XMRV integration sites with cancer cytogenetics and miRNA. The integration sites of XMRV and other retroviruses were analyzed using the NCBI's Cancer Chromosomes database, which contains more than 160,000 chromosomal breakpoints involved in structural rearrangements of various neoplastic disorders (http://www.ncbi.nlm.nih.gov/entrez?db=cancerchromosomes). The number of cancer breakpoints within each of the 320 chromosome subbands, as defined by the NCBI Genetics Review, was counted. All the subbands were ranked according to the frequency of breakpoints and divided into 10 bins, with each bin containing approximately 32 subbands and representing approximately 1/10 of the genome size (Fig. 1 and 6A). The percentages of integration sites of different retroviruses and the percentages of RefSeq genes within the 10 breakpoint bins were then determined (Fig. 6A). For the random control, the percentage of integration sites in each of the 10 breakpoint bins was ∼10%, representing a random distribution of sites into the 10 bins. For breakpoint bins with frequencies of breakpoints ranging from 55 to 687, the percentages of retroviral integration sites were largely similar to or less than those for the random control (Fig. 6A). In the two bins with the highest breakpoint frequencies (701 to 1,013 and 1,043 to 5,543), with the exception of HTLV-1 (P = 0.0525) in the bin with the highest breakpoint frequency, the percentages of integration sites for all retroviruses tested were significantly higher than those for the random control (P < 0.0001). However, none of the retroviruses tested had a percentage of integration sites significantly higher than the percentage of the RefSeq genes in the corresponding breakpoint bin, suggesting that the increase over the random control was likely a result of the preference for transcription units rather than breakpoints. A similar result was obtained when the analysis was carried out using the Mitelman recurrent-breakpoint database (data not shown).
FIG. 6.
Integration frequencies of XMRV and other retroviruses near cancer breakpoints, common fragile sites, and miRNA genes. (A) Integration near cancer breakpoints. All the known cancer breakpoints (NCBI Genetics Review) within each of the 320 chromosome subbands were counted, and the subbands were ranked according to the frequencies of breakpoints and divided into 10 bins (x axis). The percentage of random sites, the percentage of RefSeq genes, and the percentages of integration sites of different retroviruses within each of the 10 breakpoint bins were then determined. (B) Common fragile sites. The percentages of random sites, RefSeq genes, and integration sites of the indicated retroviruses within common fragile sites (NCBI Genome database) were calculated. (C) miRNA genes. The percentages of random sites, RefSeq genes, and integration sites of the indicated retroviruses within ±2 Mbp of miRNA genes were calculated.
Fragile sites occur at specific chromosome locations and are most commonly seen as nonrandom gaps or breaks during mitosis when cells are exposed to specific chemical agents or culture conditions. Depending on their frequency in the population and the tissue culture condition under which they are expressed, fragile sites are classified into rare and common types (30, 87). Rare fragile sites are familial and composed of di- and trinucleotide repeats found in less than 5% of chromosomes. Common fragile sites are normal components of chromosome structure and probably occur on all chromosomes. The cytogenetic locations of 33 rare and 88 common fragile sites are currently listed in the Human Genome Database (http://www.ncbi.nlm.nih.gov/projects/mapview/). We analyzed only common fragile sites since rare fragile sites are considered to have little importance in cancer, whereas common fragile sites are frequently affected in cancer (30, 97). The percentages of integration sites for different retroviruses and of RefSeq genes within the common fragile sites were calculated. Compared with that for the random control (20.5%), integration of all retroviruses tested showed a preference for common fragile sites (P < 0.0001), >but none of the retroviruses had a percentage of integration significantly higher than the percentage of RefSeq genes (25.0%) within the common fragile sites (Fig. 6B).
Human miRNA genes are frequently located in fragile sites and genomic regions involved in cancers (14). Additionally, miRNAs are targets for chromosome deletion and exhibit high frequencies of genomic alterations in humans (110). Altered expression of miRNAs has been documented to occur in many tumors, including prostate cancers (62, 78). We calculated the percentages of integration of the various retroviruses within ±2 Mbp of miRNA genes and compared them with that for the random control (27.5%) (Fig. 6C). The integration frequencies of all retroviruses tested were significantly higher than that of the random control (P < 0.001) (Fig. 6C). However, as with cancer breakpoints and common fragile sites, none of the retroviruses tested had a percentage of integration higher than the percentage of RefSeq genes within ±2 Mbp of miRNA genes (44.8%) (Fig. 6C).
Analysis of XMRV integration sites in human prostate cancers.
We also cloned and sequenced XMRV integration sites in prostate cancer tissues from nine different prostate cancer patients. These prostate tissue samples were selected for integration site mapping based on the detection of XMRV gag sequences by nested PCR, nested RT-PCR, or quantitative RT-PCR (data not shown). The RNASEL genotype of one prostate tissue sample, VP229, was the homozygous wild type (RR), and those of the other eight samples were the homozygous variant (QQ). From these patient samples, we cloned and sequenced a total of 14 authentic integration sites, ranging from 1 to 3 integration sites identified per positive patient sample (Table 4). Each integration site was sequenced at least twice in different experimental settings. The 14 integration sites from patient samples were referred to as XMRV-prostate cancer (XMRV-PC) to distinguish them from those obtained from acute infection of DU145 cells. The chromosomal distribution of the XMRV-PC integrations sites in the human genome was mostly unremarkable, except that three independent integrations from three different patient samples were located within 1.1 Mbp in the same cytoband, q22, on chromosome 16 (Fig. 1 and Table 4). No integration hot spot was found in this region during acute infection of DU145 cells (Table 1).
TABLE 4.
Characteristics of XMRV integration sites identified in human prostate cancer tissues
| Tissuea | Site of integration
|
Gene(s) within indicated distance of integration site (distance [kbp])c,d
|
miRNA within ±2 Mbp of integration site | Common fragile site | ||
|---|---|---|---|---|---|---|
| Cytobandb | Nucleotide position | ±50 kbp | ±100 kbp | |||
| VP29 | 3p13ζ | 73283567 | FLJ10213 (90.1), PPP4R2 | |||
| VP229 | 16q22.1† | 66531394 | PSMB10ψ (−3.1), LCATψ, CTRL*, SLC12A4ψ, PSKH1, DPEP3, DPEP2 | NRN1L, EDC4*, DDX28, DUS2L, NUTF2*, THAP11, CENPT | mir-328 | FRA16C |
| VP234 | 17q23.2e | 55946474 | APPBP2 (11.9) | PPM1Dψ, C17orf64 | mir-21 | |
| VP268 | 11q13.4† | 72182279 | STARD10ψ (0.2), ATG16L2, FCHSD2# | CENTD2ψ, | mir-139 | FRA11H |
| 16q22.1†e | 66678692 | NFATC3*# (1.8), DUS2L | DOX28, DPEP2 | mir-328 | FRA16C | |
| 7p15.1e | 28689605 | CREB5* (−2.6) | mir-196b | |||
| VP283 | 19p13.2† | 11115762 | SPC24 (11.7), ANKRD25, LDLR* | LOC55908, DOCK6, SMARCA4 | mir-199a-1 | |
| 3q29 | 198606680 | DLG1 (−96.8) | mir-570 | |||
| VP338 | 12q13.11ζ | 45110969 | SLC38A2# (−58.2) | |||
| VP363 | 16q22.1† | 67648746 | HAS3 (−48.9), TMCO7 | CTF8, CIRH1A | mir-140 | FRA16C |
| VP432 | 1q32.1ζ | 202666825 | PPPIR15B (−19.3), PIK3C2Bψ | PLEKHA6, MDM4ψ | mir-135b | |
| 6q21† | 111385428 | GTF3C6 (−1.0), BXDC1 | AMD1* | FRA6F | ||
| VP433 | 14q12 | 30803190 | HECTD1* (−56.8), HEATR5A | mir-624 | ||
| 15q22.31† | 63070335 | SPG21* (−1.0), MTFMT, ANKDD1A | OSTbeta, RASL12, LOC390594 | mir-422a | FRA15A | |
All prostate tissues, except VP229, were from patients with the QQ variant of RNase L. The RNase L genotype of VP229 is the wild type (RR).
Cytobands located within the regions with the highest frequency (1,043 to 5,543) of cancer breakpoints are denoted by †. Cytobands located within the regions with the second-highest frequency (701 to 1,013) of cancer breakpoints are denoted by ζ.
Genes are listed in the order of the distances between the transcription start site of the respective gene and the integration site. The distance in kilobase pairs of the gene nearest to the integration site is indicated within the parenthesis, with “−” or “+” denoting that the integration site is upstream or downstream, respectively, of the transcription start site. Genes hosting an integration site within the transcription unit are denoted by boldface.
Genes listed in the NCI Cancer Gene Database are denoted by ψ. Genes listed in the Affymetrix Human Cancer G110 Array are denoted by *. Genes listed in the CIS in the Retrovirus Tagged Cancer Gene Database are denoted by #.
Integration sites reported previously by Dong et al. (28).
We examined the chromosomal features associated with the XMRV-PC integration sites and compared these features with those determined earlier for DU145 cells during acute infection. We found that the percentages of XMRV-PC integration sites associated with transcription units, transcription start sites, CpG islands, DNase-hypersensitive sites, and transcription factor-binding sites were not significantly different from those of XMRV integration sites in DU145 cells (P values ranging from 0.253 to >0.999) (Fig. 7). Compared with those for the random control, XMRV-PC integration sites showed a significant preference for transcription start sites, CpG islands, and DNase-hypersensitive sites (P ≤ 0.002) (Fig. 7). However, due to the relatively small sample size, the percentages of XMRV-PC integration sites associated with transcription units and transcription factor-binding sites were not significantly different from that of the random control (P = 0.265 and 0.103, respectively).
FIG. 7.

Chromosomal features associated with XMRV integration sites identified in human prostate cancers. A total of 14 authentic XMRV integration sites from nine prostate cancer samples (XMRV-PC, black bars) were sequenced and mapped. The percentages of integration sites associated with the indicated chromosomal features were determined. The percentages of random sites (blue bars) and XMRV integration sites from DU145 cells (red bars) for each of the indicated chromosomal features were also included for comparison. CpG, CpG islands; DH, DNase-hypersensitive sites; TF, transcription factor-binding sites; TSS, transcription start sites.
Many retroviruses cause cancer by activating proto-oncogenes, and enhancer elements in the proviral LTR can influence gene expression over hundreds of kilobase pairs in distance and may possibly affect several genes at once (39, 56, 85). We examined the identity of the nearest gene and the distance from the nearest transcription start site (ranging from 0.2 to 96.8 kbp), the identities of the genes within ±50 and 100 kbp, and miRNA within ±2 Mbp of each XMRV-PC integration site (Table 4). Unlike results for the leukemia caused by an MLV-derived vector (37), we did not identify a common proto-oncogene or tumor suppressor associated with XMRV integration sites. However, 42.3% (11 of 26) and 34.5% (19 of 55) of all the known genes within 50 kbp and 100 kbp, respectively, of the integration sites matched the genes listed in the selected cancer gene databases: the National Cancer Institute (NCI) Cancer Gene Database (http://ncicb.nci.nih.gov/projects/cgdcp), the Common Integration Site (CIS) in the Retrovirus Tagged Cancer Gene Database (http://rtcgd.abcc.ncifcrf.gov) (1), and the Affymetrix Human Cancer G110 Array. Out of the 14 proviruses in the tumor samples, 7 were located within a transcription unit (the LCAT, APPBP2, STARD10, NFATc3, CREB5, TMCO7, and PIK3C2B genes), with one integration site in the first exon of the STARD10 gene, one in the last exon of the LCAT gene, and the rest within introns. Among the seven genes, the LCAT, STARD10, and PIK3C2B genes are listed in the NCI Cancer Gene Database, NFATc3 is in both the CIS and the Affymetrix Human Cancer G110 Array databases, and CREB5 is in the Affymetrix Human Cancer G110 Array database. APPBP2 and PIK3C2B are associated with androgen receptor signaling and the phosphatidylinositol 3-kinase/Akt pathway, respectively, and may play a role during prostate cancer development (47, 59, 111). Integration of the XMRV genome within exons would undoubtedly disrupt the coding sequence, but whether their insertions in exons or introns may disrupt gene function and expression was not determined.
Integration sites from prostate cancer tissues are associated with cancer breakpoints, common fragile sites, and miRNAs.
As in acute infection, we analyzed the association between XMRV-PC integration sites and cancer cytogenetics or miRNA (Fig. 8). For cancer breakpoints, distribution of the XMRV-PC integration sites was biased toward regions with high breakpoint frequencies: 50.0% of XMRV-PC integration sites were located in the bin with the highest frequency (1,043 to 5,543), and an additional 21.4% were in the bin with the second-highest frequency (701 to 1,013) (Fig. 8A). Statistical analysis of integration events in the bin with the highest breakpoint frequency showed that the percentage of XMRV-PC integration sites was significantly higher than that for the random control (P = 0.0013). However, due to the relatively small sample size, the percentage of XMRV-PC integration sites was not significantly different from that of the XMRV integration sites from acutely infected cells (P = 0.133).
FIG. 8.
Association of XMRV integration sites in prostate tumor samples with cancer breakpoints, common fragile sites, and miRNA genes. The percentages of random sites, XMRV integration sites from the prostate cancer samples, and XMRV integration sites from DU145 cells are represented by blue, black, and red bars, respectively. (A) Cancer breakpoints. The 10 cancer breakpoint bins were ranked as described in the legend to Fig. 6. (B) Common fragile sites. (C) miRNA genes.
Upon analysis for association with common fragile sites, we found that three XMRV-PC integration sites were in FRA16C, and one site each was in FRA6F, FRA11H, FRA15A, and FRA19A (Fig. 8B). The percentage of XMRV-PC integration sites in common fragile sites (43%) was significantly higher than that for the random control (20.5%; P = 0.049) and also higher than but not statistically different from that of XMRV integration sites in the cell line (26.5%; P = 0.219).
In addition to cancer cytogenetics, the percentage of XMRV-PC integration sites within ±2 Mbp of miRNA genes was analyzed (Fig. 8C). The percentage of XMRV-PC integration sites near miRNA was 78.6%, which was significantly higher than those for the random control (27.5%; P < 0.0001) and XMRV integration sites from acutely infected cells (42.6%; P < 0.0114).
DISCUSSION
We carried out a genome-wide analysis of XMRV integration sites in a prostate cell line and prostate tumor samples to determine the integration site preference of XMRV and to assess the role of proviral insertional mutagenesis mediated by XMRV infection in prostate cancer formation. In the prostate cell line, XMRV integration is characterized by a strong preference for transcription start sites, CpG islands, and DNase-hypersensitive sites, all features that are frequently associated with structurally open transcription regulatory regions of a chromosome. Integration of XMRV is also preferred in actively transcribed genes and gene-dense regions within the chromosome. In accord with this observation, XMRV integration favors the Alu repeats, which are abundant in gene-rich chromosomal domains, and disfavors L1 and LTR elements, which are scarce in gene-rich regions of the genome (52, 104).
The mechanistic basis and determinants of target site selection during retroviral DNA integration are still not fully known, but interactions between viral proteins and cellular factors are likely involved (21, 90). HIV-1 integrase is a major factor in controlling target site selection in vitro (4, 11, 40) and has been implicated as a principal determinant of integration specificity in vivo (58). The retroviruses studied thus far can be divided into three groups according to their preferences for transcription units and CpG islands (27, 67). Phylogenetic analysis showed that the three types of retroviral integration profiles correlate with the amino acid sequences of integrase and the lengths of target site duplication (27), which presumably correspond to the spacing on the target DNA between the two viral DNA ends during integrative recombination catalyzed by integrase (12). This correlation is consistent with the idea that integrase is the major determinant of integration site selection (27, 58). Based on integration site preference and the sequence identity of integrase, XMRV is most suited in the group that comprises MLV, porcine endogenous retrovirus, and foamy virus (27, 69). In terms of host determinants, the LEDGF/p75 transcription factor is a critical targeting factor during HIV-1 integration (21, 90). Transcription factor-binding sites are also favored during XMRV integration, but the involvement of a specific transcription factor for XMRV integration targeting cannot be ascertained at present. Further experiments are needed to confirm the role of integrase and to deduce the host determinants as well as the mechanisms responsible for the integration site preference observed for XMRV.
Comparisons between XMRV and MLV integration in human cells indicated that XMRV has a stronger preference for transcription start sites, CpG islands, DNase-hypersensitive sites, and gene-dense regions. The quantitative difference between XMRV and MLV may be due to the uses of different human cell lines (DU145 versus HeLa). However, analyses of HIV-1 and MLV integration indicate little or no dependency of site preference on cell types (41, 67). Alternatively, given that XMRV has evolved as a xenotropic murine retrovirus in the human population for some time and presumably has adapted to replicate in human cells, XMRV may form different interactions with human host factors than MLV even though the two viruses share a high sequence identity. Compared with that for HIV-based vectors, the higher integration preference of MLV toward transcription start sites and CpG islands has been attributed as a factor contributing to the higher genotoxicity observed with MLV-derived vectors (70). Among all retroviruses analyzed, the finding that XMRV shows the strongest preference for transcription start sites, gene-dense regions, and other features associated with open, active chromosomal regions suggests that XMRV integration may carry a significant risk, and a direct assessment of the oncogenic potential of XMRV infection is warranted.
In prostate tumor samples, analysis of XMRV integration sites also showed a preference for transcription start sites, CpG islands, and DNase-hypersensitive sites. Significantly, XMRV integration sites in tumors are commonly found within cancer breakpoints, within common fragile sites, and near miRNA genes, features that are frequently linked with human cancers. Cancer cytogenetics has been a powerful means to pinpoint the locations of cancer-initiating genes, and acquired chromosomal changes have now been reported to occur in more than 50,000 cases across all main cancer types (http://cgap.nci.nih.gov/Chromosomes/Mitelman) (50). Balanced chromosome rearrangements, particularly translocations, are strongly associated with distinct tumor entities and may represent an initial event in oncogenesis (68). The common fragile site is another cancer-associated genomic feature that is frequently altered in non-virus-associated tumors (35). Both cancer breakpoints and common fragile sites are preferential integration targets for vector DNA, hepatitis B virus, and various DNA viruses, including human papillomavirus, Epstein-Barr virus, simian virus 40, and adeno-associated virus. These integration events may contribute significantly to the development of various types of cancers by disrupting the normal activity of tumor suppressor genes or proto-oncogenes in the vicinity (35, 46, 66, 76, 77, 105). In the SCID-X1 gene-therapy trial wherein two patients received an MLV-derived vector and subsequently developed leukemia via activation of the LMO2 oncogene (37), the two integration sites targeted by the MLV-based vector reside within FRA11E, a common fragile site known to correlate with chromosomal breakpoints in tumors (7). Since XMRV integration in DU145 cells does not display a bias for cancer breakpoints and common fragile sites, the high XMRV integration preference seen in tumor samples for genomic regions with the highest frequencies of cancer breakpoints and common fragile sites is striking and likely represents a selection process. The key question of whether these integrated proviruses are an indirect consequence of genomic instability initiated by other genetic lesions or perhaps have a direct role in prostate carcinogenesis awaits further investigations.
Another remarkable finding is that three integration sites identified in three different patient samples are located within a 1.1-Mbp region in 16q22.1. Considering that analysis of this region in acutely infected cells did not reveal an integration hot spot or an increased frequency of integration than other genomic regions with similar sizes and gene densities (data not shown), the high percentage of integration sites located within 16q22.1 in the tumor samples is consistent with a selection event. The cytoband 16q22.1 has been linked to many genetic diseases (for examples, see references 42, 51, and 95), and chromosomal deletion of this region is one of the most common and frequent genetic alterations found in many solid tumors, especially breast and prostate cancers (2, 29, 72). 16q22.1 also overlaps with one of the aphidicolin-induced common fragile sites, FRA16C, and the normal allele of the rare fragile site FRA16B that harbors AT-rich minisatellite repeats (113). This region contains 115 RefSeq genes, many of these associated with cancer. Among these genes, the CDH1 (E-cadherin), DERPC (decreased expression in renal and prostate cancer), NFATc3 (nuclear factor of activated T-cell, cytoplasmic, calcineurin-dependent 3), and HAS3 (hyaluronan synthase 3) genes have been directly linked to prostate cancer development (10, 57, 93, 96). Therefore, genetic instability in this region may have an important contribution to prostate cancer. An additional integration site of interest is in 6q21, a frequently deleted region in prostate cancer (29). Two other integration sites, one each located in 11q13.4 and 19p13.2, are also in regions where high rates of chromosomal alterations have been observed in breast and prostate cancers (19, 20, 44).
Although we did not detect any significant association between XMRV integration sites and any particular proto-oncogene or tumor suppressor gene, especially those implicated in human prostate cancers, such as MYC and PTEN (26), high percentages of cellular genes near the vicinities of the integration sites from prostate cancers matched with the genes listed in the selected cancer gene databases (Table 4). Some of these cancer-related genes, such as the CREB5 (107), NFATc3 (57), PIK3C2β (47, 59), and MDM4 (99) genes, are tightly linked to prostate cancer. Additionally, several other genes were identified to have potential roles in carcinogenesis but were not listed in the selected cancer gene databases. These genes include the APPBP2 (111), HAS3 (92), DLG1 (53), and SPC24 (49) genes.
Many miRNA genes are also present within 2 Mbp of the XMRV integration sites found in tumor samples. Aberrant expression of miRNAs is involved in the initiation, progression, and metastasis of human cancer (13, 32, 60, 98). miR-21 and miR-199a-1, located near integrated XMRV in cancer tissues, are significantly overexpressed in prostate cancer (103). miR-21 is an antiapoptotic and prosurvival factor and can directly modulate the expression of PTEN, a tumor suppressor that is altered in various types of tumors, including prostate (18, 64). miR-196b, another miRNA located near integrated XMRV, has a strong association with estrogen regulation in an adult zebrafish model (22). However, since miRNA genes are closely associated with common fragile sites (14), we do not know if the association between miRNA and XMRV integration sites may just be a consequence of XMRV's preference for common fragile sites or is due to specific interactions between XMRV integration machinery and host cis or trans elements near miRNA genes.
Although the causal relationship between XMRV infection and prostate cancer has not been established, our comparative analyses of XMRV integration site preference between acutely infected cells and prostate cancer tissues are consistent with a paracrine role for XMRV (63). This proposed mechanism is based on our findings that XMRV integration sites in tumor samples are associated with frequent cancer breakpoints, common fragile sites, miRNA, and cancer-related genes, but no common integration site has been detected within or near known proto-oncogenes or tumor suppressor genes. We hypothesize that the integration preference of XMRV for the regulatory region of transcriptionally active genes confers upon the virus a propensity to disrupt or alter gene expression, and cells carrying proviral insertions that provide a selective advantage or a favorable microenvironment for cancer initiation and progression will then be enriched (8, 26). The postulated paracrine mechanism is also consistent with the previous observation that XMRV is detected in only ∼1% of prostatic stromal and hematopoietic cells rather than carcinoma cells (101). In addition to insertional mutagenesis, we have not ruled out the possibility that native proteins encoded by XMRV may have transformation potential that can alter the growth properties of infected or neighboring cells, as has been demonstrated by some oncogenic retroviruses (33, 84).
Viruses have long been associated with cancers, and an estimated 20 to 25% of human cancers worldwide have known viral etiologies (75). Altered expression of tumor suppressors or proto-oncogenes induced by retrovirus integration is one important cause of cancer induction in animal models (65). Many viruses from the gammaretrovirus genus of the Retroviridae family, such as MLV, feline leukemia virus, and koala retrovirus, are responsible for leukemogenesis and other diseases in their respective host species (83). However, until recently, evidence of authentic infections of humans by gammaretroviruses was lacking, and therefore, human cancer formation caused by such viruses has not been fully substantiated or characterized. XMRV is an authentic human gammaretrovirus and is associated with prostate cancer patients having defective RNase L. Studies for determining the casual relationship of XMRV infection to prostate cancer and the mechanism of oncogenesis will significantly affect our appreciation of the role of viral infection in human cancers and has practical applications in identifying viral or new cellular targets for cancer prevention and treatment.
Acknowledgments
We thank Hung Fan, Owen Witte, and Hong Wu for helpful discussions and Thomas A. Wilkinson for comments on the manuscript.
This work was supported by National Institutes of Health (NIH) grant CA68859 and a Transdisciplinary Cancer Research Grant from the UCLA Jonsson Comprehensive Cancer Center to S.A.C. and by grant W81XWH-07-1-338 from the U.S. Department of Defense Prostate Cancer Research Program, NIH grant CA103943, and the Mal and Lea Bank Chair to R.H.S. S.K. is partly supported by a Dissertation Year Fellowship Award from the UCLA Graduate Division.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Akagi, K., T. Suzuki, R. M. Stephens, N. A. Jenkins, and N. G. Copeland. 2004. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 32D523-D527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertson, D. G., C. Collins, F. McCormick, and J. W. Gray. 2003. Chromosome aberrations in solid tumors. Nat. Genet. 34369-376. [DOI] [PubMed] [Google Scholar]
- 3.An, D. S., S. K. Kung, A. Bonifacino, R. P. Wersto, M. E. Metzger, B. A. Agricola, S. H. Mao, I. S. Chen, and R. E. Donahue. 2001. Lentivirus vector-mediated hematopoietic stem cell gene transfer of common gamma-chain cytokine receptor in rhesus macaques. J. Virol. 753547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appa, R. S., C. G. Shin, P. Lee, and S. A. Chow. 2001. Role of the nonspecific DNA-binding region and alpha helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J. Biol. Chem. 27645848-45855. [DOI] [PubMed] [Google Scholar]
- 5.Barr, S. D., J. Leipzig, P. Shinn, J. R. Ecker, and F. D. Bushman. 2005. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 7912035-12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry, C., S. Hannenhalli, J. Leipzig, and F. D. Bushman. 2006. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bester, A. C., M. Schwartz, M. Schmidt, A. Garrigue, S. Hacein-Bey-Abina, M. Cavazzana-Calvo, N. Ben-Porat, C. Von Kalle, A. Fischer, and B. Kerem. 2006. Fragile sites are preferential targets for integrations of MLV vectors in gene therapy. Gene Ther. 131057-1059. [DOI] [PubMed] [Google Scholar]
- 8.Bhowmick, N. A., E. G. Neilson, and H. L. Moses. 2004. Stromal fibroblasts in cancer initiation and progression. Nature 432332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird, A. P. 1986. CpG-rich islands and the function of DNA methylation. Nature 321209-213. [DOI] [PubMed] [Google Scholar]
- 10.Bonilla, C., T. Mason, L. Long, C. Ahaghotu, W. Chen, A. Zhao, A. Coulibaly, F. Bennett, W. Aiken, T. Tullock, K. Coard, V. Freeman, and R. A. Kittles. 2006. E-cadherin polymorphisms and haplotypes influence risk for prostate cancer. Prostate 66546-556. [DOI] [PubMed] [Google Scholar]
- 11.Bor, Y.-C., M. D. Miller, F. D. Bushman, and L. E. Orgel. 1996. Target-sequence preferences of HIV-1 integration complexes in vitro. Virology 222283-288. [DOI] [PubMed] [Google Scholar]
- 12.Brown, P. O. 1997. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Calin, G. A., and C. M. Croce. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6857-866. [DOI] [PubMed] [Google Scholar]
- 14.Calin, G. A., C. Sevignani, C. D. Dumitru, T. Hyslop, E. Noch, S. Yendamuri, M. Shimizu, S. Rattan, F. Bullrich, M. Negrini, and C. M. Croce. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 1012999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpten, J., N. Nupponen, S. Isaacs, R. Sood, C. Robbins, J. Xu, M. Faruque, T. Moses, C. Ewing, E. Gillanders, P. Hu, P. Bujnovszky, I. Makalowska, A. Baffoe-Bonnie, D. Faith, J. Smith, D. Stephan, K. Wiley, M. Brownstein, D. Gildea, B. Kelly, R. Jenkins, G. Hostetter, M. Matikainen, J. Schleutker, K. Klinger, T. Connors, Y. Xiang, Z. Wang, A. De Marzo, N. Papadopoulos, O. P. Kallioniemi, R. Burk, D. Meyers, H. Gronberg, P. Meltzer, R. Silverman, J. Bailey-Wilson, P. Walsh, W. Isaacs, and J. Trent. 2002. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 30181-184. [DOI] [PubMed] [Google Scholar]
- 16.Carter, B. S., T. H. Beaty, G. D. Steinberg, B. Childs, and P. C. Walsh. 1992. Mendelian inheritance of familial prostate cancer. Proc. Natl. Acad. Sci. USA 893367-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casey, G., P. J. Neville, S. J. Plummer, Y. Xiang, L. M. Krumroy, E. A. Klein, W. J. Catalona, N. Nupponen, J. D. Carpten, J. M. Trent, R. H. Silverman, and J. S. Witte. 2002. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat. Genet. 32581-583. [DOI] [PubMed] [Google Scholar]
- 18.Chan, J. A., A. M. Krichevsky, and K. S. Kosik. 2005. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 656029-6033. [DOI] [PubMed] [Google Scholar]
- 19.Chen, H., W. Hernandez, M. D. Shriver, C. A. Ahaghotu, and R. A. Kittles. 2006. ICAM gene cluster SNPs and prostate cancer risk in African Americans. Hum. Genet. 12069-76. [DOI] [PubMed] [Google Scholar]
- 20.Cheng, C. K., L. W. Chow, W. T. Loo, T. K. Chan, and V. Chan. 2005. The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast cancer. Cancer Res. 658646-8654. [DOI] [PubMed] [Google Scholar]
- 21.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 111287-1289. [DOI] [PubMed] [Google Scholar]
- 22.Cohen, A., M. Shmoish, L. Levi, U. Cheruti, B. Levavi-Sivan, and E. Lubzens. 2008. Alterations in micro-ribonucleic acid expression profiles reveal a novel pathway for estrogen regulation. Endocrinology 1491687-1696. [DOI] [PubMed] [Google Scholar]
- 23.Crawford, G. E., I. E. Holt, J. C. Mullikin, D. Tai, R. Blakesley, G. Bouffard, A. Young, C. Masiello, E. D. Green, T. G. Wolfsberg, and F. S. Collins. 2004. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. USA 101992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford, G. E., I. E. Holt, J. Whittle, B. D. Webb, D. Tai, S. Davis, E. H. Margulies, Y. Chen, J. A. Bernat, D. Ginsburg, D. Zhou, S. Luo, T. J. Vasicek, M. J. Daly, T. G. Wolfsberg, and F. S. Collins. 2006. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crise, B., Y. Li, C. Yuan, D. R. Morcock, D. Whitby, D. J. Munroe, L. O. Arthur, and X. Wu. 2005. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 7912199-12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Marzo, A. M., E. A. Platz, S. Sutcliffe, J. Xu, H. Gronberg, C. G. Drake, Y. Nakai, W. B. Isaacs, and W. G. Nelson. 2007. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 7256-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derse, D., B. Crise, Y. Li, G. Princler, N. Lum, C. Stewart, C. F. McGrath, S. H. Hughes, D. J. Munroe, and X. Wu. 2007. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 816731-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong, B., S. Kim, S. Hong, J. Das Gupta, K. Malathi, E. A. Klein, D. Ganem, J. L. Derisi, S. A. Chow, and R. H. Silverman. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. USA 1041655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong, J. T. 2001. Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev. 20173-193. [DOI] [PubMed] [Google Scholar]
- 30.Durkin, S. G., and T. W. Glover. 2007. Chromosome fragile sites. Annu. Rev. Genet. 41169-192. [DOI] [PubMed] [Google Scholar]
- 31.Elgin, S. C. 1988. The formation and function of DNase I hypersensitive sites in the process of gene activation. J. Biol. Chem. 26319259-19262. [PubMed] [Google Scholar]
- 32.Esquela-Kerscher, A., and F. J. Slack. 2006. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 6259-269. [DOI] [PubMed] [Google Scholar]
- 33.Fan, H., M. Palmarini, and J. C. DeMartini. 2003. Transformation and oncogenesis by jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275139-177. [DOI] [PubMed] [Google Scholar]
- 34.Faschinger, A., F. Rouault, J. Sollner, A. Lukas, B. Salmons, W. H. Gunzburg, and S. Indik. 2008. Mouse mammary tumor virus integration site selection in human and mouse genomes. J. Virol. 821360-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feitelson, M. A., and J. Lee. 2007. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 252157-170. [DOI] [PubMed] [Google Scholar]
- 36.Hacein-Bey-Abina, S., C. von Kalle, M. Schmidt, F. Le Deist, N. Wulffraat, E. McIntyre, I. Radford, J. L. Villeval, C. C. Fraser, M. Cavazzana-Calvo, and A. Fischer. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348255-256. [DOI] [PubMed] [Google Scholar]
- 37.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302415-419. [DOI] [PubMed] [Google Scholar]
- 38.Hacker, C. V., C. A. Vink, T. W. Wardell, S. Lee, P. Treasure, S. M. Kingsman, K. A. Mitrophanous, and J. E. Miskin. 2006. The integration profile of EIAV-based vectors. Mol. Ther. 14536-545. [DOI] [PubMed] [Google Scholar]
- 39.Hanlon, L., N. I. Barr, K. Blyth, M. Stewart, P. Haviernik, L. Wolff, K. Weston, E. R. Cameron, and J. C. Neil. 2003. Long-range effects of retroviral insertion on c-myb: overexpression may be obscured by silencing during tumor growth in vitro. J. Virol. 771059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harper, A. L., M. Sudol, and M. Katzman. 2003. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. J. Virol. 773838-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hematti, P., B. K. Hong, C. Ferguson, R. Adler, H. Hanawa, S. Sellers, I. E. Holt, C. E. Eckfeldt, Y. Sharma, M. Schmidt, C. von Kalle, D. A. Persons, E. M. Billings, C. M. Verfaillie, A. W. Nienhuis, T. G. Wolfsberg, C. E. Dunbar, and B. Calmels. 2004. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishikawa, K., and H. Mizusawa. 2006. On autosomal dominant cerebellar ataxia (ADCA) other than polyglutamine diseases, with special reference to chromosome 16q22.1-linked ADCA. Neuropathology 26352-360. [DOI] [PubMed] [Google Scholar]
- 43.Jemal, A., T. Murray, E. Ward, A. Samuels, R. C. Tiwari, A. Ghafoor, E. J. Feuer, and M. J. Thun. 2005. Cancer statistics, 2005. CA Cancer J. Clin. 5510-30. [DOI] [PubMed] [Google Scholar]
- 44.Kammerer, S., R. B. Roth, R. Reneland, G. Marnellos, C. R. Hoyal, N. J. Markward, F. Ebner, M. Kiechle, U. Schwarz-Boeger, L. R. Griffiths, C. Ulbrich, K. Chrobok, G. Forster, G. M. Praetorius, P. Meyer, J. Rehbock, C. R. Cantor, M. R. Nelson, and A. Braun. 2004. Large-scale association study identifies ICAM gene region as breast and prostate cancer susceptibility locus. Cancer Res. 648906-8910. [DOI] [PubMed] [Google Scholar]
- 45.Kang, Y., C. J. Moressi, T. E. Scheetz, L. Xie, D. T. Tran, T. L. Casavant, P. Ak, C. J. Benham, B. L. Davidson, and P. B. McCray, Jr. 2006. Integration site choice of a feline immunodeficiency virus vector. J. Virol. 808820-8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kao, C., S. Q. Wu, S. DeVries, W. S. Reznikoff, F. M. Waldman, and C. A. Reznikoff. 1993. Carcinogen-induced amplification of SV40 DNA inserted at 9q12-21.1 associated with chromosome breakage, deletions, and translocations in human uroepithelial cell transformation in vitro. Genes Chromosomes Cancer 8155-166. [DOI] [PubMed] [Google Scholar]
- 47.Katso, R. M., O. E. Pardo, A. Palamidessi, C. M. Franz, M. Marinov, A. De Laurentiis, J. Downward, G. Scita, A. J. Ridley, M. D. Waterfield, and A. Arcaro. 2006. Phosphoinositide 3-kinase C2beta regulates cytoskeletal organization and cell migration via Rac-dependent mechanisms. Mol. Biol. Cell 173729-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, S., Y. Kim, T. Liang, J. S. Sinsheimer, and S. A. Chow. 2006. A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites. J. Virol. 8011313-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kline-Smith, S. L., S. Sandall, and A. Desai. 2005. Kinetochore-spindle microtubule interactions during mitosis. Curr. Opin. Cell Biol. 1735-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knutsen, T., V. Gobu, R. Knaus, H. Padilla-Nash, M. Augustus, R. L. Strausberg, I. R. Kirsch, K. Sirotkin, and T. Ried. 2005. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer 4452-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuivenhoven, J. A., H. Pritchard, J. Hill, J. Frohlich, G. Assmann, and J. Kastelein. 1997. The molecular pathology of lecithin:cholesterol acyltransferase (LCAT) deficiency syndromes. J. Lipid Res. 38191-205. [PubMed] [Google Scholar]
- 52.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 53.Laprise, P., A. Viel, and N. Rivard. 2004. Human homolog of disc-large is required for adherens junction assembly and differentiation of human intestinal epithelial cells. J. Biol. Chem. 27910157-10166. [DOI] [PubMed] [Google Scholar]
- 54.Larsen, F., G. Gundersen, R. Lopez, and H. Prydz. 1992. CpG islands as gene markers in the human genome. Genomics 131095-1107. [DOI] [PubMed] [Google Scholar]
- 55.Laufs, S., K. Z. Nagy, F. A. Giordano, A. Hotz-Wagenblatt, W. J. Zeller, and S. Fruehauf. 2004. Insertion of retroviral vectors in NOD/SCID repopulating human peripheral blood progenitor cells occurs preferentially in the vicinity of transcription start regions and in introns. Mol. Ther. 10874-881. [DOI] [PubMed] [Google Scholar]
- 56.Lazo, P. A., J. S. Lee, and P. N. Tsichlis. 1990. Long-distance activation of the Myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc. Natl. Acad. Sci. USA 87170-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehen'kyi, V., M. Flourakis, R. Skryma, and N. Prevarskaya. 2007. TRPV6 channel controls prostate cancer cell proliferation via Ca(2+)/NFAT-dependent pathways. Oncogene 267380-7385. [DOI] [PubMed] [Google Scholar]
- 58.Lewinski, M. K., M. Yamashita, M. Emerman, A. Ciuffi, H. Marshall, G. Crawford, F. Collins, P. Shinn, J. Leipzig, S. Hannenhalli, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2006. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin, H. K., Y. C. Hu, L. Yang, S. Altuwaijri, Y. T. Chen, H. Y. Kang, and C. Chang. 2003. Suppression versus induction of androgen receptor functions by the phosphatidylinositol 3-kinase/Akt pathway in prostate cancer LNCaP cells with different passage numbers. J. Biol. Chem. 27850902-50907. [DOI] [PubMed] [Google Scholar]
- 60.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435834-838. [DOI] [PubMed] [Google Scholar]
- 61.Maier, C., J. Haeusler, K. Herkommer, Z. Vesovic, J. Hoegel, W. Vogel, and T. Paiss. 2005. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br. J. Cancer 921159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattie, M. D., C. C. Benz, J. Bowers, K. Sensinger, L. Wong, G. K. Scott, V. Fedele, D. Ginzinger, R. Getts, and C. Haqq. 2006. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Memarzadeh, S., L. Xin, D. J. Mulholland, A. Mansukhani, H. Wu, M. A. Teitell, and O. N. Witte. 2007. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell 12572-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng, F., R. Henson, H. Wehbe-Janek, K. Ghoshal, S. T. Jacob, and T. Patel. 2007. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133647-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikkers, H., and A. Berns. 2003. Retroviral insertional mutagenesis: tagging cancer pathways. Adv. Cancer Res. 8853-99. [DOI] [PubMed] [Google Scholar]
- 66.Miller, D. G., L. M. Petek, and D. W. Russell. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36767-773. [DOI] [PubMed] [Google Scholar]
- 67.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitelman, F., B. Johansson, and F. Mertens. 2007. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7233-245. [DOI] [PubMed] [Google Scholar]
- 69.Moalic, Y., Y. Blanchard, H. Felix, and A. Jestin. 2006. Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus. J. Virol. 8010980-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montini, E., D. Cesana, M. Schmidt, F. Sanvito, M. Ponzoni, C. Bartholomae, L. Sergi Sergi, F. Benedicenti, A. Ambrosi, C. Di Serio, C. Doglioni, C. von Kalle, and L. Naldini. 2006. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 24687-696. [DOI] [PubMed] [Google Scholar]
- 71.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 7811656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson, W. G., A. M. De Marzo, and W. B. Isaacs. 2003. Prostate cancer. N. Engl. J. Med. 349366-381. [DOI] [PubMed] [Google Scholar]
- 73.Nowrouzi, A., M. Dittrich, C. Klanke, M. Heinkelein, M. Rammling, T. Dandekar, C. von Kalle, and A. Rethwilm. 2006. Genome-wide mapping of foamy virus vector integrations into a human cell line. J. Gen. Virol. 871339-1347. [DOI] [PubMed] [Google Scholar]
- 74.Orr-Urtreger, A., A. Bar-Shira, D. Bercovich, N. Matarasso, U. Rozovsky, S. Rosner, S. Soloviov, G. Rennert, L. Kadouri, A. Hubert, H. Rennert, and H. Matzkin. 2006. RNASEL mutation screening and association study in Ashkenazi and non-Ashkenazi prostate cancer patients. Cancer Epidemiol. Biomarkers Prev. 15474-479. [DOI] [PubMed] [Google Scholar]
- 75.Pagano, J. S., M. Blaser, M. A. Buendia, B. Damania, K. Khalili, N. Raab-Traub, and B. Roizman. 2004. Infectious agents and cancer: criteria for a causal relation. Semin. Cancer Biol. 14453-471. [DOI] [PubMed] [Google Scholar]
- 76.Pett, M., and N. Coleman. 2007. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J. Pathol. 212356-367. [DOI] [PubMed] [Google Scholar]
- 77.Popescu, N. C. 2003. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 1921-17. [DOI] [PubMed] [Google Scholar]
- 78.Porkka, K. P., M. J. Pfeiffer, K. K. Waltering, R. L. Vessella, T. L. Tammela, and T. Visakorpi. 2007. MicroRNA expression profiling in prostate cancer. Cancer Res. 676130-6135. [DOI] [PubMed] [Google Scholar]
- 79.Pruitt, K. D., K. S. Katz, H. Sicotte, and D. R. Maglott. 2000. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet. 1644-47. [DOI] [PubMed] [Google Scholar]
- 80.Rennert, H., D. Bercovich, A. Hubert, D. Abeliovich, U. Rozovsky, A. Bar-Shira, S. Soloviov, L. Schreiber, H. Matzkin, G. Rennert, L. Kadouri, T. Peretz, Y. Yaron, and A. Orr-Urtreger. 2002. A novel founder mutation in the RNASEL gene, 471delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am. J. Hum. Genet. 71981-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rohdewohld, H., H. Weiher, W. Reik, R. Jaenisch, and M. Breindl. 1987. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J. Virol. 61336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rokman, A., T. Ikonen, E. H. Seppala, N. Nupponen, V. Autio, N. Mononen, J. Bailey-Wilson, J. Trent, J. Carpten, M. P. Matikainen, P. A. Koivisto, T. L. Tammela, O. P. Kallioniemi, and J. Schleutker. 2002. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am. J. Hum. Genet. 701299-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-586. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 84.Ross, T. M., S. M. Pettiford, and P. L. Green. 1996. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J. Virol. 705194-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sauvageau, M., M. D. Miller, S. Lemieux, J. Lessard, J. Hebert, and G. Sauvageau. 2008. Quantitative expression profiling guided by common retroviral insertion sites reveals novel and cell type specific cancer genes in leukemia. Blood 111790-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110521-529. [DOI] [PubMed] [Google Scholar]
- 87.Schwartz, M., E. Zlotorynski, and B. Kerem. 2006. The molecular basis of common and rare fragile sites. Cancer Lett. 23213-26. [DOI] [PubMed] [Google Scholar]
- 88.Seggewiss, R., S. Pittaluga, R. L. Adler, F. J. Guenaga, C. Ferguson, I. H. Pilz, B. Ryu, B. P. Sorrentino, W. S. Young III, R. E. Donahue, C. von Kalle, A. W. Nienhuis, and C. E. Dunbar. 2006. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood 1073865-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shook, S. J., J. Beuten, K. C. Torkko, T. L. Johnson-Pais, D. A. Troyer, I. M. Thompson, and R. J. Leach. 2007. Association of RNASEL variants with prostate cancer risk in Hispanic Caucasians and African Americans. Clin. Cancer Res. 135959-5964. [DOI] [PubMed] [Google Scholar]
- 90.Shun, M. C., N. K. Raghavendra, N. Vandegraaff, J. E. Daigle, S. Hughes, P. Kellam, P. Cherepanov, and A. Engelman. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 211767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Silverman, R. H. 2003. Implications for RNase L in prostate cancer biology. Biochemistry 421805-1812. [DOI] [PubMed] [Google Scholar]
- 92.Simpson, M. A. 2006. Concurrent expression of hyaluronan biosynthetic and processing enzymes promotes growth and vascularization of prostate tumors in mice. Am. J. Pathol. 169247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simpson, M. A., C. M. Wilson, and J. B. McCarthy. 2002. Inhibition of prostate tumor cell hyaluronan synthesis impairs subcutaneous growth and vascularization in immunocompromised mice. Am. J. Pathol. 161849-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith, J. R., D. Freije, J. D. Carpten, H. Gronberg, J. Xu, S. D. Isaacs, M. J. Brownstein, G. S. Bova, H. Guo, P. Bujnovszky, D. R. Nusskern, J. E. Damber, A. Bergh, M. Emanuelsson, O. P. Kallioniemi, J. Walker-Daniels, J. E. Bailey-Wilson, T. H. Beaty, D. A. Meyers, P. C. Walsh, F. S. Collins, J. M. Trent, and W. B. Isaacs. 1996. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 2741371-1374. [DOI] [PubMed] [Google Scholar]
- 95.Sprecher, E., R. Bergman, G. Richard, R. Lurie, S. Shalev, D. Petronius, A. Shalata, Y. Anbinder, R. Leibu, I. Perlman, N. Cohen, and R. Szargel. 2001. Hypotrichosis with juvenile macular dystrophy is caused by a mutation in CDH3, encoding P-cadherin. Nat. Genet. 29134-136. [DOI] [PubMed] [Google Scholar]
- 96.Sun, M., L. Ma, L. Xu, J. Li, W. Zhang, G. Petrovics, M. Makarem, I. Sesterhenn, M. Zhang, E. J. Blanchette-Mackie, J. Moul, S. Srivastava, and Z. Zou. 2002. A human novel gene DERPC on 16q22.1 inhibits prostate tumor cell growth and its expression is decreased in prostate and renal tumors. Mol. Med. 8655-663. [PMC free article] [PubMed] [Google Scholar]
- 97.Sutherland, G. R. 2003. Rare fragile sites. Cytogenet. Genome Res. 10077-84. [DOI] [PubMed] [Google Scholar]
- 98.Tavazoie, S. F., C. Alarcon, T. Oskarsson, D. Padua, Q. Wang, P. D. Bos, W. L. Gerald, and J. Massague. 2008. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Toledo, F., and G. M. Wahl. 2006. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6909-923. [DOI] [PubMed] [Google Scholar]
- 100.Trobridge, G. D., D. G. Miller, M. A. Jacobs, J. M. Allen, H. P. Kiem, R. Kaul, and D. W. Russell. 2006. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. USA 1031498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. DeRisi. 2006. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Vijaya, S., D. L. Steffen, and H. L. Robinson. 1986. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J. Virol. 60683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Volinia, S., G. A. Calin, C.-G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 1032257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang, G. P., A. Ciuffi, J. Leipzig, C. C. Berry, and F. D. Bushman. 2007. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 171186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wentzensen, N., S. Vinokurova, and M. von Knebel Doeberitz. 2004. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 643878-3884. [DOI] [PubMed] [Google Scholar]
- 106.Wiklund, F., B. A. Jonsson, A. J. Brookes, L. Stromqvist, J. Adolfsson, M. Emanuelsson, H. O. Adami, K. Augustsson-Balter, and H. Gronberg. 2004. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin. Cancer Res. 107150-7156. [DOI] [PubMed] [Google Scholar]
- 107.Wu, D., H. E. Zhau, W. C. Huang, S. Iqbal, F. K. Habib, O. Sartor, L. Cvitanovic, F. F. Marshall, Z. Xu, and L. W. Chung. 2007. cAMP-responsive element-binding protein regulates vascular endothelial growth factor expression: implication in human prostate cancer bone metastasis. Oncogene 265070-5077. [DOI] [PubMed] [Google Scholar]
- 108.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 3001749-1751. [DOI] [PubMed] [Google Scholar]
- 109.Xiang, Y., Z. Wang, J. Murakami, S. Plummer, E. A. Klein, J. D. Carpten, J. M. Trent, W. B. Isaacs, G. Casey, and R. H. Silverman. 2003. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 636795-6801. [PubMed] [Google Scholar]
- 110.Zhang, L., J. Huang, N. Yang, J. Greshock, M. S. Megraw, A. Giannakakis, S. Liang, T. L. Naylor, A. Barchetti, M. R. Ward, G. Yao, A. Medina, A. O'Brien-Jenkins, D. Katsaros, A. Hatzigeorgiou, P. A. Gimotty, B. L. Weber, and G. Coukos. 2006. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 1039136-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang, Y., Y. Yang, S. Yeh, and C. Chang. 2004. ARA67/PAT1 functions as a repressor to suppress androgen receptor transactivation. Mol. Cell. Biol. 241044-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72753-765. [DOI] [PubMed] [Google Scholar]
- 113.Zlotorynski, E., A. Rahat, J. Skaug, N. Ben-Porat, E. Ozeri, R. Hershberg, A. Levi, S. W. Scherer, H. Margalit, and B. Kerem. 2003. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 237143-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]