Abstract
Differential allelic expression has been shown to be common in mice, humans and maize, and variability in the expression of polymorphic alleles has been associated with human disease. Here, we describe the differential expression pattern of Paraoxonase-1, a gene involved in lipid metabolism and implicated in the formation of atherosclerotic lesions. We measured the expression of the murine Paraoxonase-1 gene (Pon1) in livers at different stages of embryonic development using F1 hybrid crosses and quantified the transcriptional level of both parental alleles. Using human foetal tissues, we analysed the expression of the human orthologue (PON1) and found monoallelic or preferential allelic expression in 6/7 and 4/4 samples from liver and pancreas, respectively. We observed that Pon1 does not show a parent-of-origin preference in its allelic expression, but has dramatic variations in allele-specific expression occurring throughout development. This study has important repercussions in the analysis of haplotypes at disease loci, since it implies that the expression of polymorphic alleles can be unequal and dynamic.
INTRODUCTION
Allele-specific differences in gene expression dispute the century-old Mendelian paradigm of equal contribution from the two inherited alleles. Allele-specific gene expression patterns have been classically characterized only for genes controlled by epigenetic mechanisms, such as genomic imprinting and X-chromosome inactivation. However, studies focusing on non-random allele-specific gene expression have found conclusive evidence of the unequal expression of allelic copies in autosomal non-imprinted genes and have demonstrated that it is a common occurrence in human, mouse and maize (1–4). The physiological importance of these differences is gradually being unravelled and is highlighted by the fact that altered levels of expression of polymorphic alleles have been implicated in disease pathogenesis and have functional significance (5–8). The difference in expression between two alleles has been shown to be as high as 90%, and variations in cis-acting DNA regulatory elements are considered to be primarily responsible for such allele-specific gene expressions (9,10). Consequently, the systematic characterization of such regulatory variations will facilitate the identification of disease-associated polymorphisms.
To determine the frequency of unequal allelic expression, Lo et al. (1) analysed 602 transcripts using microarray technology and determined that 54% of these showed preferential expression of one gene copy in at least one of the seven individuals studied. Similar results were obtained by Bray et al. (2), who found that the expression of 7/15 of the cerebral genes analysed varied by ≥20% in at least one of the 19 individuals studied. Analyses of gene expression levels between monozygotic twins, siblings and unrelated individuals have suggested that variability in expression levels is genetically determined (11). In support of this, recent studies have shown that differential allelic expression is a heritable trait, which may be inherited in a Mendelian manner (10,12,13).
In this study, we have identified preferential allelic expression in the Paraoxonase-1 gene (mouse gene Pon1; human gene PON1; OMIM: 168820). Paraoxonase-1 (EC 3.1.1.2), known to hydrolyse organophosphate compounds, plays an important role in lipid metabolism by associating with high-density lipoproteins and preventing the oxidation of low-density lipoproteins (14–16). As such, Pon1-knockout mice are more susceptible to atherosclerotic lesions than wild-type controls (17). Studies have identified various common polymorphisms in Pon1's sequence and promoter region, which have been shown to correlate with enzymatic activity or gene expression levels (18–22).
PON1 is a member of a family of at least three genes, all of which map to human chromosome 7q21.3. The mouse orthologues of two of these genes, Pon2 and Pon3, were recently identified as being part of a large cluster of imprinted genes located on chromosome 6A1 (Fig. 1) (23). The gene centromeric to Pon1, Ppp1r9a, has also been shown to be imprinted in both human and mouse (23,24). On the basis of the observation that imprinted genes are located in clusters throughout the genome (25,26), we considered Pon1 to be a potential candidate for imprinting. Therefore, its imprinting status was examined using F1 reciprocal hybrids of inbred mice strains. Although we did not identify a parent-of-origin pattern of allelic expression, we discovered that Pon1 shows preferential allelic expression in the liver and that this expression level changes dynamically in a strain-dependent manner throughout embryonic development. To our knowledge, this is the first demonstration that allele-specific effects on gene expression can be dependent on developmental stage.
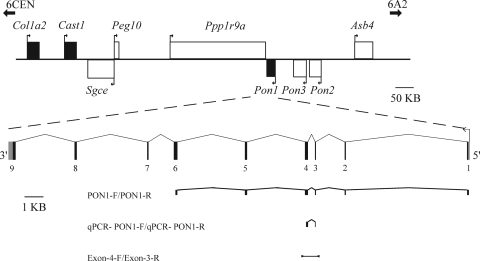
Figure 1.
Overview of the murine Pon1 locus. The 1 Mb region contains several imprinted genes (white boxes), as well as genes showing preferential or biallelic expression (black boxes). Transcriptional direction of each gene is shown by arrows. The intron–exon structure of Pon1 is depicted below, as well as its non-coding or putative non-coding region (grey segments). Primers used for amplifying cDNA fragments and primers used for amplification of genomic DNA in SNP identification are indicated by fragments and a black bar, respectively.
RESULTS
Allelic expression analysis of murine Pon1 gene
To determine the allelic expression pattern of Pon1, a single-nucleotide polymorphism (SNP) (T/C) was identified in its cDNA sequence at nucleotide 280 of NM_011134. The T allele was present in C57BL/6J (BL6) inbred strains, whereas the C allele was present in both JF1/Ms (JF1) and CAST/Ei (CAST) strains. Tissues were extracted at different stages of development [12.5 days post-coitum (dpc), 15.5 dpc and P0 (postnatal day 0)] from the F1 embryos of hybrid strains BxC and BxJ (where B is the female), as well as their reciprocal crosses. Pon1 expression was determined by RT-PCR in the lung, liver, yolk sac, placenta, intestine, limb and brain. A detectable level of Pon1 expression at all three stages was observed only in the liver. The transcript was also found to be expressed in brain, but at low levels, and was consequently excluded from further analyses.
Allelic expression analysis of Pon1 was performed by direct sequencing of RT-PCR products, which span exons 1–6 (Fig. 1). A preferential expression pattern, independent of parent of origin, was observed in the liver samples and was subsequently confirmed by pyrosequencing (Fig. 2). This method has been previously shown to be a quantitative method of determining allelic ratios (27,28). These analyses revealed a higher expression of the CAST and JF1 alleles at 12.5 dpc in each cross and a marked decrease in its allelic expression level in the later stages examined (Fig. 3, Supplementary Material, Table S1). This decrease was most notable in crosses involving CAST, where the expression of the BL6 allele surpassed that of the CAST allele in P0 samples. In JF1 hybrid crosses, however, the JF1 allele always had a stronger expression than BL6, which is consistent with the allelic preference at P0 observed by Ono et al. (23). Notably, the decrease in CAST and JF1 alleles was more drastic when BL6 was maternally inherited. This disparity in the expression was best observed at 15.5 dpc in crosses involving CAST, where this strain’s allelic frequency is measured at 45.3% in BxC, whereas it is 67.6% in CxB. The observed biased allelic expression patterns were reproduced in a separate set of embryonic livers by direct sequencing of RT-PCR samples at each stage of development, indicating the possibility that the allelic expression patterns are not random but developmentally regulated (Fig. 2). In addition, the findings were replicated using SNaPshot™, a fluorescent-based primer extension method which allows the quantification of differences in peak heights between two alleles (29), suggesting that the findings are not dependent on the methodology (Supplementary Material, Fig. S1).
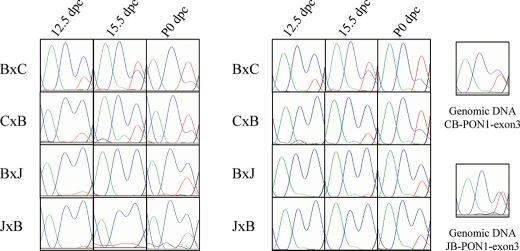
Figure 2.
Expression pattern of Pon1. Electropherograms from sequencing of two sets of liver cDNA samples from hybrid crosses at different developmental stages are shown. Results from amplification of genomic DNA are indicated on the right. The blue and red peaks indicate C and T, respectively, where T is the allele inherited from BL6 in Pon1. Letters B, C and J refer to species BL6, CAST and JF1, respectively, with the first letter of each cross representing the mother. The results show reproducibility in the two sample sets and indicate a dynamic pattern of expression throughout development.
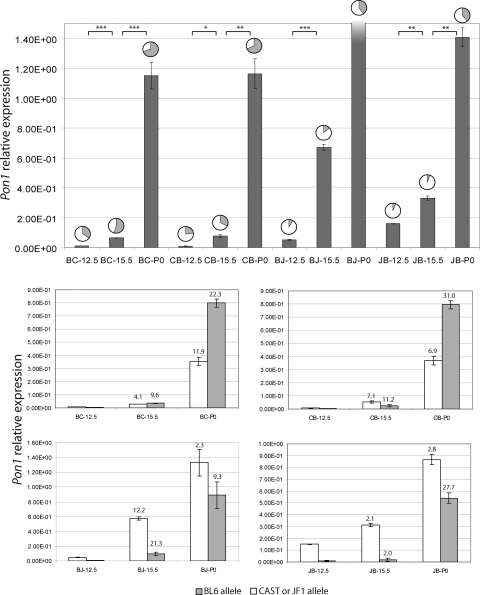
Figure 3.
Real-time quantitative PCR analysis of Pon1. Transcript abundance of hepatic Pon1 at various stages of embryonic development in F1 hybrid mice was quantified by SYBR Green detection method in triplicate. Values in the uppermost section represent total average Pon1 measurements normalized by β-2-microglobulin expression in each sample. Frequencies of CAST or JF1 and BL6 alleles measured by pyrosequencing are represented by pie charts, where the white portions represent the CAST or JF1 allele and the grey portions represent the BL6 allele. The shading in BJ-P0 expression indicates that it goes beyond the upper limit of the graph. The total expression at this developmental time point was found to be 22.2 ± 3.00 × 10−1. The four charts in the lower section represent the relative contribution of each allele towards the total expression of Pon1, where the cross and developmental time point is indicated at the bottom of each bar. Fold increase in the expression of each allele from the previous time point is also indicated at the top of the bar. Error bars were calculated using the standard deviation of both pyrosequencing and quantitative PCR results. The results indicate a disproportionate increase in the expression of each allele. JB, CB, BC and BJ refer to JxB, CxB, BxC and BxJ F1 hybrids, respectively.
To identify splice variants whose increased expression may account for the differential pattern observed, 5′ RACE was performed and the whole length of Pon1 (from exons 1–9) was also amplified. Additional 5′ ends or splice variants were not identified.
Quantitative RT-PCR analysis of Pon1 expression during liver development
To determine whether the dynamic allelic patterns of the expression observed in Pon1 were due to a static expression of the CAST and JF1 alleles with increases in BL6 allele or whether they were caused by decreases in the CAST and JF1 alleles, quantitative PCR was performed using the SYBR Green detection method. The expression of Pon1 was quantified in each liver samples for every developmental stage and hybrid cross. The values were normalized by β-2-microglobulin gene expression quantities at the corresponding stage, and a final relative expression was obtained (Fig. 3). The analysis revealed that Pon1 expression increased in an exponential pattern during embryonic development. This increase in total gene expression was far more striking in crosses involving JF1. Similar results were obtained using an independent set of liver samples and by β-actin normalization (data not shown).
Upon incorporation of allele frequencies obtained from pyrosequencing analysis into the quantitative measurements, it was apparent that both alleles increased, yet in disproportionate amounts as shown in Figure 3. The BL6 allele shows a higher fold increase in expression than the CAST or JF1 allele. This is best illustrated in the JxB hybrid crosses, where a 27.7-fold increase in BL6 allelic expression is measured between 15.5 dpc and P0. In contrast, only a 2.8-fold increase in the expression of the JF1 allele was observed between the same two developmental stages (Fig. 3, Supplementary Material, Table S1).
Screening of candidate SNPs responsible for allele-specific gene expression and promoter methylation analysis
Several SNPs identified upstream from the human PON1 gene have been associated with paraoxonase protein and transcript levels. On the basis of this observation, ∼1 kb of the region upstream from the murine Pon1 start codon was sequenced, as well as exon 1, in order to identify SNPs in the regulatory region. Two polymorphisms were identified at −227 (G/T) and −413 (C/T), where the nucleotide prior to the ATG is −1. At the −227 polymorphism, in silico analysis using TESS (www.cbil.upenn.edu/tess) identified a TFIID-binding site present in BL6 and absent in CAST and JF1. Further sequencing of the region 2 kb upstream from the start codon identified an average of one SNP every 57 bp, indicating relaxed selective constraint and, consequently, a lower probability of the presence of regulatory elements in the region.
In addition, the possibility of differential methylation of the Pon1 promoter was investigated. DNA was extracted from livers at four developmental time points (12.5, 16.5, 17.5 dpc and P10) from JF1 × BL6 hybrid embryos. The samples were treated with bisulphite and PCR-amplified to analyse the methylation of cytosines in the promoter. Since our expression analyses had revealed an increase in BL6 allele frequency throughout development in this strain, we hypothesized that we would observe a loss of methylation on this allele. However, no discernable difference was seen in the methylation pattern between the developmental time points (data not shown).
Allelic expression analysis of murine Pon2 and Pon3 genes
Pon2 and Pon3 expression in embryonic liver was analysed to determine whether these genes are co-regulated with Pon1. The allelic ratio of SNPs was observed by sequencing electropherograms obtained from the gene-specific amplification of the cDNA used for Pon1 analysis. This analysis revealed that Pon2 and Pon3 show allelic preference in embryonic liver gene expression with a strain-specific effect (unpublished data). However, unlike Pon1, this preference varies little throughout development, indicating that these genes are not co-regulated.
Allelic expression analysis for the human PON1 gene
PON1 expression was analysed in the placenta, muscle, stomach, kidney, intestine, liver, pancreas and lung by RT-PCR. High expression was observed in the liver and weaker expression in the pancreas (Fig. 4A). Consequently, only these two tissues were used in subsequent analyses.
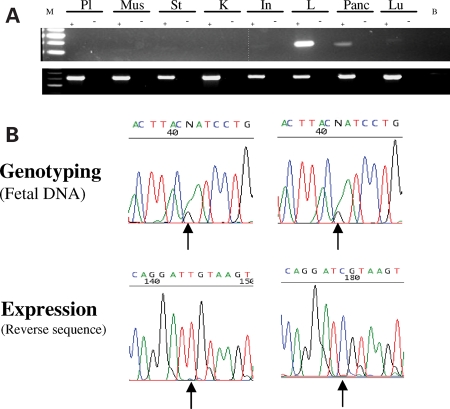
Figure 4.
Human PON1 expression. (A) PON1 tissue expression. RT-PCR was performed on placenta (Pl), muscle (Mus), stomach (St), kidney (K), intestine (In), liver (L), pancreas (Panc) and lung (Lu). B, blank; M, marker. Upper panel shows PON1 expression, lower panel shows GAPDH control expression. (B) Monoallelic expression of human PON1 in the liver and pancreas. Biallelic sequencing results of a human PON1 polymorphism from foetal samples are shown in the top panels. cDNA sequencing of PON1 in the liver (left panel) and pancreas (right panel) showing monoallelic expression of the polymorphism (bottom panels).
Thirteen foetal liver samples were genotyped at an A/G polymorphic position in exon 6 of PON1. Seven of these samples, five of which were second trimester foetuses and two of which were of unknown gestational stages, were found to be heterozygous and were used for subsequent analyses. Sequencing of cDNA extracted from these seven tissues revealed monoallelic expression of PON1 in four samples, two displayed preferential expression and the remaining was biallelically expressed (Fig. 4B). It must be noted that the latter was from an unknown gestational stage.
Ten pancreatic samples were genotyped for the same SNP, and four were found to be heterozygous. cDNA analyses revealed that three of these samples showed preferential allelic expression of PON1 and one sample showed monoallelic expression of the gene (Fig. 4B). Since parental samples corresponding to the foetal samples analysed were unavailable, we were unable to determine the parent of origin in the expression of PON1.
DISCUSSION
Our study determined that Pon1 exhibits preferential allelic expression in the liver and that the allelic expression level can change in a dynamic manner throughout embryonic development and is also dependent on genetic background. The allelic expression pattern of Pon1 was examined by Ono et al. (23), but only in neonatal liver and lung tissues. The allelic difference we detected in gene expression changed throughout development in a strain-dependent manner and was found to be biased against the allele carried by CAST and JF1, although the bias was stronger in crosses involving CAST and when the BL6 allele was maternally inherited. Our observations illustrate the first case of a developmentally regulated dynamic allelic expression pattern. A recent study performed by Wilkins et al. (30) demonstrated that differences in allelic expression can occur between different regions of a single-tissue sample from the same individual. Consequently, owing to the fact that whole-liver samples were used in our study, its findings may reflect differences in allelic expression occurring in a subset of cells within the liver.
An analysis of the human PON1 protein in infants revealed an increased expression level of 2 to 7-fold from birth until 6 to 12 months, at which point it reached a plateau (31). The equivalent stage is reached at 3 weeks of age in mice and rats. We found that Pon1 transcript levels increased substantially during embryonic formation, providing evidence that Pon1 protein activity escalates during hepatic development and plateaus at postnatal stages. Future studies may determine whether such an increase in expression correlates with Pon1 enzymatic activity.
Monoallelic and preferential expression of human PON1 was observed in various liver and pancreatic foetal samples, yet we were unable to determine the parent-of-origin. The differences in human PON1 expression may be attributed to polymorphic imprinting, which has been observed in several human genes (32,33). However, it is important to note that preferential monoallelic expression can occur in a non-parent-of-origin pattern (10), as was seen in Pon1 at 12.5 dpc in BJ and JB hybrids (Fig. 2). Consequently, it is plausible to hypothesize that PON1 is expressed monoallelically at early gestational stages, preferentially later in development, and biallelically expressed neonatally. Such a dynamic pattern of expression would account for the preferential and biallelic expression patterns seen in several of the human samples. However, the differences in PON1 expression may also be due to polymorphisms in cis-acting regulatory regions between the biallelically and monoallelically expressed samples.
Variation in allelic expression dependent on developmental stage has been suggested to be a regulatory mechanism in the decline of lactase activity in mammalian maturation. Cis-acting regulatory variants have been identified upstream of the lactase gene, and these have been associated with hereditary lactase persistence (34,35). These variants have been associated with increased/decreased levels of lactase transcript and have been shown to exhibit differential binding of nuclear proteins (36,37). Human studies have also shown a decline in the expression from the allele associated with hypolactasia with age (38). Such a developmentally regulated pattern of preferential allelic expression may also occur in Pon1, where the expression from one allele varies with developmental stage, due to differential binding of transcription factors.
Two common coding polymorphisms have been identified in human PON1, L55M and Q192R, where the former has been associated with a greater production of mRNA (L allele) (22) and greater serum paraoxonase levels (18). Brophy et al. (21) found that this polymorphism is in linkage disequilibrium with a polymorphism in the promoter region (–108C/T). To determine whether sequence variations in regulatory regions may likewise account for the preferential pattern of the expression observed in mice, we sequenced the 1 kb region upstream from the coding region and identified two SNPs. In silico analysis of the SNPs identified a putative binding site for TFIID, a TATA-box-binding protein required for RNA polymerase II activity, at SNP –227, implicating a possible role for this general transcription factor in the expression pattern of Pon1. However, this analysis does not exclude the possibility that cis-acting regulatory elements may be located in other regions of the gene, including introns and 3′ regulatory regions.
An analysis of IL10 production and promoter cis-acting variations within the locus identified specific haplotypes which were associated with higher allelic transcription (13). In a similar analysis, an intronic SNP in the lymphotoxin-α gene was found to be correlated to the protein’s production owing to the haplotype-specific binding of a bHLH protein which gave rise to allele-specific regulation (39). Such variations may account for the difference in Pon1 gene expression levels seen between JF1 and CAST mice strains. Further analyses using reporter-based promoter studies may corroborate the impact of cis-acting variations on Pon1 allele-specific expression.
The results of this study emphasize the importance of determining expression levels, not only in reciprocal crosses as shown here (3,23), but also at different developmental stages when analysing preferential patterns of expression. This is highlighted by the fact that each gene in the paraoxonase family shows a different pattern of expression, although located within a cluster of imprinted genes. Such a finding could only be proved by reciprocal analyses of hybrid samples, which allows one to distinguish between monoallelic or preferential expression and true imprinting which must be parent-of-origin specific. In addition, our results were duplicated on an independent set of liver samples, further stressing the absence of random expression.
Our findings further stress the need to determine allelic levels at disease loci, since differences in these levels are sources of phenotypic variation in human genetic diseases. If the mechanism of Pon1 regulation is conserved, it may imply that polymorphisms in human PON1 may not be a clear indicator of increased risk for atherosclerosis, since the expression of both alleles is neither equivalent nor static. However, further experiments need to be performed to determine levels of human PON1 allelic variance.
MATERIALS AND METHODS
Allelic expression analysis of murine tissues
Hybrid crosses were performed between C57BL/6J and CAST/Ei, and C57BL/6J and JF1/Ms, and tissues were obtained from the F1 generations and their reciprocal crosses at 12.5 and 15.5 dpc and P0. Total RNA was extracted using TRIzol reagent (Invitrogen) as outlined by the manufacturer. Two micrograms of DNase-treated total RNA was used for cDNA synthesis using random primers (SuperScript II reverse transcriptase, Invitrogen).
To identify SNPs in Pon1, a 607 bp fragment was sequenced from cDNA using primers Pon1-F and Pon1-R whose sequences are 5′-ACAAGAACCATCGGTCTTCC-3′ and 5′-CCTTCTGCTACCACCTGGAC-3′ (23), respectively. The identified SNP at nucleotide 280 of NM_011134 was confirmed in genomic DNA by PCR amplification using primers Exon-4F (5′-TGATGTCTCGAGAGCAATGG-3′) and Exon-3R (5′-TGCACCACGTTTGAAACAAT-3′). Genomic SNPs were identified in the promoter using primers Promoter-F 5′-ATGGCCTGAGACAGATGGAC-3′ and Promoter-R 5′-CCTCCTTCCACCACACAAGT-3′, which amplified an 803 bp fragment. The cycling conditions were initial denaturation at 94°C for 5 min, followed by 35 cycles for cDNA and 40 cycles for genomic DNA, denaturation at 94°C for 30 s, annealing at 57.5°C for 30 s and extension at 72°C for 90 s. Products were purified using microCLEAN (Microsone Ltd) and were subsequently sequenced on an ABI 3730XL using BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems), combined with Half BigDye sequencing buffer (Sigma Aldrich). Biased expression levels were confirmed in at least two embryos at each stage of development and for each hybrid cross.
Pyrosequencing analysis
Pon1 cDNA was amplified for pyrosequencing using Taq2000 (Stratagene) following the manufacturer’s protocol and using primers (forward) 5′-GACGGGTGCTGAAGACTTAGA-3′ and (reverse) 5′- AGGCTTACTGGGATCGAAACT-3′. The biotinylated products were purified using streptavidin sepharose (GE Healthcare), following the PSQ 96 sample preparation guide. The sequencing primer used in the pyrosequencing reaction was 5′-GGACTAACTTTC TTTAGCAC-3′ at a concentration of 0.4 μm per reaction. Pyrosequencing was performed using the Pyro Gold Enzyme Mixture (Biotage) and analysed using PSQ 96MA 2.1 ID system.
SNaPshot analysis
Unincorporated PCR primers and dNTPs were removed from 25 μl of PCR products by the addition of 8.3 U of shrimp alkaline phosphatase (SAP) and 3.3 U of Exonuclease I (Exo) (USB). Mixtures were incubated at 37°C for 1 h followed by 75°C for 15 min to inactivate the enzymes. SNP genotyping was performed using SNaPshot single-basepair extension reactions (Applied Biosystems). Primers for the extension reactions were designed according to the manufacturer’s instructions: SNP-g-mPON1-ss2 (5′-GACTTATAATAAATGTCAATAAACACTCAC-3′) was used to quantify the allelic ratios of genomic samples, and mPON1 (5′-GACTAATGGACTAACTTTCTTTAGCAC-3′) was used for cDNA.
Reactions contained 7 µl of cleaned PCR product, 2 µl of SNaPshot multiplex enzyme mix and 2 pmol of primer for a total volume of 10 µl. Cycling conditions for the extension reaction were 25 cycles of 96°C for 10 s, 55°C for 5 s and 60°C for 30 s. Following the extension reaction, unincorporated ddNTPs were removed by adding 1 U of SAP and incubating products at 37°C for 1 h followed by 75°C for 15 min.
Volumes of 1–2 µl of SNaPshot reactions were suspended in 9 µl of Hi-Di formamide (ABI) and run on an ABI3100 genetic analyser (Applied Biosystems) using the POP4 polymer and dye set E5. Results were analysed using the GENESCAN v. 3.1 software.
Real-time quantitative PCR
To compare Pon1 levels at different stages of embryonic development, quantitative PCR was performed using the Brilliant SYBR Green qPCR Master Mix (Stratagene). The primers used for the reaction were qPCR-Pon1-F (5′-CGGGTGCTGAAGACTTAGAGA-3′) and qPCR-Pon1-R (5′-CTCTGACACTGCTGGCTCCT-3′). The PCR reactions were performed in triplicate and in separate tubes. Absolute quantitation of Pon1 was obtained from cDNA from hybrid mice at various stages of growth. Results were normalized by β-2-microglobulin and β-actin, which were quantified using the following primers: β-2-microglobulin-F (5′-ATGGGAAGCCGAACATACTG-3′) and β-2-microglobulin-R (5′-CAGTCTCAGTGGGGGTGAAT-3′); β-actin-F (5′-TTGTTACCAACTGGGACGAC-3′) and β-actin-R (5′- TCTCAGCTGTGGTGGTGAAG-3′). Results were analysed using the standard curve method according to the manufacturer's instructions using the Mx3005P quantitative PCR system (Stratagene, La Jolla, USA). The system’s default PCR conditions were used, with annealing temperature at 58°C. The standard curve for all three transcripts quantified was developed using dilutions of a single-liver cDNA sample.
Methylation analysis
DNA was extracted from livers of hybrid embryo from the offspring of JF1/Ms (JF1) and C57BL/6J (BL6) using DNeasy blood and tissue kit (Qiagen). Bisulphite treatment was performed using the EZ methylation protocol (Zymo Research). The bisulphite-treated promoter region of Pon1 was amplified using the following primers: (forward) 5′-GTTAGAGTTTTTTAGAGGTATTTTGTTGG-3′ and (reverse) 5′-CATAACTAACACTCAATAAACCTCAATC-3′. One microlitre of the reaction was used for a semi-nested reaction where the same forward primer was used with the following reverse primer: 5′-CCTCAATCACATAAAAAAAATACTAAATAA-3′. The amplified product was purified using microCLEAN (Microzone Ltd) and was subcloned using the TOPO TA-cloning system (Invitrogen).
Human foetal tissues
Foetal tissues were obtained from the MRC Tissue Bank at Imperial College London, Hammersmith hospital site. Local ethical approval for the use of the collection was granted by the Hammersmith, Queen Charlotte’s and Chelsea and Acton Hospitals Research Ethics Committee (2001/6028).
Expression analysis on human foetal samples
Total RNA was extracted from foetal tissues using TRIzol (Life Technologies). RT-PCR of PON1 was performed on cDNA synthesised from the total RNA following reverse transcription using the primer pair 1F (5′-TATTGTTGCTGTGGGACCTGAG-3′) and 1R (5′-CCACAGATATGTTATCCACG-3′). For each RNA sample, an RT-negative control was used and standard GAPDH primers were used to confirm RNA integrity.
A 97 bp DNA fragment was amplified from foetal genomic DNA using primers ex6 1F (5′-TATTGTTGCTGTGGGACCTGAG-3′) and 2R (5′-CAGGCTAAACCCAAATACATCTC-3′) and was sequenced to assay for SNP rs662 on exon 6. The cycling conditions were initial denaturation at 94°C for 3 min, followed by 35 cycles denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 90 s. The PCR products were purified and sequenced using primer 5′-TATTGTTGCTGTGGGACCTGAG-3′. Allelic expression pattern was determined by comparing the peak heights of two alleles at an SNP site in sequence electropherograms. When the peak height for one allele was lower than one-tenth of that of the other allele, it was interpreted as a monoallelic expression. When one peak was equal or higher than one-tenth of the other and lower than the half of the other, it was assigned to be a preferential expression. When one peak was equal or higher than the half of the other, it was regarded as a biallelic expression.
SUPPLEMENTARY MATERIAL
FUNDING
The research is supported by grants from the Canadian Institutes of Health Research (CIHR) by grant MOP-74527, the McLaughlin Centre for Molecular Medicine, Genome Canada/Ontario Genomics Institute and the Hospital for Sick Children Foundation. L.P.-K. is supported by the Natural Sciences and Engineering Research Council (NSERC). Research by D.M., G.E.M and E.B. is supported by the Wellcome Trust and the MRC. S.W.S. is Scholar of the Canadian Institute for Advanced Research (CIFAR) and holds the GlaxoSmithKline Pathfinder Chair in Genetics and Genomics at the University of Toronto.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the technical assistance of Dr Tara Paton, Adam Smith, Ka Ki Michelle Lee, Katharine Sansom, Jennifer Skaug and others in The Centre for Applied Genomics (http://tcag.bioinfo.sickkids.on.ca/) at The Hospital for Sick Children.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray N.J., Buckland P.R., Owen M.J., O’Donovan M.C. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- 3.Cowles C.R., Hirchhorn J.N., Altshuler D., Lander E. Detection of regulatory variation in mouse genes. Nat. Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- 4.Guo M., Rupe M.A., Zinselmeier C., Habben J., Bowen B.A., Smith O.S. Allelic variation of gene expression in maize hybrids. Plant Cell. 2004;16:1707–1716. doi: 10.1105/tpc.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H., Dobbie Z., Gruber S.B., Markowitz S., Romans K., Giardiello F.M., Kinzler K.W., Vogelstein B. Small changes in expresion affect predisposition to tumorigenesis. Nat. Genet. 2001;30:25–26. doi: 10.1038/ng799. [DOI] [PubMed] [Google Scholar]
- 6.Yang W.-S., Tsou P.-L., Lee W.-J., Tseng D.-L., Chen C.-L., Peng C.-C., Lee K.-C., Chen M.-J., Huang C.-J., Tai T.-Y., et al. Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. J. Mol. Med. 2003;81:428–434. doi: 10.1007/s00109-002-0409-4. [DOI] [PubMed] [Google Scholar]
- 7.Bray N.J., Buckland P.R., Williams N.M., Williams H.J., Norton N., Owen M.J., O’Donovan M.C. A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am. J. Hum. Genet. 2003;73:152–161. doi: 10.1086/376578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laitinen T., Polvi A., Rydman P., Vendelin J., Pulkkinen V., Salmikangas P., Siru M., Rehn M., Pirskanen A., Rautanen A., et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004;304:300–304. doi: 10.1126/science.1090010. [DOI] [PubMed] [Google Scholar]
- 9.Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genomic-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastinen T., Sladek R., Gurd S., Sammak A.A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brändström H., et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol. Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- 11.Cheung V.G., Conlin L.K., Weber T.M., Arcaso M., Jen K.-Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 12.Yan H., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 13.Kurreeman F.A., Schonkeren J.J., Heijmans B.T., Toes R.E., Huizinga T.W. Transcription of the IL10 gene reveals allele-specific regulation at the mRNA level. Hum. Mol. Genet. 2004;13:1755–1762. doi: 10.1093/hmg/ddh187. [DOI] [PubMed] [Google Scholar]
- 14.Mackness M., Arrol S., Durrington P. Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Lett. 1991;29:152–154. doi: 10.1016/0014-5793(91)80962-3. [DOI] [PubMed] [Google Scholar]
- 15.Mackness M., Arrol S., Abbott C., Durrington P. Protection of low-density lipoprotein against oxidative modification by high-density lipoprotein associated paraoxonase. Atherosclerosis. 1993;104:129–135. doi: 10.1016/0021-9150(93)90183-u. [DOI] [PubMed] [Google Scholar]
- 16.Watson A., Berliner J., Hama S., La Du B., Faull K., Fogelman A.M., Navab M. Protective effect of high density lipoprotein associated paraoxonase. Inhibition of the biological activity of minimally oxidized low density lipoprotein. J. Clin. Invest. 1995;96:2882–2891. doi: 10.1172/JCI118359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih D.M., Gu L., Xia Y.-R., Navab M., Li W.-F., Hama S., Castellani L.W., Furlong C.E., Costa L.G., Fogelman A.M., et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;364:284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- 18.Garin M.-C.B., James R.W., Dussoix P., Blanche H., Passa P., Froguel P., Ruiz J. Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J. Clin. Invest. 1997;99:62–66. doi: 10.1172/JCI119134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brophy V.H., Hastings M.D., Clendenning J.B., Richter R.J., Jarvik G.P., Furlong C.E. Polymorphisms in the human paraoxonase (PON1) promoter. Pharmacogenetics. 2001;11:77–84. doi: 10.1097/00008571-200102000-00009. [DOI] [PubMed] [Google Scholar]
- 20.Suehiro T., Nakamura T., Inoue M., Shiinoki T., Ikeda Y., Kumon Y., Shindo M., Tanaka H., Hashimoto K. A polymorphism upstream from the human paraoxonase (PON1) gene and its association with PON1 expression. Atherosclerosis. 2000;150:295–298. doi: 10.1016/s0021-9150(99)00379-2. [DOI] [PubMed] [Google Scholar]
- 21.Brophy V.H., Jampsa J.B., Clendenning J.B., McKinstry L.A., Jarvik G.P., Furlong C.E. Effects of 5′ regulatory-region polymorphisms on paraoxonse-gene (PON1) expression. Am. J. Hum. Genet. 2001;68:1428–1436. doi: 10.1086/320600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leviev I., Negro F., James R.W. Two alleles of the human paraoxonase gene produce different amounts of mRNA: an explanation for differences in serum concentrations of paraoxonase associated with the (Leu-Met54) polymorphism. Arterioscler. Thromb. Vasc. Biol. 1997;17:2935–2939. doi: 10.1161/01.atv.17.11.2935. [DOI] [PubMed] [Google Scholar]
- 23.Ono R., Shiura H., Aburatani H., Kohda T., Kaneko-Ishino T., Ishino F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 2003;13:1696–1705. doi: 10.1101/gr.906803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakabayashi K., Makino S., Minagawa S., Smith A.C., Bamforth J.S., Stanier P., Preece M., Parker-Katiraee L., Paton T., Oshimura M., et al. Genomic imprinting of PPP1R9A encoding neurabin I in skeletal muscle and extra-embryonic tissues. J. Med. Gen. 2004;41:601–608. doi: 10.1136/jmg.2003.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walter J., Paulsen M. The potential role of gene duplications in the evolution of imprinting mechanisms. Hum. Mol. Genet. 2003;12:215R–220R. doi: 10.1093/hmg/ddg296. [DOI] [PubMed] [Google Scholar]
- 26.Verona R.I., Mann M.R., Bartolomei M.S. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 27.Sun A., Ge J., Siffert W., Frey U.H. Quantification of allele-specific G-protein beta3 subunit mRNA transcripts in different human cells and tissues by pyrosequencing. Eur. J. Hum. Genet. 2005;13:361–369. doi: 10.1038/sj.ejhg.5201334. [DOI] [PubMed] [Google Scholar]
- 28.Wittkopp P.J., Haerum B.K., Clark A.G. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 29.Norton N., Williams N.M., Williams H.J., Spurlock G., Kirov G., Morris D.W., Hoogendoorn B., Owen M.J., O’Donovan M.C. Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum. Genet. 2002;110:417–478. doi: 10.1007/s00439-002-0706-6. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins J.M., Southam L., Price A.J., Mustafa Z., Carr A., Loughlin J. Extreme context specificity in differential allelic expression. Hum. Mol. Genet. 2007;16:537–546. doi: 10.1093/hmg/ddl488. [DOI] [PubMed] [Google Scholar]
- 31.Cole T.B., Jampsa J.B., Walter B.J., Arndt T.L., Richter R.J., Shih D.M., Tward A., Lusis A.J., Jack R.M., Costa L.G., et al. Expression of human paraoxonase (PON1) during development. Pharmacogenetics. 2003;13:357–364. doi: 10.1097/00008571-200306000-00007. [DOI] [PubMed] [Google Scholar]
- 32.Bunzel R., Blumcke I., Cichon S., Normann S., Schramm J., Propping P., Nothen M.M. Polymorphic imprinting of the serotonin-2A receptor gene in human adult brain. Mol. Brain Res. 1998;59:90–92. doi: 10.1016/s0169-328x(98)00146-6. [DOI] [PubMed] [Google Scholar]
- 33.Jinno Y., Yun K., Nishiwaki K., Kubota T., Ogawa O., Reeve A., Niikawa N. Mosaic and polymorphic imprinting of the WT1 gene in humans. Nat. Genet. 1994;6:305–309. doi: 10.1038/ng0394-305. [DOI] [PubMed] [Google Scholar]
- 34.Enattah N.S., Sahi T., Savilahti E., Terwilliger J.D., Peltonen L., Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Harvey C.B., Pratt W.S., Sams V.R., Sarner M., Rossi M., Auricchio S., Swallow D.M. The lactase persistence/non-persistence polymorphism is controlled by a cis-acting element. Hum. Mol. Genet. 1995;4:657–662. doi: 10.1093/hmg/4.4.657. [DOI] [PubMed] [Google Scholar]
- 36.Kuokkanen M., Enattah N.S., Oksanen A., Savilahti E., Orpana A., Järvelä I. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut. 2003;52:647–652. doi: 10.1136/gut.52.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troelsen J.T., Olsen J., Moller J., Sjostrom H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003;125:1686–1694. doi: 10.1053/j.gastro.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 38.Rasinperä H., Kuokkanen M., Kolho K.L., Lindahl H., Enattah N.S., Savilahti E., Orpana A., Järvelä I. Transcriptional downregulation of the lactase (LCT) gene during childhood. Gut. 2005;54:1660–1661. doi: 10.1136/gut.2005.077404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight J.C., Keating B.J., Kwiatkowski D.P. Allele-specific repression of lymphotoxin-a by activated B cell factor-1. Nat. Genet. 2004;36:394–399. doi: 10.1038/ng1331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.