Abstract
The clinical overlap between monogenic Familial Hemiplegic Migraine (FHM) and common migraine subtypes, and the fact that all three FHM genes are involved in the transport of ions, suggest that ion transport genes may underlie susceptibility to common forms of migraine. To test this leading hypothesis, we examined common variation in 155 ion transport genes using 5257 single nucleotide polymorphisms (SNPs) in a Finnish sample of 841 unrelated migraine with aura cases and 884 unrelated non-migraine controls. The top signals were then tested for replication in four independent migraine case–control samples from the Netherlands, Germany and Australia, totalling 2835 unrelated migraine cases and 2740 unrelated controls. SNPs within 12 genes (KCNB2, KCNQ3, CLIC5, ATP2C2, CACNA1E, CACNB2, KCNE2, KCNK12, KCNK2, KCNS3, SCN5A and SCN9A) with promising nominal association (0.00041 < P < 0.005) in the Finnish sample were selected for replication. Although no variant remained significant after adjusting for multiple testing nor produced consistent evidence for association across all cohorts, a significant epistatic interaction between KCNB2 SNP rs1431656 (chromosome 8q13.3) and CACNB2 SNP rs7076100 (chromosome 10p12.33) (pointwise P = 0.00002; global P = 0.02) was observed in the Finnish case–control sample. We conclude that common variants of moderate effect size in ion transport genes do not play a major role in susceptibility to common migraine within these European populations, although there is some evidence for epistatic interaction between potassium and calcium channel genes, KCNB2 and CACNB2. Multiple rare variants or trans-regulatory elements of these genes are not ruled out.
INTRODUCTION
Migraine (OMIM no. 157300) is a common neurovascular disorder with a strong genetic component. Although genetic variants have been well established for Mendelian forms of migraine, no susceptibility variants associated with common forms of migraine have been consistently reported. So far, most association studies for headache traits have been performed in relatively small study samples and typically for only one or two genes per study. Thus, comprehensive candidate gene association studies saturating large gene families or whole genome association studies have not been reported for headache traits as yet.
The best insight to understand the genetic background underlying pathogenetic mechanisms in migraine is provided by studies in Familial Hemiplegic Migraine (FHM), a Mendelian subtype of migraine with aura (MA) associated with hemiparesis. Families with FHM type 1 (FHM1) (OMIM no. 141500) have missense mutations in CACNA1A (OMIM no. 601011) (1), which encodes the pore-forming α1 subunit of neuronal voltage-gated Cav2.1 (P/Q-type) Ca2+ channels, type 2 (FHM2) (OMIM no. 602481) patients show mutations in ATP1A2 (OMIM no. 182340) (2), which encodes the α2 subunit of Na+/K+ ATPases that are expressed in astrocytes in the adult brain, whereas type 3 (FHM3) (OMIM no. 609634) patients have mutations in the α1 subunit of the neuronal voltage-gated Na+ channel Nav1.1 (SCN1A) (OMIM no. 182389) (3).
All three FHM genes are involved in the transport of ions and suggest that disturbances of ions and neurotransmitter levels in the synaptic cleft play a key role in migraine pathophysiology by influencing neuronal excitability (4,5). Experimental evidence points to cortical spreading depression (CSD)—a wave of neuronal and glial depolarization followed by long-lasting suppression of neuronal activity that propagates slowly over the cerebral cortex—as the key event for episodic activation of the trigeminovascular system, resulting in migraine headache (6,7). CACNA1A FHM1 mutations are predicted to cause increased release of glutamate (the predominant excitatory amino acid transmitter in brain) in the cortex, thus increasing neuronal cortical network excitability rendering the cortex more susceptible to CSD (8). ATP1A2 FHM2 mutations result in reduced activity or decreased affinity for K+ of the Na+/K+ pump and thus impaired clearance of K+ and glutamate from the synaptic cleft. SCN1A FHM3 mutations are believed to have effects similar to CACNA1A mutations, in the sense that they predict increased neuronal excitability.
A relevant hypothesis is that the same pathways involved in rare Mendelian forms of migraine might also be involved in the pathogenesis of more common forms of migraine. This is especially appropriate as the main symptoms of headache and aura (as well as the accompanying symptoms of nausea, photophobia and phonophobia) of FHM attacks are very similar to those of MA, and both types of attack occur in FHM patients (9). In fact, migraine without aura (MO) and MA, frequently co-occur. A Dutch study found that 42% of active migraineurs with aura also reported having migraine attacks without aura (10). Moreover, MO and MA frequently coexist within a family; a Headache Centre in Italy reported that 45% of MA families also had MO cases (11) and the co-occurrence of FHM, MA and MO has been reported in families (12,13). Furthermore, changes in the presenting symptoms of migraine attacks from hemiplegic to severe headache with or without aura in later life (12), as well as the development of aura among subjects with MO and the converse (12,14), suggest overlapping aetiology.
The overlap in symptomatology among migraine subtypes and the fact that all three known FHM genes are involved in the transport of ions, strongly suggests that ion channels and ion transporters, may also underlie susceptibility to the more common forms of MO and MA. The relevance of ion channels and transporters is also apparent from the fact that effective migraine prophylactic agents such as valproate and topiramate act on ion channels or transporters (15).
There are several reports that rare mutations in FHM genes may also play a role in families with MA and/or MO (16,17). A systematic investigation of rare predisposing variants in common migraine has become feasible because of recently improved technology to perform extensive sequencing in a large numbers of samples, but has not been performed. However, an equally relevant hypothesis is that common variants in either coding or non-coding regions of ion transport genes might predispose to migraine. Association of common variants to common traits can be studied with tools provided by HapMap (18) and modern genotyping techniques.
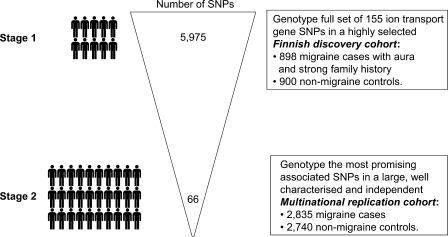
Our multicentre collaboration (the Migraine Genetics Consortium; MGC) assembled the largest cohort of patients with common migraine for a genetic mapping study to date. To investigate the potential role of genes involved in the transport of ions in common migraine, we performed the first comprehensive screen of common genetic variants in highly plausible candidate ion transport genes. A total of 5257 single nucleotide polymorphisms (SNPs) were examined in 155 genes in a large well-characterized Finnish case–control sample consisting of 841 unrelated MA cases and 884 unrelated non-migraine controls. Twelve genes producing nominally significant (P < 0.005) association signals in the Finnish sample were subsequently tested for replication in four independent migraine case–control samples from the Netherlands, Germany and Australia involving altogether 2835 migraine cases and 2740 controls.
Also, given the reported functional interaction between M-type KCNQ2 and KCNQ3 potassium channels (19) and their modulation by the KCNE2 subunit (20), plus an account of compound heterozygosity for two novel allelic ATP1A2 missense mutations in the proband of a FHM family (21), we also tested for interactions between SNPs with nominally significant (P < 0.005) association signals in the Finnish sample.
RESULTS
Of the 5269 SNPs passing Perlegen's quality control (QC) criteria analyzed in the Finnish case–control cohort, PLINK identified 12 SNPs with MAF < 0.01 and subsequently excluded them from further analysis. Importantly, the PLINK identity-by-state (IBS)-based cluster analysis determined that one cluster best fit the data, thus providing no evidence for population stratification within the Finnish cohort. Absence of stratification was further supported with PLINK reporting a genomic inflation factor (based on median chi-squared) of 1 (values >1 indicate population structure) and mean chi-squared statistic of 0.945 (a mean of 1 is expected under the null hypothesis of no stratification).
No evidence for association with MA was observed for SNPs within the footprints of the three FHM genes. Briefly, none of the 25 ATP1A2 and 26 SCN1A SNPs produced point-wise P-values <0.05. Although five of the 87 CACNA1A SNPs (rs10416717, rs2419233, rs7249323, rs11882861, rs2074880) did produce nominal point-wise P-values ≤0.05 (0.0131, 0.01822, 0.04142, 0.04257, 0.04915, respectively), they were not significant after adjustment for testing multiple SNPs.
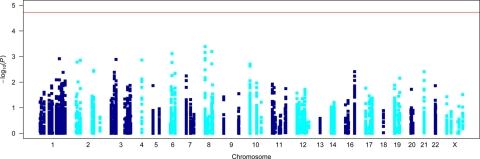
Allelic association results −log10(P) for all 5257 SNPs are presented in Figure 1 (also see Supplementary Material, Table S1). The smallest observed point-wise P-value from allelic association analysis of 5257 SNPs in the Finnish 841 MA cases and 884 controls was obtained for SNP rs13276133 (χ12 = 12.49; point-wise P = 0.00041) residing in the potassium voltage-gated channel, Shab-related, member 2 (KCNB2) gene on chromosome 8q13.3. The next smallest P-value obtained for SNPs within KCNQ3 on 8q24.22 (rs13267466; P = 0.00064) and CLIC5 on 6p12.3 (rs2095771; P = 0.00074) also showed encouraging evidence for association. An additional nine genes (ATP2C2, CACNA1E, CACNB2, KCNE2, KCNK12, KCNK2, KCNS3, SCN5A and SCN9A) produced promising association signals (P < 0.005) (Table 1). However, after multiple test correction for the analysis of 5257 SNPs via permutation, our smallest point-wise P-value (rs13276133; P = 0.00041) was not globally significant with a global EMP2 P-value of 0.7713. Hence, none of the tested SNPs reached global statistical significance (EMP2 P ≤ 0.05) (Table 1).
Figure 1.
Allelic association results for Finnish MA cases versus controls. Horizontal red line indicates the required global significance threshold [−log10(P) = 4.7286] to ensure an overall type I error rate of 5%.
Table 1.
Single SNP association results for single nucleotide polymorphisms selected for replication from Finnish case–control cohort
| Chr | NCBI build 36 position (bp) | dbSNP rs no. | Primary gene HGNC symbol | Case minor allele | Case MAF | Control MAF | Allelic P | Allelic OR | Allelic EMP2 P |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 179680697 | rs12073221 | CACNA1E | C | 0.3165 | 0.275 | 0.008168 | 1.22 | 1 |
| 1 | 179691156 | rs3856087 | CACNA1E | A | 0.3356 | 0.2837 | 0.001215 | 1.28 | 0.9839 |
| 1 | 179702677 | rs644796 | CACNA1E | C | 0.467 | 0.4255 | 0.01537 | 1.18 | 1 |
| 1 | 179709099 | rs505738 | CACNA1E | C | 0.4619 | 0.4268 | 0.0382 | 1.15 | 1 |
| 1 | 213353044 | rs4300189 | KCNK2 | A | 0.1391 | 0.1599 | 0.08803 | 0.85 | 1 |
| 1 | 213365431 | rs11120499 | KCNK2 | T | 0.2218 | 0.2576 | 0.01368 | 0.82 | 1 |
| 1 | 213395890 | rs12567520 | KCNK2 | A | 0.09869 | 0.1179 | 0.06979 | 0.82 | 1 |
| 1 | 213398386 | rs2363556 | KCNK2 | G | 0.1391 | 0.1612 | 0.0698 | 0.84 | 1 |
| 1 | 213428215 | rs1556905 | KCNK2 | A | 0.3254 | 0.3502 | 0.129 | 0.90 | 1 |
| 1 | 213428830 | rs10494994 | KCNK2 | A | 0.1663 | 0.2043 | 0.004157 | 0.78 | 1 |
| 1 | 213458463 | rs12143625 | KCNK2 | C | 0.1901 | 0.2244 | 0.01314 | 0.81 | 1 |
| 1 | 213467066 | rs6670661 | KCNK2 | G | 0.3061 | 0.333 | 0.09116 | 0.88 | 1 |
| 1 | 213476729 | rs6686529 | KCNK2 | G | 0.1902 | 0.2254 | 0.01113 | 0.81 | 1 |
| 2 | 17908169 | rs12996816 | KCNS3 | T | 0.1177 | 0.08541 | 0.001656 | 1.43 | 0.9952 |
| 2 | 17916155 | rs10178489 | KCNS3 | G | 0.2996 | 0.2661 | 0.02894 | 1.18 | 1 |
| 2 | 47660997 | rs17568951 | KCNK12 | C | 0.1946 | 0.1534 | 0.001409 | 1.33 | 0.9909 |
| 2 | 166879155 | rs11890824 | SCN9A | A | 0.4512 | 0.4745 | 0.1714 | 0.91 | 1 |
| 2 | 166889773 | rs11898284 | SCN9A | G | 0.1795 | 0.1433 | 0.003754 | 1.31 | 1 |
| 2 | 166902637 | rs12622743 | SCN9A | G | 0.1798 | 0.1433 | 0.003573 | 1.31 | 1 |
| 3 | 38599347 | rs3922843 | SCN5A | A | 0.2429 | 0.2036 | 0.005644 | 1.26 | 1 |
| 3 | 38602829 | rs7645358 | SCN5A | G | 0.1611 | 0.1799 | 0.1435 | 0.88 | 1 |
| 3 | 38609146 | rs7374540 | SCN5A | C | 0.4007 | 0.4274 | 0.1114 | 0.90 | 1 |
| 3 | 38653505 | rs7427106 | SCN5A | G | 0.1397 | 0.1753 | 0.004134 | 0.76 | 1 |
| 3 | 38656222 | rs9311195 | SCN5A | G | 0.1534 | 0.1878 | 0.007321 | 0.78 | 1 |
| 3 | 38672343 | rs9861643 | SCN5A | T | 0.1974 | 0.22 | 0.1033 | 0.87 | 1 |
| 6 | 45993676 | rs11758298 | CLIC5 | T | 0.1308 | 0.1471 | 0.1677 | 0.87 | 1 |
| 6 | 45995399 | rs1416167 | CLIC5 | T | 0.2903 | 0.2571 | 0.02875 | 1.18 | 1 |
| 6 | 45996051 | rs3729618 | CLIC5 | G | 0.2905 | 0.2576 | 0.03071 | 1.18 | 1 |
| 6 | 46008544 | rs3777567 | CLIC5 | A | 0.2655 | 0.2197 | 0.001715 | 1.28 | 0.9966 |
| 6 | 46012610 | rs2095771 | CLIC5 | A | 0.25 | 0.2017 | 0.0007735 | 1.32 | 0.9358 |
| 6 | 46018442 | rs3777580 | CLIC5 | C | 0.321 | 0.2778 | 0.005546 | 1.23 | 1 |
| 6 | 46026947 | rs3777585 | CLIC5 | T | 0.1871 | 0.1586 | 0.0265 | 1.22 | 1 |
| 8 | 73598648 | rs12550268 | KCNB2 | A | 0.382 | 0.4189 | 0.02711 | 0.86 | 1 |
| 8 | 73601624 | rs1431659 | KCNB2 | A | 0.2271 | 0.2704 | 0.003311 | 0.79 | 0.9999 |
| 8 | 73609424 | rs5007872 | KCNB2 | G | 0.2187 | 0.2669 | 0.001311 | 0.77 | 0.9876 |
| 8 | 73614029 | rs349358 | KCNB2 | C | 0.1249 | 0.1489 | 0.04 | 0.82 | 1 |
| 8 | 73618324 | rs349355 | KCNB2 | A | 0.1244 | 0.1493 | 0.03353 | 0.81 | 1 |
| 8 | 73651525 | rs349335 | KCNB2 | C | 0.1013 | 0.131 | 0.006741 | 0.75 | 1 |
| 8 | 73653731 | rs1431656 | KCNB2 | G | 0.2525 | 0.3056 | 0.0006998 | 0.77 | 0.9172 |
| 8 | 73659142 | rs1542709 | KCNB2 | A | 0.2368 | 0.2773 | 0.00673 | 0.81 | 1 |
| 8 | 74006869 | rs7006287 | KCNB2 | G | 0.4006 | 0.4552 | 0.001205 | 0.80 | 0.983 |
| 8 | 74012757 | rs11782118 | KCNB2 | A | 0.365 | 0.4184 | 0.001352 | 0.80 | 0.9892 |
| 8 | 74016115 | rs922772 | KCNB2 | C | 0.4518 | 0.4072 | 0.008148 | 1.20 | 1 |
| 8 | 74017814 | rs13276133 | KCNB2 | C | 0.3556 | 0.4145 | 0.000409 | 0.78 | 0.7713 |
| 8 | 133408579 | rs13267466 | KCNQ3 | C | 0.1198 | 0.1602 | 0.0006373 | 0.71 | 0.897 |
| 10 | 18458186 | rs7897594 | CACNB2 | G | 0.3719 | 0.4117 | 0.01682 | 0.85 | 1 |
| 10 | 18460390 | rs11596974 | CACNB2 | A | 0.3641 | 0.3991 | 0.03481 | 0.86 | 1 |
| 10 | 18799543 | rs7076100 | CACNB2 | A | 0.419 | 0.4559 | 0.02933 | 0.86 | 1 |
| 10 | 18800641 | rs11598027 | CACNB2 | T | 0.147 | 0.1117 | 0.001975 | 1.37 | 0.9986 |
| 10 | 18803689 | rs1409202 | CACNB2 | A | 0.147 | 0.1122 | 0.002354 | 1.36 | 0.9998 |
| 10 | 18822886 | rs8181477 | CACNB2 | C | 0.4613 | 0.504 | 0.01226 | 0.84 | 1 |
| 10 | 18823994 | rs11014504 | CACNB2 | C | 0.4457 | 0.4819 | 0.03356 | 0.86 | 1 |
| 16 | 82964929 | rs17740111 | ATP2C2 | C | 0.1942 | 0.2347 | 0.003894 | 0.79 | 1 |
| 16 | 82971665 | rs7350833 | ATP2C2 | C | 0.2643 | 0.2333 | 0.03534 | 1.18 | 1 |
| 16 | 82974143 | rs10459853 | ATP2C2 | G | 0.1285 | 0.1619 | 0.005822 | 0.76 | 1 |
| 16 | 83002938 | rs247820 | ATP2C2 | C | 0.4388 | 0.4791 | 0.01756 | 0.85 | 1 |
| 16 | 83040732 | rs247889 | ATP2C2 | A | 0.4214 | 0.3776 | 0.008578 | 1.20 | 1 |
| 16 | 83055346 | rs429313 | ATP2C2 | A | 0.2811 | 0.2474 | 0.02536 | 1.19 | 1 |
| 16 | 83062088 | rs394533 | ATP2C2 | T | 0.3214 | 0.2874 | 0.03043 | 1.17 | 1 |
| 16 | 83063868 | rs400922 | ATP2C2 | G | 0.2935 | 0.256 | 0.01439 | 1.21 | 1 |
| 16 | 83065318 | rs390208 | ATP2C2 | A | 0.2843 | 0.2471 | 0.01406 | 1.21 | 1 |
| 21 | 34612656 | rs2834442 | KCNE2 | T | 0.2771 | 0.3222 | 0.003897 | 0.81 | 1 |
| 21 | 34620260 | rs1013063 | KCNE2 | T | 0.3526 | 0.3907 | 0.0205 | 0.85 | 1 |
| 21 | 34636414 | rs1467847 | KCNE2 | G | 0.35 | 0.3815 | 0.05496 | 0.87 | 1 |
| 21 | 34639061 | rs2834462 | KCNE2 | T | 0.2762 | 0.3171 | 0.008623 | 0.82 | 1 |
| 21 | 34643430 | rs8128316 | KCNE2 | C | 0.3909 | 0.4304 | 0.01872 | 0.85 | 1 |
Exploratory analysis of haplotypes spanning two to six contiguous SNPs also did not produce significant evidence for association (data not shown), with the smallest observed point-wise P-value (χ12 = 13.82; point-wise P = 0.00020) obtained for the specific 3-allele G-T-T haplotype of SNPs rs1998822–rs7076100–rs11598027 within the CACNB2 gene on chromosome 10p12.33. The omnibus 3-allele haplotype test for this three SNP window (correcting for haplotype tests for all observed 3-allele combinations) only produced a χ12 of 18.12 with point-wise P = 0.0012, compared with the single SNP point-wise P = 0.00198 for rs11598027. Given that these results are not corrected for testing, all other two to six SNP haplotypes, analysis of haplotypes in these data did not produce stronger evidence for association compared with the single SNP analyses.
In addition to the primary MA phenotype, we also performed exploratory association analyses with strict International Headache Society (IHS) migraine criteria (i.e. excluding the 114 MA individuals with aura symptoms not strictly fulfilling the IHS aura criteria, but diagnosed as MA by our clinical neurologist; Table 2) and the key trait components of pulsation, photophobia and phonophobia (Table 1, Supplementary Material, Table S1). Although these exploratory analyses did not provide increased evidence for association between MA and SNPs within the three FHM genes, they did produce the smallest observed P-value (χ12 = 16.00; point-wise P = 0.000063), obtained for SNP rs12996816 residing in the KCNS3 gene on chromosome 2p24.2 with the pulsation phenotype; however, this result does not surpass the primary analysis global significance threshold of point-wise P ≤ 0.000019. Furthermore, given an additional global multiple test correction is required for examining four additional traits; these exploratory analyses do not alter the overall (negative) interpretation of our individual SNP association results.
Table 2.
Demographic factors of the Finnish case and control groups
| Cases |
Controls | ||
|---|---|---|---|
| Twins (population-based sample) | Familial patients (clinic-based sample) | Twins | |
| n | 279 | 619 | 900 |
| Migraine status | MA 260 (93%) | MA 524 (85%) | No migraine |
| Other 19 (7%) | Other 95 (15%) | No family history of migraine | |
| Gender distribution % female | 81.7 | 79.2 | 75.6 |
| Mean age (years) | 56 | 48 | 57 |
| Age range (years) | 47–80 | 13–91 | 48–66 |
| Age range at the time of assessment (years) | 36–69 | 10–88 | 39–57 |
Other; migraine aura symptoms not fulfilling the International Headache Society aura criteria, but classified as migraine with aura by a clinical neurologist.
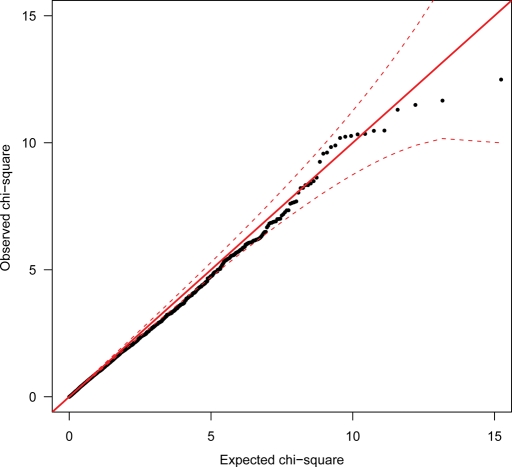
Although no single SNP or haplotype reached statistical significance, multiple small effect association signals in multiple genes may nonetheless be present. To examine this possibility, we plotted the observed distribution of the 5257 rank-ordered chi-square (χ12) values obtained from the single SNP allelic association analysis using MA as trait, against the distribution of χ12 values expected under the null hypothesis of no association. The resulting QQ-plot (Fig. 2) shows a definite, although non-significant preponderance of χ12 values ranging from around 9.573 (point-wise P = 0.00198) to 10.19 (point-wise P = 0.00141) [i.e. the top 11–15 χ12 values].
Figure 2.
QQ-plot of Finnish case–control allelic association results for clinical migraine with aura end-diagnosis (closed black circles). The solid red line represents the expected (reference) distribution under the null. The broken red lines indicate the point-wise 95% CI envelope based on the standard errors of order statistics for an independent sample from the reference distribution.
Applying the false discovery rate (FDR) approach to the 5257 rank-ordered allelic association P-values produced q-values (FDRs) of 0.673 and 0.692 for the 1st–11th and 12th–15th smallest P-values, respectively. Assigned q-values quickly and substantially increased in magnitude for the 16th (q-value = 0.773), 17th–23rd (q-value = 0.950) and remaining 24th–5257th ranked P-values (q-value = 0.9997). These results indicate that there are a maximum of four to five likely true-positives contained within the top 15 most significant allelic associations [i.e. true positive probability (1 − q-value) multiplied by the total number in the set with equivalent or smaller q-values: (1– 0.6922) × 15 = 4.62]. Also, our stage-1 nominal significance threshold for allelic association with MA in the Finnish cohort demarcates q-values of 0.950 and 0.9997, thus supporting P < 0.005 as a reasonable compromise between following-up true risk loci and the total number of loci to follow-up (i.e. minimizing the multiple-test burden while maintaining high replication power).
A total of 66 SNPs within 12 genes (Table 1) were selected for follow-up in the replication cohorts (see Supplementary Material, Tables S2a and S2b for estimates of power to replicate the Finnish association results). The 66 SNPs were primarily selected upon the basis of having the smallest allelic association results among all the Finnish MA cases and controls (P < 0.005). The SNPs within these 12 genes were also selected if they produced nominal evidence (point-wise P < 0.05) for MA and/or produced more significant P-values, compared with the primary MA phenotype, for association with the traits examined in the exploratory association analyses (Table 1, Supplementary Material, Table S1). In addition to the investigation of multiple effects within each gene, we selected multiple associated SNPs within the 12 most promising genes to help guard against potential assay failure in the follow-up replication studies. Counts of case–control individuals successfully genotyped are provided in Table 3.
Table 3.
Replication cohort sizes for individuals successfully genotyped
| Replication cohort | Cases |
Controls |
||||
|---|---|---|---|---|---|---|
| Total | Male | Female | Total | Male | Female | |
| Leiden | 800 | 192 | 608 | 946 | 520 | 426 |
| Cologne | 601 | 136 | 465 | 651 | 159 | 492 |
| Munich | 288 | 40 | 248 | 0 | 0 | 0 |
| Brisbane | 1146 | 203 | 943 | 1143 | 691 | 452 |
| Total | 2835 | 571 | 2264 | 2740 | 1370 | 1370 |
All 66 SNPs, except those which failed, were tested first in the Leiden sample with MA and MO (one SNP failed). Following the Leiden results, 60 SNPs were successfully tested in the population-based Brisbane sample with MA and MO. Finally, 44 SNPs were successfully tested in the Cologne and Munich clinic-based German MA samples. We note that due to the Munich cohort consisting of only migraine cases, two German-specific analyses were performed; (i) Cologne cases versus Cologne controls and (ii) combined Cologne cases and Munich cases versus Cologne controls. Post hoc analyses (not presented) in the replication cohorts containing a mixture of migraine sub-groups did not identify significant allele frequency differences between MO, MA and the latent class analysis (LCA)-based migraine groups, thus supporting the combination of these sub-groups. After QC checks, some genotype assays were failed in each cohort.
The results from association analysis of the 66 replication SNPs are presented in Table 4. Although a small number of SNPs showed significant evidence for replication (EMP2 P ≤ 0.05) in the Brisbane [CLIC5 SNP rs3729618 (EMP2 P = 0.0459)] and Cologne cohort [KCNE2 SNPs rs1467847 (EMP2 P = 0.0139) and rs1013063 (EMP2 P = 0.0193)], none of the SNPs produced consistent evidence for replication across the cohorts. Furthermore, none of the SNPs were significant in the combined replication cohort Cochran-Mantel-Haenszel (CMH) test after correction for testing 66 SNPs. Analysis of the Leiden, Brisbane and/or combined replication MA subset did not alter the overall results. Due to the associated CLIC5 alleles in the Finnish and Brisbane cohorts being in opposite directions, the KCNE2 associations remain as the sole result which could justify further investigation. See Supplementary Material, Table S3 for point-wise replication association analysis results for all 66 replication SNPs in the four individual replication cohorts and combined replication sample.
Table 4.
Replication association analysis results
| Chr | NCBI build 36 position (bp) | dbSNP rs no. | Primary gene HGNC symbol | Finnish allelic P (MA) | Finnish allelic EMP2 P (MA) | Leiden replication allelic EMP2 P | Cologne replication allelic EMP2 P | Cologne + Munich replication allelic EMP2 P | Brisbane replication allelic EMP2 P | Replication CMH pointwise P | Replication CMH EMP2 P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 179680697 | rs12073221 | CACNA1E | 0.00817 | 1 | 0.9985 | 0.1543 | 0.9993 | |||
| 1 | 179691156 | rs3856087 | CACNA1E | 0.00122 | 0.9839 | 1 | 1 | 0.9999 | 0.4142 | 1 | |
| 1 | 179702677 | rs644796 | CACNA1E | 0.01537 | 1 | 1 | 1 | 1 | 1 | 0.7915 | 1 |
| 1 | 179709099 | rs505738 | CACNA1E | 0.0382 | 1 | 1 | 1 | 1 | 1 | 0.872 | 1 |
| 1 | 213353044 | rs4300189 | KCNK2 | 0.08803 | 1 | 1 | 1 | 0.8259 | 1 | ||
| 1 | 213365431 | rs11120499 | KCNK2 | 0.01368 | 1 | 1 | 1 | 1 | 1 | 0.8649 | 1 |
| 1 | 213395890 | rs12567520 | KCNK2 | 0.06979 | 1 | 1 | 0.3703 | 1 | |||
| 1 | 213398386 | rs2363556 | KCNK2 | 0.0698 | 1 | 1 | 1 | 0.7654 | 1 | ||
| 1 | 213428215 | rs1556905 | KCNK2 | 0.129 | 1 | 1 | 1 | 0.664 | 1 | ||
| 1 | 213428830 | rs10494994 | KCNK2 | 0.00416 | 1 | 1 | 1 | 1 | 1 | 0.7921 | 1 |
| 1 | 213458463 | rs12143625 | KCNK2 | 0.01314 | 1 | 1 | 1 | 0.9547 | 1 | ||
| 1 | 213467066 | rs6670661 | KCNK2 | 0.09116 | 1 | 1 | 1 | 0.9523 | 1 | ||
| 1 | 213476729 | rs6686529 | KCNK2 | 0.01113 | 1 | 0.9967 | 1 | 0.696 | 1 | ||
| 2 | 17908169 | rs12996816 | KCNS3 | 0.00166 | 0.9952 | 1 | 1 | 1 | 1 | 0.3029 | 1 |
| 2 | 17916155 | rs10178489 | KCNS3 | 0.02894 | 1 | 1 | 1 | 1 | 1 | 0.9674 | 1 |
| 2 | 47660997 | rs17568951 | KCNK12 | 0.00141 | 0.9909 | 1 | 1 | 1 | 1 | 0.6712 | 1 |
| 2 | 166879155 | rs11890824 | SCN9A | 0.1714 | 1 | 1 | 0.2164 | 1 | |||
| 2 | 166889773 | rs11898284 | SCN9A | 0.00375 | 1 | 1 | 1 | 0.8456 | 1 | ||
| 2 | 166902637 | rs12622743 | SCN9A | 0.00357 | 1 | 1 | 0.3085 | 1 | |||
| 3 | 38599347 | rs3922843 | SCN5A | 0.00564 | 1 | 1 | 0.9948 | 0.9995 | 1 | 0.08824 | 0.9832 |
| 3 | 38602829 | rs7645358 | SCN5A | 0.1435 | 1 | 1 | 1 | 0.3676 | 1 | ||
| 3 | 38609146 | rs7374540 | SCN5A | 0.1114 | 1 | 1 | 1 | 0.6739 | 1 | ||
| 3 | 38653505 | rs7427106 | SCN5A | 0.00413 | 1 | 0.998 | 1 | 1 | 1 | 0.4155 | 1 |
| 3 | 38656222 | rs9311195 | SCN5A | 0.00732 | 1 | 1 | 1 | 1 | 1 | 0.8305 | 1 |
| 3 | 38672343 | rs9861643 | SCN5A | 0.1033 | 1 | 1 | 1 | 0.4628 | 1 | ||
| 6 | 45993676 | rs11758298 | CLIC5 | 0.1677 | 1 | 1 | 1 | 0.2692 | 1 | ||
| 6 | 45995399 | rs1416167 | CLIC5 | 0.02875 | 1 | 1 | 0.06909 | 0.03557 | 0.8129 | ||
| 6 | 45996051 | rs3729618 | CLIC5 | 0.03071 | 1 | 1 | 0.8306 | 0.9745 | 0.0459 | 0.3258 | 1 |
| 6 | 46008544 | rs3777567 | CLIC5 | 0.00172 | 0.9966 | 1 | 0.6888 | 0.938 | 0.2892 | 0.9135 | 1 |
| 6 | 46012610 | rs2095771 | CLIC5 | 0.00077 | 0.9358 | 1 | 0.7314 | 0.941 | 0.3037 | 0.6519 | 1 |
| 6 | 46018442 | rs3777580 | CLIC5 | 0.00555 | 1 | 1 | 1 | 1 | 0.5138 | 0.09749 | 0.9884 |
| 6 | 46026947 | rs3777585 | CLIC5 | 0.0265 | 1 | 1 | 0.1563 | 0.03622 | 0.8174 | ||
| 8 | 73598648 | rs12550268 | KCNB2 | 0.02711 | 1 | 1 | 0.9129 | 0.9347 | 0.9683 | 0.7111 | 1 |
| 8 | 73601624 | rs1431659 | KCNB2 | 0.00331 | 0.9999 | 1 | 0.6798 | 0.9853 | 0.9953 | 0.6447 | 1 |
| 8 | 73609424 | rs5007872 | KCNB2 | 0.00131 | 0.9876 | 1 | 0.7153 | 0.9728 | 1 | 0.7553 | 1 |
| 8 | 73614029 | rs349358 | KCNB2 | 0.04 | 1 | 1 | 0.9995 | 1 | 0.9996 | 0.5346 | 1 |
| 8 | 73618324 | rs349355 | KCNB2 | 0.03353 | 1 | 1 | 1 | 1 | 1 | 0.5737 | 1 |
| 8 | 73651525 | rs349335 | KCNB2 | 0.00674 | 1 | 1 | 1 | 1 | 1 | 0.6819 | 1 |
| 8 | 73653731 | rs1431656 | KCNB2 | 0.0007 | 0.9172 | 1 | 0.8643 | 0.9822 | 1 | 0.978 | 1 |
| 8 | 73659142 | rs1542709 | KCNB2 | 0.00673 | 1 | 1 | 0.9223 | 0.9938 | 1 | 0.9925 | 1 |
| 8 | 74006869 | rs7006287 | KCNB2 | 0.00121 | 0.983 | 0.9863 | 1 | 1 | 1 | 0.1335 | 0.998 |
| 8 | 74012757 | rs11782118 | KCNB2 | 0.00135 | 0.9892 | 0.9891 | 1 | 1 | 1 | 0.04141 | 0.8557 |
| 8 | 74016115 | rs922772 | KCNB2 | 0.00815 | 1 | 1 | 0.9997 | 0.9999 | 1 | 0.06849 | 0.9585 |
| 8 | 74017814 | rs13276133 | KCNB2 | 0.00041 | 0.7713 | 0.9975 | 1 | 1 | 0.9998 | 0.04397 | 0.8723 |
| 8 | 133408579 | rs13267466 | KCNQ3 | 0.00064 | 0.897 | 1 | 1 | 1 | 0.9996 | 0.5143 | 1 |
| 10 | 18458186 | rs7897594 | CACNB2 | 0.01682 | 1 | 0.9998 | 1 | 1 | 0.3177 | 0.003368 | 0.1659 |
| 10 | 18460390 | rs11596974 | CACNB2 | 0.03481 | 1 | 1 | 0.7107 | 0.05995 | 0.939 | ||
| 10 | 18799543 | rs7076100 | CACNB2 | 0.02933 | 1 | 0.9984 | 0.8954 | 0.6093 | 1 | ||
| 10 | 18800641 | rs11598027 | CACNB2 | 0.00198 | 0.9986 | 0.9998 | 1 | 1 | 1 | 0.9374 | 1 |
| 10 | 18803689 | rs1409202 | CACNB2 | 0.00235 | 0.9998 | 0.9328 | 1 | 0.9999 | 1 | 0.9421 | 1 |
| 10 | 18822886 | rs8181477 | CACNB2 | 0.01226 | 1 | 1 | 0.9947 | 1 | 1 | 0.6363 | 1 |
| 10 | 18823994 | rs11014504 | CACNB2 | 0.03356 | 1 | 1 | 1 | 0.2729 | 1 | ||
| 16 | 82964929 | rs17740111 | ATP2C2 | 0.00389 | 1 | 1 | 1 | 1 | 0.6789 | 1 | |
| 16 | 82971665 | rs7350833 | ATP2C2 | 0.03534 | 1 | 1 | 1 | 1 | 1 | 0.5818 | 1 |
| 16 | 82974143 | rs10459853 | ATP2C2 | 0.00582 | 1 | 1 | 0.9994 | 0.4499 | 1 | ||
| 16 | 83002938 | rs247820 | ATP2C2 | 0.01756 | 1 | 1 | 1 | 1 | 1 | 0.5331 | 1 |
| 16 | 83040732 | rs247889 | ATP2C2 | 0.00858 | 1 | 0.7355 | 0.9927 | 0.4468 | 1 | 0.5936 | 1 |
| 16 | 83055346 | rs429313 | ATP2C2 | 0.02536 | 1 | 1 | 0.4746 | 0.1494 | 0.9999 | 0.323 | 1 |
| 16 | 83062088 | rs394533 | ATP2C2 | 0.03043 | 1 | 1 | 0.2917 | 0.1177 | 0.859 | 0.54 | 1 |
| 16 | 83063868 | rs400922 | ATP2C2 | 0.01439 | 1 | 1 | 0.9851 | 0.8241 | 0.06808 | 0.9575 | |
| 16 | 83065318 | rs390208 | ATP2C2 | 0.01406 | 1 | 1 | 0.9896 | 0.7866 | 1 | 0.4388 | 1 |
| 21 | 34612656 | rs2834442 | KCNE2 | 0.0039 | 1 | 1 | 0.2738 | 0.4455 | 1 | 0.3635 | 1 |
| 21 | 34620260 | rs1013063 | KCNE2 | 0.0205 | 1 | 1 | 0.0193 | 0.1565 | 1 | 0.1872 | 1 |
| 21 | 34636414 | rs1467847 | KCNE2 | 0.05496 | 1 | 1 | 0.0139 | 0.146 | 1 | 0.1605 | 0.9995 |
| 21 | 34639061 | rs2834462 | KCNE2 | 0.00862 | 1 | 1 | 0.708 | 0.9251 | 1 | 0.2167 | 1 |
| 21 | 34643430 | rs8128316 | KCNE2 | 0.01872 | 1 | 1 | 0.7774 | 1 | 0.7774 | 1 |
Interestingly, a SNP × SNP epistasis analysis of the 66 replication SNPs in the Finnish case–control sample detected an interaction between KCNB2 SNP rs1431656 on chromosome 8q13.3 and CACNB2 SNP rs7076100 on chromosome 10p12.33 (point-wise P = 0.0000199), which remained significant after conservatively correcting for all 2145 possible interaction tests [i.e. treating the 66 replication SNPs as independent] (global P = 0.042). Accounting for non-independence between SNPs due to intermarker linkage disequilibrium (LD) (22), the global P-value for the rs1431656 × rs7076100 interaction is P = 0.022. This interaction was driven by the rs1431656–rs7076100 G-A haplotype providing increased protection against migraine occurrence (Table 5). An interaction between these two SNPs was not observed in any of the replication samples (Table 5 for comparison of population sample allele frequencies). However, other SNPs within these same genes, KCNB2 and CACNB2, provided suggestive evidence for interaction in the Brisbane cohort and marginally significant evidence for interaction in the combined Cologne and Munich cohort (i.e. between CACNB2 SNP rs8181477 and KCNB2 SNPs rs7006287 and rs11782118, point-wise P = 0.004017 and P = 0.004454, respectively). A more detailed description of the interaction analysis is provided in the Supplementary Material, Appendix.
Table 5.
Allele frequency data for rs1431656 and rs7076100 interaction
| Single nucleotide polymorphism | Allele | Finland |
Leiden |
Brisbane |
|||
|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | ||
| rs1431656 | G | 0.25 | 0.31 | 0.27 | 0.26 | 0.28 | 0.27 |
| A | 0.75 | 0.69 | 0.73 | 0.74 | 0.72 | 0.73 | |
| rs7076100 | A | 0.42 | 0.46 | 0.40 | 0.38 | 0.41 | 0.44 |
| T | 0.58 | 0.54 | 0.60 | 0.62 | 0.59 | 0.56 | |
| Haplotype | G–A | 0.08 | 0.17 | 0.10 | 0.10 | 0.12 | 0.12 |
| G–T | 0.17 | 0.14 | 0.17 | 0.17 | 0.16 | 0.15 | |
| A–A | 0.34 | 0.29 | 0.30 | 0.28 | 0.29 | 0.32 | |
| A–T | 0.41 | 0.40 | 0.43 | 0.45 | 0.43 | 0.41 | |
DISCUSSION
Triggered by the observation that all three genes for FHM, a monogenic subtype of MA, encode ion transporters, we investigated the role of ion transport genes in the common forms of migraine. To this end, we performed the first comprehensive screen of common genetic variants in highly plausible candidate ion transport genes. We tested a total of 5257 SNPs covering 155 of such genes initially in a large well-characterized Finnish case–control sample. The most significantly associated 66 SNPs in 12 genes were subsequently tested in four additional independent cohorts, but no consistent replication was detected across all cohorts.
Supplementary Material, Tables S2a and S2b contain estimates of the power to replicate association for the four most significantly associated SNPs [rs13276133, rs1431656 (KCNB2); rs13267466 (KCNQ3); rs2095771 (CLIC5)] in the Finnish cohort, plus the single CLIC5 SNP (rs3729618) and two KCNE2 SNPs (rs1013063, rs1467847) producing significant evidence for replication (EMP2 P ≤ 0.05) in the individual Brisbane and Cologne cohorts, respectively. The combined replication sample had very high (>99%) power to replicate the four most significant Finnish association signals [rs13276133, rs1431656 (KCNB2); rs13267466 (KCNQ3); rs2095771 (CLIC5)], while the individual Leiden, Cologne, combined Cologne and Munich, and Brisbane cohorts each had 68–71%, 54–57%, 61–65% and 79–82% power to replicate at a nominal point-wise threshold of P ≤ 0.05. At a globally significant replication threshold (adjusted for testing all 66 SNPs selected for replication) of P ≤ 0.001 [estimated using SNPSpD (22)], the individual cohorts' power to replicate dropped considerably to 19–22%, 11–12%, 14–17% and 30–34%, respectively. However, the power to replicate (P ≤ 0.001) remains high (85–89%) in the combined replication sample.
Despite our use of large and well-characterized migraine case–control cohorts (total of 2835 cases and 2740 controls), only the potassium voltage-gated channel, Isk-related, member 2 (KCNE2) gene on chromosome 21q22.11 produced marginally convincing evidence for replication in the Cologne cohort. However, the lack of association evidence in the remaining three replication cohorts, including a similar sample from Munich, cast doubt on the robustness of this finding. Furthermore, analysis of the KCNE2 SNPs (rs1013063, rs1467847) in the combined replication sample produced non-significant CMH point-wise P-values of 0.187 and 0.161, and global EMP2 P-values of 1 and 0.9995, respectively (see Supplementary Material, Table S3).
While no single SNP produced significant evidence for association in the Finnish case–control sample, SNP × SNP epistasis analysis of the 66 SNPs selected for replication detected a significant interaction between KCNB2 SNP rs1431656 on chromosome 8q13.3 and CACNB2 SNP rs7076100 on chromosome 10p12.33 (point-wise P = 0.0000199; global P = 0.022). Although this specific interaction was not observed in our replication cohorts, the significance of the rs1542709 × rs7076100 interaction in the Finnish sample and supporting evidence for inter-genic SNP × SNP interaction in the combined Cologne and Munich replication cohort, indicate further study of KCNB2 and CACNB2 variants and their combined contribution to migraine susceptibility may be warranted.
To investigate how the reported results would compare to those potentially obtained in a high-density genome-wide association (GWA) study, we examined the SNP coverage across the gene footprints provided by the Illumina HumanCNV370-Duo and HumanHap550-Duo BeadChips (array marker lists were downloaded from http://illumina.com.au on 19 December 2007). These two arrays were selected owing to both their popularity and design. More specifically, in contrast to the Affymetrix arrays 5.0 and 6.0 (containing 440 794 and 906 600 SNPs, respectively), which are not designed with reference to LD structure, the Illumina arrays contain SNPs selected to tag LD bins—analogous to the approach employed in the design of our ion transport Perlegen array. However, although the Affymetrix 6.0 array was not designed with reference to LD structure, it is worth noting that it has been shown to provide similar LD coverage to the Illumina HumanHap550 [http://www.ashg.org/genetics/ashg07s/index.shtml; (23)].
Of the 370 405 SNPs present on the HumanCNV370 array, 3272 are located within the 155 gene ‘footprints’ (includes extension of 20 kb upstream of transcription start and 10 kb downstream of transcription end). Of the 561 467 HumanHap550 SNPs, 5270 are located within the 155 gene ‘footprints’. Given the usable number of Illumina array SNPs would likely be reduced after QC, these counts indicate that our study provides at least as much coverage of common variation as that provided by the high resolution HumanHap550 array [which captures 90% of all HapMap Phase I + II loci (MAF > 0.05) with r2 > 0.8] and far greater coverage than the HumanCNV370 array. Hence, the results of our detailed association screen of 155 ion transport genes are highly relevant to the burgeoning field of whole GWA studies. Indeed, the results of our study may be readily compared with GWA studies as they come to pass.
Furthermore, although our study was well-powered and had excellent SNP coverage, the absence of significant association within these 155 highly plausible migraine candidate genes at a moderate significance threshold (approximately P < 10−5) compared to that required for GWA studies (recent guidelines recommend P < 10−7) (24,25), indicates that common variation of moderate effect in these candidate genes does not play a major role in common migraine susceptibility within the samples examined. These results therefore suggest that either common variation of smaller effect in these candidate genes, or other ‘less obvious’ genes and pathways likely underlie the genetic susceptibility of the common forms of migraine. Alternatively, the investigated ion transport genes contribute to migraine through multiple, relatively rare and/or possibly interacting variants, or variation in distant cis- or trans-regulatory elements not tested in this study design. Although sample sizes in excess of 10 000 cases and 10 000 controls will be required to detect rare variants (MAF < 0.1) (26), a GWA study in large, well-characterized samples would be an unbiased way to test the hypothesis whether common variants in any part of the genome are associated with common forms of migraine. Indeed, efforts are now underway within the MGC to perform a GWA study utilizing at least 2000 unrelated MA cases. The results of the MGC GWA study should ultimately determine whether common genetic variants underlie susceptibility to common migraine and this approach would not restrict our thinking to previously known or predicted pathways. It enables testing of less predictable regions of the genome and allows examination of SNP × SNP interaction on a true genome-wide scale.
In addition, pathway-based approaches can be readily applied to GWA studies to yield biological insights that are otherwise undetectable by focusing only on individual genes and/or regions that have the strongest evidence for association (27). For example, a typical pathway-based approach might rank all genes by their significance of association and then investigate whether a particular group of genes is enriched at the significant end of the ranked list more than that expected by chance. Application of such pathway-based approaches, where multiple genes in the same pathway contribute to disease aetiology, but common variations in each of the causal genes make modest contributions to disease risk, has enormous potential to both detect novel and confirm hypothesized causal pathways and disease mechanisms other than ion transportation (e.g. dopaminergic and serotonergic pathways).
MATERIALS AND METHODS
Finnish discovery cohort
For this cohort, patients with a diagnosis of MA were drawn from the ongoing Finnish Migraine Study. MA was chosen as the principal diagnosis, as the Finnish patient pool has been primarily ascertained for this disorder and because the inclusion of aura as a diagnostic criterion reduces the possibility that other headache disorders would be included in the case sample (28). Patients were recruited from four headache clinics across Finland. Index cases were selected based on clinical examination by a neurologist from patients with at least three affected first-degree relatives; all available family members were also recruited into the study. Index cases and family members were diagnosed based on the IHS criteria (29,30) using the validated Finnish Migraine-Specific Questionnaire for Family studies (31). For the Finnish study, all participants gave informed consent and approval to conduct the research was obtained from the Helsinki University Central Hospital Ethics Committee. Over 6000 individuals in more than 800 families have taken part in this study and recruitment is under progress. From the total cohort of migraineurs in the Finnish family study, 898 unrelated individuals with MA were selected for this case–control study. Control subjects for the study were 900 unrelated individuals drawn from the Finnish Twin Cohort (32). One twin was selected from each pair, and these subjects were matched with respect to gender and age to cases. Control subjects have all responded negatively to the question of lifetime occurrence of migraine and also report no family history of migraine. Table 2 details key characteristics of patient findings for the Finnish study (33).
Replication cohorts
Leiden cohort
The Leiden sample is a well-phenotyped population-based migraine cohort from the Genetic Epidemiology of Migraine (GEM) study from the Netherlands. DNA was available from 823 unrelated migraine cases (555 MO; 268 MA) and 946 unrelated non-migraine controls. The GEM cohort is embedded within the ‘MORGEN’ project—a population-based study designed to monitor risk factors for and the prevalence of chronic diseases of public health importance in Dutch adults 20–65 years of age. The sample for the MORGEN project was selected randomly within equal-size strata of a five-year age group and gender from two Dutch county population registries. During 1995–1996, a total of 6491 individuals (52.7%) participated in the study. Respondents signed a general informed consent for the MORGEN project, and a specific informed consent for the GEM Study. For case-finding, MORGEN participants were mailed an extensive self-administered questionnaire that included questions about sociodemographic characteristics, medical history, psychosocial functioning and five migraine screening questions [adapted from Stewart et al. (34)]. Screen-positive participants completed a more detailed questionnaire that focused on signs and symptoms of migraine headache and aura as outlined in the IHS criteria. Very special care was given to diagnose aura and those reporting visual aura symptoms were also asked to draw what they saw. A semi-structured telephone interview was obtained in a random sample of (83%) screen-positive and (5%) screen-negative participants to clarify the signs and symptoms of migraine headache and aura reported in the stage 2 questionnaire, and to validate the earlier questionnaire. After a screening procedure, final (lifetime) diagnosis of migraine was made in 863 participants, 620 of whom had active migraine (10). The lifetime prevalence of migraine in women was 33% and the 1-year prevalence was 25%. In men, the lifetime prevalence was 13.3% and the 1-year prevalence was 7.5%. Among patients with migraine in the past year, 63.9% had MO, 17.9% had MA and 13.1% had both MA and MO. The prevalence of migraine was significantly higher in women and not associated with socioeconomic status. Migraine patients suffered a median of 12 migraine attacks per year; 25% had at least two attacks per month. The Leiden control sample was drawn from participants who were screened negative on the five migraine screening questions and matched the cases with respect to age and gender. The GEM sample has been used for genetic and clinical research, including assessing the cardiovascular risk profile and impact of migraine on quality of life (35–37).
Cologne/Kiel cohort
All participants were diagnosed by the revised criteria of the IHS (30) (601 MA patients and 651 matched, healthy control individuals) and have provided their written informed consent; the study was approved by the Local University Ethics Committees. All patients were interviewed personally or by telephone by an experienced neurologist and a detailed questionnaire was completed for each of them. The complete patient ascertainment was performed by a single highly specialized headache centre. The questionnaire included a comprehensive assessment of (i) the type, frequency, localization and duration of aura symptoms, (ii) the possible existence of motor symptoms such as weakness or hemiplegia, (iii) the properties, frequency, localization and duration of the headache attack, (iv) a history of medication, (v) the existence of other possible diseases and (vi) the family history. All control individuals were unrelated, of German origin and were also gender-matched. Control individuals were interviewed to exclude those who suffer from frequent headaches or migraine. From each family, the index case, provided that he/she had a positive family history, has been included in the case–control sample [for more detailed cohort information see, for example, Netzer et al. (38)].
Munich cohort
All subjects were unrelated and were diagnosed with MA based on the revised criteria of the IHS and have given their written informed consent. All patients were personally seen and examined by a headache specialist. The majority of subjects were sampled through a headache clinic (approximately 60%) or a headache specialist in private practice (30%). The remaining patients were recruited at the Department of Neurology (Klinikum Großhadern, Munich). All participants received an extensive, validated questionnaire (31) used in studies on the genetics of migraine. The questionnaire includes a comprehensive assessment of (i) premonitory symptoms, (ii) the type, frequency, localization and duration of aura symptoms, (iii) the presence/absence of motor symptoms, (iv) the properties, frequency, localization and duration of the headache attack, (v) a history of medication, (vi) comorbidities, (vii) family history and (viii) ethnic origin. The study was approved by the Local Ethics Committee in Munich.
Brisbane cohort
The Australia-wide sample is a large population-based migraine cohort which has been used in a number of studies addressing various aspects of migraine genetics (39). The Brisbane sample is made up of two cohorts drawn from the Australian Twin Registry. In 1993–1995, a telephone interview comprising a diagnostic assessment of psychiatric disorders, including alcohol use and abuse, anxiety, depression and phobias, was conducted to an older cohort of twins, born 1902–1964. A total of 5996 individual twins completed the 1993 interview (40). Between 1996 and 2000, a younger cohort of twins (born 1964–1975) undertook a similar extensive semi-structured telephone interview, designed to assess physical, psychological and social manifestations of alcoholism and related disorders (41); 6265 individual twins completed the interview. For the Australian study, all participants gave informed consent and approval to conduct the research was obtained from the Queensland Institute of Medical Research (QIMR) Human Research Ethics Committee. Both cohort samples were not selected with regard to personal or family history of alcoholism or other psychiatric or medical disorders including migraine. Migraine symptom data were obtained during the course of the telephone interviews. The wording of questions was identical between cohorts. However, the younger cohort were asked questions relating to all IHS symptoms, whereas the older cohort had a somewhat more restricted number of questions (no questions regarding attack frequency, duration and pain intensity). The two cohorts were combined, thus allowing LCA to impute class membership in the older cohort, based on the pattern of all 10 symptoms observed in the younger cohort. To examine the accuracy of imputed class memberships, we compared the classification results for the younger dataset utilizing all the 10 available IHS symptoms, to the classification results for the younger dataset utilizing only the seven IHS symptoms that were available for the older cohort. Compared to using all 10 symptoms, when only the seven symptoms were used, these analyses found 98.5% and 96.0% of individuals were correctly classified as unaffected and affected, respectively; thus indicating that the three missing symptoms in the older cohort have negligible effect on the accuracy of individual LCA classifications (42). Given that we have previously shown LCA classified migraine to be a valid and accurate measure for genetic research (39), we combined Brisbane cases satisfying IHS criteria (64%) with cases classified as affected via our LCA approach (36%).
The Brisbane case–control replication sample consists of 1152 unrelated migrainous headache (39,42) cases of which 971 report visual aura and 1152 unrelated Australian twin controls screened negative for migraine. All controls have participated in the same survey and indicated that neither the study subject nor any first-degree relative had migraine.
Candidate gene selection
We targeted a comprehensive listing of ion channels and ion-channel modulators expressed in human brain for the association screen. Microarrays for genotyping were designed with the goal that they will have utility in a number of disorders beyond migraine in which ion-transporters are also predicted to play an important role, including epilepsy and cardiac arrhythmias. Therefore, we targeted all the relevant ion channels with sufficient SNP marker density to have high power to detect association. During the month of October 2005, we selected an inclusive list of ion-transporters from publicly available databases, including the ion-channel categorization on the Kyoto Encyclopedia of Genes and Genomes (KEGG) website http://www.genome.jp/kegg/ a text-based search of ion channels, ATPases and related proteins at the University of California Santa Cruz (‘Golden Path’) database http://genome.ucsc.edu/ and a screen of the Ensembl database http://www.ensembl.org/index.html for other ion-transport genes (Biological Process Gene Ontology term ID: 0030001). These genes were then further screened for expression in brain or heart using the eGenetics/SANBI databases via the Ensembl website. Additional screening for molecular function was performed using the data available in the National Center for Biotechnology Information (NCBI) Online Mendelian Inheritance in Man (OMIM) database http://www.ncbi.nlm.nih.gov/omim. This iterative process yielded a comprehensive listing of 155 ion channel and ion-channel modulators (see Supplementary Material, Table S1). Table 6 shows a breakdown of the major categories.
Table 6.
Breakdown of candidate gene categories
| Genes (n) | Gene category |
|---|---|
| 26 | Voltage-gated calcium channels |
| 74 | Voltage-gated potassium channels |
| 14 | Voltage-gated sodium channels |
| 21 | Chloride channels |
| 20 | ATPase ion transporters |
Polymorphism selection
Polymorphism selection for the ion-transport screen was primarily based on Perlegen's ‘LD bin-map’ of SNPs that are validated in their assay, have a minor allele frequency (MAF) >0.1, and have been tested in their screen of almost 1.6 million SNPs in 71 Americans of European, African and Asian ancestry to define whole genome patterns of variability (43). Perlegen has identified tag SNPs within each bin that are in LD with every other SNP in the bin with r2 > 0.8 using the algorithm of Carlson et al. (44). Such representative tag SNPs were selected from LD bins spanning each of the 155 gene ‘footprints’ (transcriptional footprint extended 20 kb upstream of transcription start and 10 kb downstream of transcription end). The mean and median gene footprint was 136 280 bp and 67 100 bp, respectively.
In designing the microarrays, one-tag SNP was selected from bins containing one to two SNPs. For bins containing three or more SNPs, two-tag SNPs were selected to guard against possible assay failure of a single-tag SNP, thus minimizing the potential for loss of the substantial allelic variation contained within larger LD bins. A total of 5975 SNPs were selected for genotyping. These 5975 tag SNPs provide an overall mean and median per-gene resolution of one SNP every 5034 bp and 3552 bp, respectively; and effectively test a total of 11 148 SNPs with r2 > 0.8, thus providing a very high resolution to test all common alleles within these genes.
Genotyping
Genotyping of the ion-transport SNPs utilized microarrays designed by Perlegen Sciences, Inc. (Mountain View, CA, USA) and manufactured by Affymetrix, Inc. (Santa Clara, CA, USA). These assays have been documented to have a >99% concordance with other genotyping methods (43,45). Genotyping assays and QC was performed by Perlegen. A total of 59 individuals were excluded from further analysis due to an overall call rate of <80% and the data quality remaining poor after repeat genotyping of the sample. An additional 14 individuals were excluded from further analysis due to evidence of familial relatedness after examination of allele sharing among pairs of individuals using the genotypic relative risk program (46).
A total of 706 SNPs were excluded from further analysis according to at least one of the following QC criteria: (i) call rate of <95%, (ii) P < 10−4 for deviation from Hardy–Weinberg equilibrium and (iii) X and Y chromosome SNP performance (i.e. for non-pseudoautosomal X chromosome SNPs, females can have up to two alleles, whereas males can have only one allele; for Y chromosome SNPs, females should have zero alleles while males should have one allele). The resulting 5269 tag-SNPs still provide a very high resolution to test all common alleles within the 155 ion-transport genes with an overall mean and median per-gene resolution of one SNP every 5968 bp and 4323 bp, respectively.
For the replication study, SNP genotypes were determined in a 384-well format using the Sequenom iPLEX system (Sequenom, San Diego, CA, USA) in which the distinction between genotypes is based on the mass differences between the primer extension products of the two alleles. Assays were designed using the AssayDesign software (Sequenom); primer information is available from the corresponding author upon request. The multiplex polymerase chain reaction and primer extension reactions were performed under standard conditions according to the manufacturer's instructions using 20 ng of genomic DNA as a template.
Power of Finnish case–control sample to detect association
To account for non-independence between SNPs due to intermarker LD, we determined the approximate effective number of independent SNPs (Meff) following the SNPSpD approach of Nyholt (22) and applying equation (5) of Li and Ji (47). Utilizing observed genotype data for the final 5269 SNPs passing Perlegen's QC protocols, we calculated the intermarker LD (pair-wise r2) for SNPs located on the same chromosome. Examination of the eigenvalues after spectral decomposition of the resulting chromosome-specific correlation matrices (√r2) determined an equivalent of 2677 total independent SNPs to be tested for association. Consequently, to ensure an overall type I error rate of 5%, we require a Bonferroni-adjusted significance level (α) of P = 0.05/2677 = 0.000019 [−log10(α) = 4.7386].
Power calculations were performed for a case–control study assuming a disease prevalence for MA of 5% using the Genetic Power Calculator (48). In short, power calculations for our Finnish discovery cohort assumed loci with dominant and multiplicative modes of inheritance and were based on our post-QC sample of 841 cases and 884 controls. The Finnish cohort has over 80% power to detect dominant disease-predisposing alleles of frequency 0.10, 0.25 and 0.5 contributing a GRR of 1.78, 1.70 and 2.32, respectively. For the general multiplicative model, there is 80% power to detect risk alleles of frequency 0.10, 0.25 and 0.5 contributing a GRR of 1.65, 1.45 and 1.40, respectively. These calculations demonstrate our Finnish discovery cohort to have high power to detect ion-transport gene associations of moderate effect.
Statistical analysis
The following statistical analyses were performed using the PLINK analysis program (version 1.00) (http://pngu.mgh.harvard.edu/purcell/plink/) (49). PLINK QC options were utilized with default thresholds: maximum missing genotypes per person (‐ ‐mind option) ≤0.10, maximum missing genotypes per SNP (‐ ‐geno option) ≤0.10, MAF (‐ ‐maf option) ≥0.01 and Hardy–Weinberg disequilibrium (exact) P-value ≥0.001 in controls (‐ ‐hwe option). Cluster analysis based on pair-wise IBS distance (‐ ‐cluster option) of the SNPs passing both Perlegen and PLINK QC criteria was utilized to identify possible population structure within the Finnish case–control cohort. Single marker basic allelic association (χ12) tests (‐ ‐assoc option) were performed for each of the post-QC SNPs. PLINK's max(T) permutation procedure (‐ ‐mperm option) was used with 10 000 iterations to obtain accurate global empirical P-values (i.e. point-wise P-values are for the single test only, whereas global P-values are adjusted for testing all analyzed SNPs) (‘EMP2’ column of ‐ ‐mperm output). Genome-wide significant association is therefore obtained at a global EMP2 P-value of 0.05 or smaller. Although the primary analysis in the Finnish cohort tested for association to MA, we also performed exploratory association analysis to strict IHS criteria, and the individual migraine trait components of ‘pulsation’, ‘photophobia’ and ‘phonophobia’ to help select SNPs for replication.
The FDR approach (50–52), which allows a controlled proportion of positive results to be false, while detecting more true-positives was also used to help select and interpret the most promising SNP associations for replication. This was done using the default settings of the QVALUE computer program (version 1.1) (53). A preponderance of χ12 values suggestive of association was also examined via a QQ-plot, by plotting the distribution of observed chi-square (χ12) values obtained from the single SNP allelic association analysis of MA in Finns against the distribution of χ12 values expected under the null hypothesis of no association.
Given that recent results from two-stage association studies indicate the most significant SNP in the total sample is not necessarily the most significant SNP in the first stage, to balance guarding against not following-up a true risk locus with the number of loci to follow-up (i.e. minimizing the multiple-test burden to maintain high replication power), SNPs producing nominally significant (P < 0.005) allelic association with MA in the Finnish cohort, together with supporting evidence for association to strict IHS migraine and individual trait components were selected for genotyping in the replication cohorts. The nominal significance threshold of P < 0.005 was selected after examination of the QQ-plot and FDR results.
Given PLINK's clustering procedure (‐ ‐cluster option) requires whole-genome level data to give accurate results (probably >10 000 SNPs), to allow for potential population differences between the replication cohorts, a CMH analysis (‐ ‐mh option) was used to test for overall association in the replication samples, controlling for cohort. The CMH test for 2 × 2 × K-stratified tables is based on an ‘average’ odds ratio (OR) that controls for the potential confounding due to the cluster (cohort) variable. To account for differences in ascertainment and diagnoses in addition to potential genetic differences at the population level, the Leiden (Dutch), Cologne and Munich (German), and Brisbane (Australian) cohorts were each assigned to the separate clusters, resulting in a CMH test that controls for four separate clusters. A schematic overview of our study design is provided in Figure 3.
Figure 3.
Overview of study design.
To further examine peak association results and guard against missing a strong association signal by only analyzing observed/single SNP data, exploratory association analysis of haplotypes spanning two to six contiguous SNPs were also performed using a sliding window approach via PLINK's ‐ ‐hap-window and ‐ ‐hap-assoc options.
Given the strong prior hypothesis for involvement of these genes in migraine, we also tested for SNP × SNP epistasis using PLINK's ‐ ‐epistasis option. For our total 5257 SNPs there are 13 815 396 possible SNP × SNP interactions. Therefore, although it is possible for loci with no individual effects to still have a component of genetic variance attributable to epistatic effects, following the recommendation of Blangero et al. (54), to minimize epistasis multiple-testing burden, analyses were restricted to SNPs producing nominally significant (P < 0.005) allelic association with MA in the Finnish cohort.
Finally, the PS program (55) was used to perform power calculations in the replication cohorts. Briefly, the power to replicate promising association results was estimated in the individual and combined replication cohorts utilizing the control allele frequency and OR observed in the Finnish case–control cohort.
SUPPLEMENTARY MATERIAL
FUNDING
The Helsinki component of this study was supported by the Sigrid Juselius Foundation, the Academy of Finland (200923 to A.P.; 00213 to M.W.), the Helsinki University Central Hospital, the EuroHead project (LSHM-CT-2004-504837), the GenomEUtwin project (QLG2-CT-2002-01254 and to V.A.), the Oxnard Foundation, the Helsinki Biomedical Graduate School (to V.A.), the Biomedicum Helsinki Foundation, the Finnish Cultural Foundation (to V.A.), the Finnish Neurology Foundation and National Institutes of Health (RO1 NS37675 to A.P.), National Institute on Alcohol Abuse and Alcoholism (United States) grants AA007535, AA013320, AA013326, AA014041, AA07728, AA10249 and AA11998. Academy of Finland Center of Excellence in Complex Disease Genetics (to L.P., A.P. and J.K.). The Brisbane component was supported by National Health and Medical Research Council (NHMRC) (Australia) grants 941177, 951023, 241916 and 442981. D.R.N. and G.W.M were supported by NHMRC R.D. Wright and Senior Research Fellowships. The Cologne component was supported by Deutsche Forschungsgemeinschaft (DFG, FOR423), National Genome Network (NGFN) by the Bundesministerium für Bildung und Forschung (BMBF), Koeln Fortune Program/Faculty of Medicine, University of Cologne. The Leiden component was supported by the Netherlands Organization for Scientific Research (NWO) (903-52-291 to M.D.F, R.R.F. and Vici 918.56.602 to M.D.F), the Migraine Trust (R.R.F., M.D.F.), the EuroHead project (LSHM-CT-2004-504837) and the Centre for Medical Systems Biology (CMSB) in the framework of the Netherlands Genomics Initiative (NGI). The Munich component was supported by the Deutsche Forschungsgemeinschaft (DFG D1 722/8-1 and DI 722/8-2).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kauko Heikkilä for database management of the Finnish Twin Cohort and the participating twins. We thank Dixie Statham and Clare Redfern of QIMR, for project coordination; Anjali Henders and Megan Campbell, for managing sample processing; David Smyth and Scott Gordon for data management; and the twins for their generous participation. The Cologne component thanks Mrs. I. Goebel for technical assistance. The Leiden component: IRP, NIA (LJL) thank Dr Kaate Vanmolkot and Judith van Vark for technical assistance.
Conflict of Interest statement. None of the authors have any conflicts of interest to declare.
REFERENCES
- 1.Ophoff R.A., Terwindt G.M., Vergouve M.N., van Eijk R., Oefner P.J., Hoffman S.M., Lamerdin J.E., Mohrenweiser H.W., Bulman D.E., Ferrari M., et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutation in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/s0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- 2.De Fusco M., Marconi R., Silvestri L., Atorino L., Rampoldi L., Morgante L., Ballabio A., Aridon P., Casari G. Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat. Genet. 2003;33:192–196. doi: 10.1038/ng1081. [DOI] [PubMed] [Google Scholar]
- 3.Dichgans M., Freilinger T., Eckstein G., Babini E., Lorenz-Depiereux B., Biskup S., Ferrari M.D., Herzog J., van den Maagdenberg A.M., Pusch M., et al. Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet. 2005;366:371–377. doi: 10.1016/S0140-6736(05)66786-4. [DOI] [PubMed] [Google Scholar]
- 4.Moskowitz M.A., Bolay H., Dalkara T. Deciphering migraine mechanisms: clues from familial hemiplegic migraine genotypes. Ann. Neurol. 2004;55:276–280. doi: 10.1002/ana.20035. [DOI] [PubMed] [Google Scholar]
- 5.Pietrobon D., Striessnig J. Neurobiology of migraine. Nat. Rev. Neurosci. 2005;4:386–398. doi: 10.1038/nrn1102. [DOI] [PubMed] [Google Scholar]
- 6.Bolay H., Reuter U., Dunn A.K., Huang Z.H., Boas D.A., Moskowitz M.A. Intrinsic brain activity triggers trigeminal meningeal afferents in a migraine model. Nat. Med. 2002;8:136–142. doi: 10.1038/nm0202-136. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari M.D., Goadsby P.J. Migraine as a cerebral ionopathy with abnormal central sensory processing. In: Gilman S. (ed), editor. Neurobiology of Disease. New York: Elsevier; 2006. pp. 333–348. [Google Scholar]
- 8.van den Maagdenberg A.M., Haan J., Terwindt G.M., Ferrari M.D. Migraine: gene mutations and functional consequences. Curr. Opin. Neurol. 2007;20:299–305. doi: 10.1097/WCO.0b013e3281338d1f. [DOI] [PubMed] [Google Scholar]
- 9.Thomsen L.L., Eriksen M.K., Roemer S.F., Andersen I., Olesen J., Russell M.B. A population-based study of familial hemiplegic migraine suggests revised diagnostic criteria. Brain. 2002;125:1379–1391. doi: 10.1093/brain/awf132. [DOI] [PubMed] [Google Scholar]
- 10.Launer L.J., Terwindt G.M., Ferrari M.D. The prevalence and characteristics of migraine in a population-based cohort: the GEM study. Neurology. 1999;53:537–542. doi: 10.1212/wnl.53.3.537. [DOI] [PubMed] [Google Scholar]
- 11.Mochi M., Sangiorgi S., Cortelli P., Carelli V., Scapoli C., Crisci M., Monari L., Pierangeli G., Montagna P. Testing models for genetic determination in migraine. Cephalalgia. 1993;13:389–394. doi: 10.1046/j.1468-2982.1993.1306389.x. [DOI] [PubMed] [Google Scholar]
- 12.Ophoff R., van Eijk R., Sandkuijl L., Terwindt G., Grubben C., Haan J., Lindhout D., Ferrari M., Frants R. Genetic heterogeneity of familial hemiplegic migraine. Genomics. 1994;22:21–26. doi: 10.1006/geno.1994.1340. [DOI] [PubMed] [Google Scholar]
- 13.Joutel A., Ducros A., Vahedi K., Labauge P., Delrieu O., Pinsard N., Mancini J., Ponsot G., Gouttiere F., Gastaut J.L., et al. Genetic heterogeneity of familial hemiplegic migraine. Am. J. Hum. Genet. 1994;55:1166–1172. [PMC free article] [PubMed] [Google Scholar]
- 14.Kallela M., Wessman M., Havanka H., Palotie A., Färkkilä M. Familial migraine with and without aura: clinical characteristics and co-occurrence. Eur. J. Neurol. 2001;8:441–449. doi: 10.1046/j.1468-1331.2001.00260.x. [DOI] [PubMed] [Google Scholar]
- 15.Goadsby P.J., Lipton R.B., Ferrari M.D. Migraine–current understanding and treatment. N. Engl. J. Med. 2002;346:257–270. doi: 10.1056/NEJMra010917. [DOI] [PubMed] [Google Scholar]
- 16.de Vries B., Freilinger T., Vanmolkot K.R., Koenderink J.B., Stam A.H., Terwindt G.M., Babini E., van den Boogerd E.H., van den Heuvel J.J., Frants R.R., et al. Systematic analysis of three FHM genes in 39 sporadic patients with hemiplegic migraine. Neurology. 2007;69:2170–2176. doi: 10.1212/01.wnl.0000295670.01629.5a. [DOI] [PubMed] [Google Scholar]
- 17.Todt U., Dichgans M., Jurkat-Rott K., Heinze A., Zifarelli G., Koenderink J.B., Goebel I., Zumbroich V., Stiller A., Ramirez A., et al. Rare missense variants in ATP1A2 in families with clustering of common forms of migraine. Hum. Mutat. 2005;26:315–321. doi: 10.1002/humu.20229. [DOI] [PubMed] [Google Scholar]
- 18.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W.P., Levesque P.C., Little W.A., Conder M.L., Ramakrishnan P., Neubauer M.G., Blanar M.A. Functional expression of two KvLQT1-related potassium channels responsible for an inherited idiopathic epilepsy. J. Biol. Chem. 1998;273:19419–19423. doi: 10.1074/jbc.273.31.19419. [DOI] [PubMed] [Google Scholar]
- 20.Tinel N., Diochot S., Lauritzen I., Barhanin J., Lazdunski M., Borsotto M. M-type KCNQ2-KCNQ3 potassium channels are modulated by the KCNE2 subunit. FEBS Lett. 2000;480:137–141. doi: 10.1016/s0014-5793(00)01918-9. [DOI] [PubMed] [Google Scholar]
- 21.Vanmolkot K.R., Stam A.H., Raman A., Koenderink J.B., de Vries B., van den Boogerd E.H., van Vark J., van den Heuvel J.J., Bajaj N., Terwindt G.M., et al. First case of compound heterozygosity in Na,K-ATPase gene ATP1A2 in familial hemiplegic migraine. Eur. J. Hum. Genet. 2007;15:884–888. doi: 10.1038/sj.ejhg.5201841. [DOI] [PubMed] [Google Scholar]
- 22.Nyholt D.R. A simple correction for multiple testing for SNPs in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe C.E., Cooper J.D., Todd J.A. Post genome-wide association challenges at the complex-disease associated locus CD25 on chromosome 10p15 [abstract 216]. Presented at the Annual Meeting of The American Society of Human Genetics; 26 October 2007; San Diego, CA, USA. [Google Scholar]
- 24.Balding D.J. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y.S.W., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 26.Nannya Y., Taura K., Kurokawa M., Chiba S., Ogawa S. Evaluation of genome-wide power of genetic association studies based on empirical data from the HapMap project. Hum. Mol. Genet. 2007;16:2494–2505. doi: 10.1093/hmg/ddm205. [DOI] [PubMed] [Google Scholar]
- 27.Wang K., Li M., Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wessman M., Kallela M., Kaunisto M.A., Marttila P., Sobel E., Hartiala J., Oswell G., Leal S.M., Papp J.C., Hamalainen E., et al. A susceptibility locus for migraine with aura, on chromosome 4q24. Am. J. Hum. Genet. 2002;70:652–662. doi: 10.1086/339078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Headache Classification Committee of the International Headache Society. Classification and diagnostic criteria for headache disorders, cranial neuralgias and facial pain. Cephalalgia. 1988;8(Suppl. 7):1–96. [PubMed] [Google Scholar]
- 30.Headache Classification Subcommittee of the International Headache Society. The international classification of headache disorders: 2nd edition. Cephalalgia. 2004;24(Suppl. 1):9–160. doi: 10.1111/j.1468-2982.2003.00824.x. [DOI] [PubMed] [Google Scholar]
- 31.Kallela M., Wessman M., Färkkilä M. Validation of a migraine-specific questionnaire for use in family studies. Eur. J. Neurol. 2001;8:61–66. doi: 10.1046/j.1468-1331.2001.00165.x. [DOI] [PubMed] [Google Scholar]
- 32.Kaprio J., Koskenvuo M. Genetic and environmental factors in complex diseases: the older Finnish Twin Cohort. Twin Res. 2002;5:358–365. doi: 10.1375/136905202320906093. [DOI] [PubMed] [Google Scholar]
- 33.Kaunisto M.A., Kallela M., Hamalainen E., Kilpikari R., Havanka H., Harno H., Nissila M., Sako E., Ilmavirta M., Liukkonen J., et al. Testing of variants of the MTHFR and ESR1 genes in 1798 Finnish individuals fails to confirm the association with migraine with aura. Cephalalgia. 2006;26:1462–1472. doi: 10.1111/j.1468-2982.2006.01228.x. [DOI] [PubMed] [Google Scholar]
- 34.Stewart W.F., Lipton R.B., Celentano D.D., Reed M.L. Prevalence of migraine headache in the United States. Relation to age, income, race, and other sociodemographic factors. JAMA. 1992;267:64–69. [PubMed] [Google Scholar]
- 35.Scher A.I., Terwindt G.M., Picavet H.S., Verschuren W.M., Ferrari M.D., Launer L.J. Cardiovascular risk factors and migraine: the GEM population-based study. Neurology. 2005;64:614–620. doi: 10.1212/01.WNL.0000151857.43225.49. [DOI] [PubMed] [Google Scholar]
- 36.Scher A.I., Terwindt G.M., Verschuren W.M., Kruit M.C., Blom H.J., Kowa H., Frants R.R., van den Maagdenberg A.M., van Buchem M., Ferrari M.D., et al. Migraine and MTHFR C677T genotype in a population-based sample. Ann. Neurol. 2006;59:372–375. doi: 10.1002/ana.20755. [DOI] [PubMed] [Google Scholar]
- 37.Terwindt G.M., Ferrari M.D., Tijhuis M., Groenen S.M., Picavet H.S., Launer L.J. The impact of migraine on quality of life in the general population: the GEM study. Neurology. 2000;55:624–629. doi: 10.1212/wnl.55.5.624. [DOI] [PubMed] [Google Scholar]
- 38.Netzer C., Freudenberg J., Toliat M.R., Heinze A., Heinze-Kuhn K., Thiele H., Goebel I., Nürnberg P., Ptácek L.J., Göbel H., et al. Genetic association studies of the chromosome 15 GABA-A receptor cluster in migraine with aura. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147:37–41. doi: 10.1002/ajmg.b.30560. [DOI] [PubMed] [Google Scholar]
- 39.Nyholt D.R., Gillespie N.A., Heath A.C., Merikangas K.R., Duffy D.L., Martin N.G. Latent class analysis does not support migraine with aura and migraine without aura as separate entities. Genet. Epidemiol. 2004;26:231–244. doi: 10.1002/gepi.10311. [DOI] [PubMed] [Google Scholar]
- 40.Heath A.C., Bucholz K.K., Madden P.A., Dinwiddie S.H., Slutske W.S., Bierut L.J., Statham D.J., Dunne M.P., Whitfield J.B., Martin N.G. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol. Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 41.Heath A.C., Howells W., Kirk K.M., Madden P.A., Bucholz K.K., Nelson E.C., Slutske W.S., Statham D.J., Martin N.G. Predictors of non-response to a questionnaire survey of a volunteer twin panel: findings from the Australian 1989 twin cohort. Twin Res. 2001;4:73–80. doi: 10.1375/1369052012182. [DOI] [PubMed] [Google Scholar]
- 42.Nyholt D.R., Morley K.I., Ferreira M.A.R., Medland S.E., Boomsma D.I., Heath A.C., Merikangas K.R., Montgomery G.W., Martin N.G. Genomewide significant linkage to migrainous headache on chromosome 5q21. Am. J. Hum. Genet. 2005;77:500–512. doi: 10.1086/444510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 44.Carlson C.S., Eberle M.A., Rieder M.J., Yi Q., Kruglyak L., Nickerson D.A. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The GAIN Collaborative Research Group. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat. Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 46.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:741–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 47.Li J., Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 49.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 51.Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 52.Storey J.D. A direct approach to false discovery rates. J. R. Stat. Soc. B Stat. Methodol. 2002;64:479–498. [Google Scholar]
- 53.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blangero J., Williams J.T., Almasy L. Quantitative trait locus mapping using human pedigrees. Hum. Biol. 2000;72:35–62. [PubMed] [Google Scholar]
- 55.Dupont W.D., Plummer W.D.J. Power and sample size calculations. A review and computer program. Control. Clin. Trials. 1990;11:116–128. doi: 10.1016/0197-2456(90)90005-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.