SUMMARY
The nearly 600 proteases in the human genome regulate a diversity of biological processes, including programmed cell death. Comprehensive characterization of protease signaling in complex biological samples is limited by available proteomic methods. We have developed a general approach for global identification of proteolytic cleavage sites based on enzymatic biotinylation of free protein N-termini and positive enrichment of corresponding N-terminal peptides. Using this method to study apoptosis, we have sequenced 333 caspase-like cleavage sites distributed among 292 protein substrates. These sites are generally not predicted by in vitro caspase substrate specificity, but can be used to predict other physiological caspase cleavage sites. Structural bioinformatic studies show that caspase cleavage sites often appear in surface accessible loops and even occasionally in helical regions. Strikingly, we also find that a disproportionate number of caspase substrates physically interact, suggesting that these dimeric proteases target protein complexes and networks to elicit apoptosis.
INTRODUCTION
Apoptosis is a non-inflammatory form of cell death that regulates tissue differentiation and homeostasis in higher eukaryotes (for a review, see Taylor et al., 2008). Since apoptotic turnover of cells lies in direct opposition to the uncontrolled growth of tumor cells, a strong link exists between apoptosis and cancer. Indeed, the terminal cellular effect of most chemotherapeutic compounds is induction of apoptosis (Kaufmann and Earnshaw, 2000). The widespread intracellular proteolysis that is a hallmark of apoptosis is predominantly mediated by a family of dimeric aspartate-specific proteases termed caspases. Apoptosis can be induced by extracellular death ligands, such as Fas ligand, TNF-α, or TRAIL, via the extrinsic pathway to activate caspase-8. It can also be induced by agents such as cytotoxic compounds, radiation, and other environmental stresses via the intrinsic pathway with release of proapoptotic factors from mitochondria to activate caspase-9. Initiator caspases-8 and -9 in turn activate executioner caspases, among them caspases-3 and -7. Caspases then catalyze a multitude of proteolytic events to inactivate prosurvival/antiapoptotic proteins and activate antisurvival/proapoptotic proteins. This proteolysis results in apoptotic cell death and clearance of apoptotic bodies by phagocytes.
Because the study of apoptotic pathways has ramifications for development of therapies for treatment of cancer, there is significant interest in gaining a better understanding of caspase activity during apoptosis. For example, identification of new targets of proteolysis in apoptosis can lead to the discovery of prosurvival/antiapoptotic factors, which can lead to identification of novel chemotherapeutic targets. Over 300 publications describing a wide variety of cell types and apoptotic inducers have reported the proteolysis of approximately 360 human proteins in apoptosis (Lüthi and Martin, 2007). Adding to this complexity, the nature of the apoptotic response varies widely in a cell-dependent and stimulus-dependent manner that cannot be easily predicted (Fulda et al., 2001; Stepczynska et al., 2001; Wiegand et al., 2001). Thus, combined datasets of caspase substrates from studies using varied inducers and cell types have limited use for understanding how a single inducer can cause apoptosis in a particular cell type.
We have developed an enzymatic approach for global profiling of proteolysis and sequencing of cleavage sites in complex mixtures that is based on positive selection of protein fragments containing unblocked α-amines, characteristically produced in proteolysis. This positive selection is enabled by use of an engineered peptide ligase termed subtiligase to selectively biotinylate unblocked protein α-amines with absolute selectivity over ε-amines of lysine side chains. We have used this method to sequence 333 cleavage sites in 292 different protein substrates targeted by caspase-like proteolysis in Jurkat cells following intrinsic induction of apoptosis with the classic chemotherapeutic etoposide. In profiling the proteolysis that is induced by a single agent in a single cell line, this work reveals the vastness of caspase-like proteolysis that takes place during apoptosis, sheds light on determinants of specificity for this activity in a cellular context, and demonstrates the utility of a powerful degradomic technology to study proteolysis in biological samples.
RESULTS
A degradomic technology for positive selection of protease substrates
Direct and selective labeling of protein α-amines or α-carboxylates is a powerful approach for profiling proteolysis in complex mixtures since it permits direct identification of cleavage sites in protein substrates. Approximately 80% of mammalian proteins are known to be N-terminally acetylated (Brown and Roberts, 1976). Thus, greater signal over background can be achieved through N-terminal instead of C-terminal labeling. However, such labeling must still be extremely selective for α-amines over lysine ε-amines, which are approximately 25 times more abundant in an average protein. To achieve this selectivity, we have adopted an enzymological approach that makes use of the rationally designed protein ligase subtiligase. This engineered enzyme exhibits absolute selectivity for modification of α-amines (Abrahmsén et al., 1991; Chang et al., 1994).
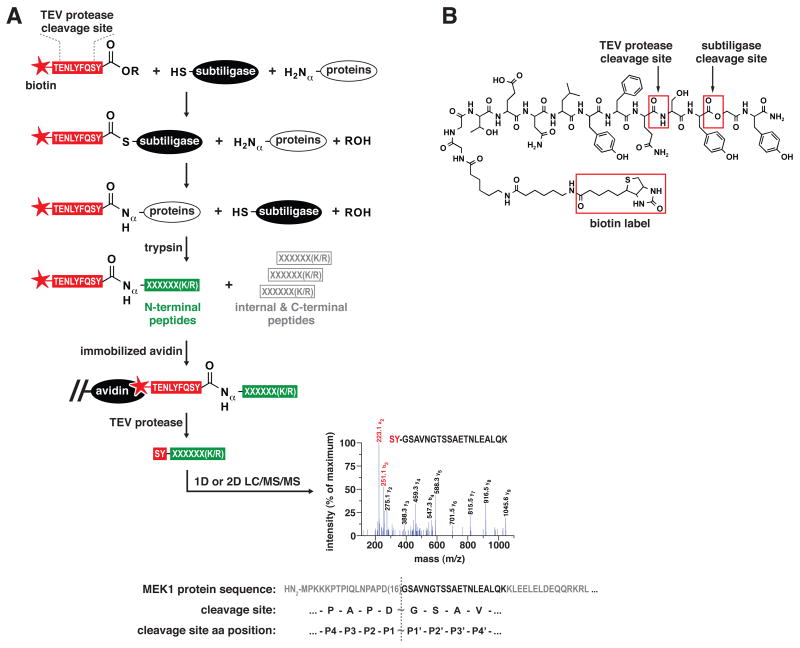
We have developed a proteomic method utilizing subtiligase that enables capture and sequencing of N-terminal peptides found in complex biochemical mixtures (Figure 1A). Proteins in biological samples are N-terminally biotinylated by treatment with subtiligase and a peptide glycolate ester substrate specially tailored to our proteomic workflow (Figure 1B). Biotinylated samples are exhaustively digested with trypsin, and N-terminal peptides are captured using avidin affinity media. The peptide ester substrate contains a tobacco etch virus (TEV) protease cleavage site to permit facile recovery of captured peptides. An important aspect of our workflow is that recovered peptides retain an N-terminal SY-dipeptide modification, providing a key hallmark to distinguish labeled peptides from contaminating unlabeled peptides using tandem mass spectrometry (LC/MS/MS). In standard protease nomenclature, substrates are cleaved between the P1 (N-terminal) and P1′ (C-terminal) residues, with Pn and Pn′ residues increasing in count by one in both directions away from the scissile bond (Schechter and Berger, 1968). Thus, the Pn′ residues of a cleavage site correspond to N-terminal residues of the labeled peptide identified, while the Pn residues of a cleavage site can be inferred from the protein sequence preceding the identified peptide.)
Figure 1.
A subtiligase-based method for positive selection of peptides corresponding to N-termini of proteins from complex mixtures. (A) Workflow for the biotinylation of protein N-termini in complex mixtures using subtiligase and a biotinylated peptide ester that contains a TEV protease cleavage site, trypsinization of labeled proteins, capture of biotinylated N-terminal peptides with immobilized avidin, recovery of captured peptides using TEV protease, and analysis of N-terminal peptides by 1D or 2D LC/MS/MS for identification of corresponding proteins and cleavage sites. The representative MS/MS spectrum corresponds to semi-tryptic peptide GSAVNGTSSAETNLEALQK from MEK1 (MP2K1_HUMAN) and identifies a previously unknown caspase-like cleavage site at Asp 16. The a2 and b2 ions at m/z 223 and 251 are characteristic hallmarks of a ligated, SY-bearing, N-terminal peptide. (B) Structure of the biotinylated peptide glycolate ester used in the proteomic workflow.
As a validation of this method, we analyzed endogenous N-termini in non-apoptotic Jurkat cells in two small-scale experiments using one-dimensional reversed-phase (1D) LC/MS/MS and two large-scale experiments using two-dimensional strong cation exchange/reversed-phase (2D) LC/MS/MS (summarized in Supplemental Tables 1 and 2). Comparison of data obtained in both types of experiments is informative since 1D LC/MS/MS typically results in identification of abundant N-termini, whereas the increased proteomic coverage afforded by 2D LC/MS/MS results in additional identification of lower abundance N-termini. Of the combined 131 unique N-termini identified in small-scale experiments, 72% are either annotated in Swiss-Prot as native protein N-termini, or correspond to cleavages within the first 50 residues of proteins as would be expected for N-terminal signal or transit peptide processing (Supplemental Figure 1A). The remaining 28% correspond to cleavages outside the first 50 residues and may be derived from constitutive protein degradation. In support of this notion, 51% of the combined 661 unique N-termini identified in large-scale experiments correspond to cleavages outside the first 50 residues (Supplemental Figure 1B). The increased frequency of such N-termini in large-scale experiments is consistent with the expected lower abundance for products of constitutive protein degradation.
Degradomic analysis of apoptotic Jurkat cells
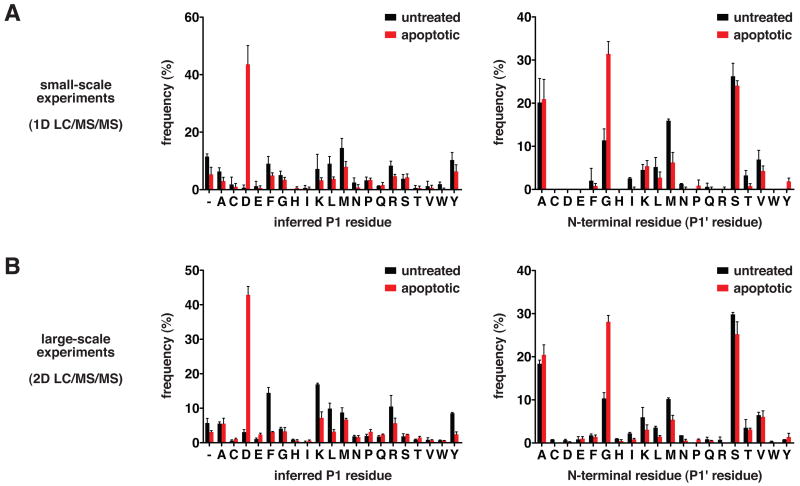
For analysis of apoptosis in Jurkat cells, we conducted several small-scale (1D) and large-scale (2D) LC/MS/MS experiments (representatives are summarized in Supplemental Tables 3 and 4) with cells treated with the topoisomerase II poison etoposide. The experiments with untreated cells described above serve as respective controls for the small- and large-scale experiments with apoptotic cells, in which a combined 244 and 733 unique N-termini, respectively, were identified. Combined datasets of all N-terminal peptides identified in untreated and apoptotic Jurkat cells, respectively, are included as supplemental data (Supplemental Table 5 and Table 6). Caspases are known to exhibit strict substrate specificity for aspartate at P1, and for glycine > serine > alanine at P1′ (Schilling and Overall, 2008; Stennicke et al., 2000). In small-scale experiments, 43% of N-termini identified in apoptotic cells were derived from P1 aspartate cleavages, in contrast to less than 1% in untreated cells (Figure 2A). In large-scale experiments, 43% of N-termini identified in apoptotic cells were derived from P1 aspartate cleavages, in contrast to 3% in untreated cells (Figure 2B). An increased frequency of glycine at the first position of N-termini is also observed in apoptotic cells relative to untreated cells at both experimental scales (Figure 2A and 2B). The N-termini uniquely identified in apoptotic Jurkat cells are thus consistent with induction of caspase-like activity.
Figure 2.
N-termini derived from caspase-like proteolytic processing are a hallmark of apoptotic cells. (A) Frequencies of P1 and P1′ amino acid residues corresponding to non-homologous N-termini identified in small-scale 1D LC/MS/MS experiments with untreated and apoptotic Jurkat cells. Data are represented as mean ± SD (n=2 for untreated and n=4 for apoptotic). (B) Frequencies of P1 and P1′ amino acid residues corresponding to non-homologous N-termini identified in large-scale 2D LC/MS/MS experiments with untreated and apoptotic Jurkat cells. Data are represented as mean ± SD (n=2 for untreated and n=3 for apoptotic). “ – “ indicates lack of a putative P1 residue in cases where the P1′ residue is an initiator methionine.
Of the 3% P1 aspartate N-termini detected in large-scale experiments with untreated cells (Figure 2B), 55% correspond to reported caspase substrates (Lüthi and Martin, 2007). Thus, it is likely that these originate from the small number of apoptotic cells typically present in untreated cultures. The detection of 3% P1 aspartate N-termini in large-scale experiments with untreated cells and less than 1% in small-scale experiments is consistent with the low abundance of such N-termini in cultures of normal cells. Additionally, if one considers that N-termini annotated in Swiss-Prot are representative of native N-termini in healthy cells, it is notable that < 1% are derived from proteolytic processing following an aspartate residue (Supplemental Figure 2). In apoptotic samples, we find that the increased frequency of N-termini located beyond the first 50 residues is solely attributable to P1 aspartate N-termini (Supplemental Figure 1B and 1C). Thus, the vast majority of proteolysis we observe in apoptosis is attributable to caspases or proteases with caspase-like substrate specificity.
Among the total 1099 SY-labeled peptides identified in etoposide-treated Jurkat cells, 418 follow aspartate in corresponding protein sequences (Supplemental Table 4 and Table 6). These peptides correspond to 333 P1 aspartate N-termini and caspase-like cleavage sites (identified cleavage sites are listed in Supplemental Table 7). In turn, these cleavage sites map to 282 unique substrates and 10 additional others that cannot be distinguished from homologs containing the same identified cleavage site (identified substrates are listed in Supplemental Table 8). The average overlap between datasets obtained in separate experiments is 55% at the peptide level and 58% at the protein level (Supplemental Figure 3A and 3B). Similar overlap levels (~ 67%) have been previously observed for replicate analyses of complex mixtures of peptides using LC/MS/MS (Elias et al., 2005). Using immunoblotting, we have verified that 16 of the proteins identified as caspase substrates in our studies are cleaved during apoptosis (Supplemental Figure 4A = 8 proteins, Figure 6 = 5 proteins, and unpublished data of Crawford & Wells = 4 proteins). We have also determined that the proteolysis of a representative set of substrates is blocked by the broad-spectrum caspase inhibitor Z-VAD(OMe)-fmk, consistent with this proteolysis being caspase-dependent (Supplemental Figure 4B).
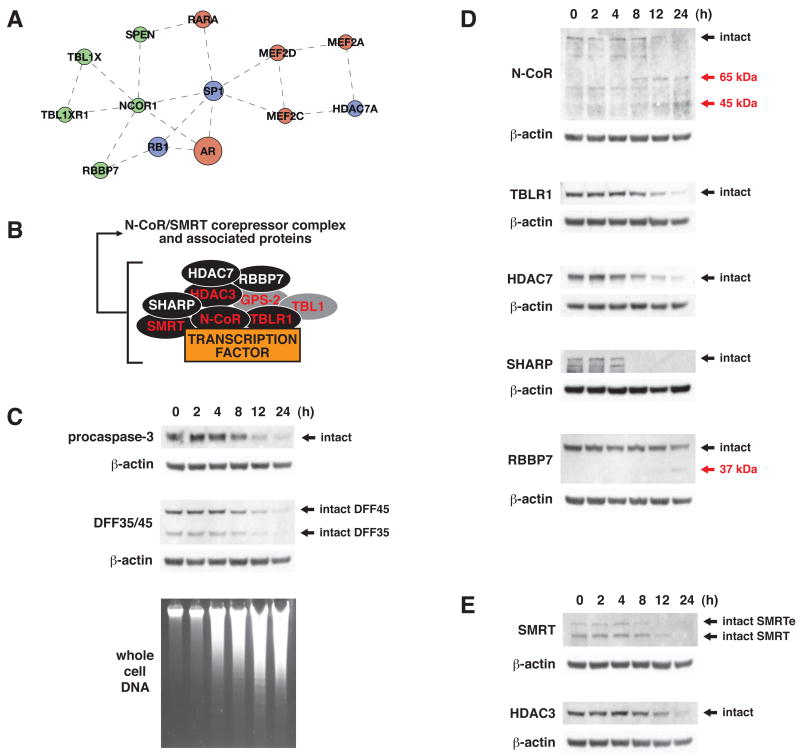
Figure 6.
Analysis of proteolysis of N-CoR/SMRT corepressor complex components during apoptosis in Jurkat cells following treatment with etoposide (50 μM). (A) Caspase substrate protein interaction subnetwork encompassing components of the N-CoR/SMRT corepressor complex (N-CoR, SPEN, TBLR1, RBBP7, and HDAC7), and transcription factors such as retinoic acid receptor, androgen receptor, and SP1 (green = from this work, red = from literature, blue = in both datasets). (B) Schematic representation of N-CoR/SMRT corepressor complex resident components and visiting interactors (red label = resident component, white label = visiting interactor, black fill = target of proteolysis in apoptosis). (C) Time courses for the proteolysis of procaspase-3 and DFF35/45, and for oligonucleosomal DNA fragmentation. (D) Full cleavage of N-CoR, HDAC7, SHARP, and TBLR1, and partial cleavage of RBBP7. (E) Full cleavage of SMRT and of HDAC-3, a previously identified caspase substrate. Black arrows indicate full-length proteins. Red arrows indicate expected cleavage products for cleavage at the sites identified in our studies (cleavage products were not detected in all cases).
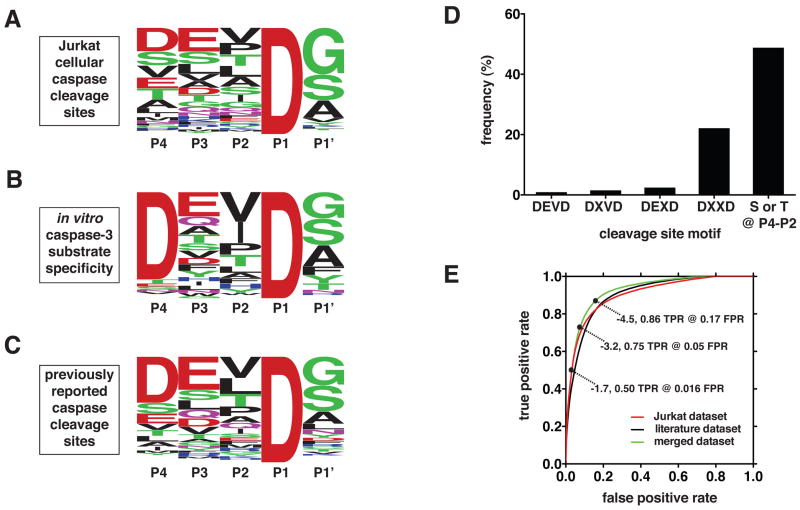
The most frequent residues at the P4, P3, P2, and P1′ positions of the caspase-like cleavage sites identified in apoptotic Jurkat cells are aspartate, glutamate, valine, and glycine, respectively (Figure 3A). Thus, an averaged composite of these cleavage sites indicates that the most common caspase activity in apoptotic cells exhibits a specificity that is most similar to the substrate specificity of executioner caspases-3 and -7, as determined using peptide substrates (Figure 3B) (Thornberry et al., 1997). However, there are significant differences between the cellular cleavage sites and the in vitro specificity profiles. Notably, the canonical DEVD cleavage site motif is found in less than 1% of the caspase-like cleavage sites observed in apoptotic Jurkat cells, and the broader DXXD motif is still only found in 22% of the identified cleavage sites (Figure 3D). A distinct difference in the composite cellular profile is the high frequency of serine and threonine residues at P4, P3, and P2, which is not observed in vitro for any of the caspases (Supplemental Figure 5). Interestingly, a composite of all previously reported human caspase cleavage sites (Lüthi and Martin, 2007) is very similar to the Jurkat cellular profile reported here (Figure 3C).
Figure 3.
Substrate specificity of the caspase-like proteolytic activity in etoposide-stimulated Jurkat cells. (A) Sequence logo representation (Crooks et al., 2004) of the frequency of amino acid residues in the identified caspase cleavage sites. (B) Sequence logo representation of the in vitro substrate specificity of caspase-3 (Stennicke et al., 2000; Thornberry et al., 1997). (C) Sequence logo representation of the frequency of amino acid residues in known human caspase cleavage sites (Lüthi and Martin, 2007). (D) Frequency of P4-P1 motifs in the identified caspase cleavage sites. (E) Receiver operator characteristic curves showing the discrimination ability of HMMs constructed from three different cleavage site training sets (Jurkat, literature, and merged). Three representative HMM score threshold values for the merged dataset are indicates (TPR = true positive rate, FPR = false positive rate).
These observations suggest that caspase substrate specificity determined using peptide substrates has limited value as a predictor of physiological caspase cleavage sites. To investigate the predictive value of a large set of known physiological caspase cleavage sites, we constructed three profile hidden Markov models (HMMs) using the cleavage sites identified in our studies, previously reported cleavage sites, and the union of these two datasets (a detailed description of this analysis is found in Supplemental Experimental Procedures). The accuracy of these HMMs was estimated using jacknifing and plotted in a receiver operator characteristic (ROC) plot, showing the true positive rate versus the false positive rate at different HMM score thresholds. While all three HMMs predict caspase cleavage sites relatively accurately, the HMM built from the merged substrate set performed most accurately (Figure 3E). Its true positive rate was 0.86 at the false positive rate of 0.15, compared to the average true positive rate of 0.84 at the false positive rate of 0.17 for the other two HMMs.
Analysis of structural determinants of caspase substrate specificity
The combined dataset of the 333 caspase cleavage sites identified in our work and the approximately 300 previously identified human caspase cleavage sites (Lüthi and Martin, 2007) allows an opportunity to expand our understanding of caspase substrate specificity from primary structure to the level of secondary and higher order structures. To accomplish this goal, we mapped the known caspase cleavage sites onto experimentally determined atomic structures in the Protein Data Bank (PDB) (Berman et al., 2002), as well as comparative protein structure models in the ModBase database (Pieper et al., 2006). Stringent filters were applied so that only models likely to be sufficiently accurate for the analysis were used.
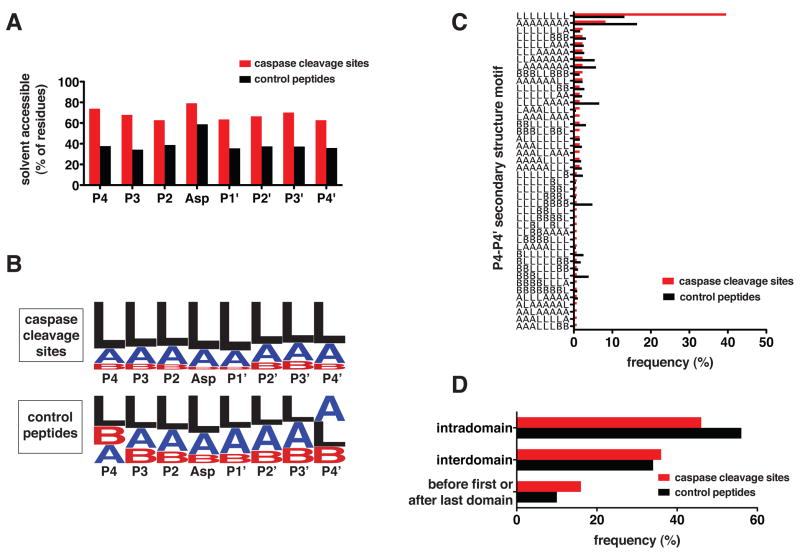
We identified 18 cleavage sites in known structures and 116 sites in comparative models. Depending on P4 through P4′ position, between 60% to 80% of cleavage site residues are solvent accessible, as defined by solvent exposure of greater than 33% total surface area (Figure 4A). Averaged across P4 through P4′, cleavage site residues are 76% more exposed than a reference control of all octapeptide sequences in the PDB containing an aspartate residue at the fourth position. The type of secondary structure was assigned using DSSP (Kabsch and Sander, 1983) for P4 through P4′ positions. The frequency of secondary structure types at each position reveals that caspases most frequently cleave protein substrates at loops relative to the octapeptide reference control described above (Figure 4B). Surprisingly, proteolysis at α-helical regions is not uncommon. Binning of cleavage sites into secondary structure motifs reveals that while an all-loop motif is most common secondary structure motif, the second most common one is an all-helix motif (Figure 4C). The finding that some cleavages occur at solvent inaccessible and α-helical regions likely reflects structural dynamics of these regions. Structural examples of cleavages identified in our studies are included as supplemental data (Supplemental Figure 6).
Figure 4.
Structural determinants of caspase substrate specificity. (A) Solvent accessibility (> 30% surface area exposed) at each position of all known P4-P4′ caspase cleavage sites and each position of all eight residue sequences containing aspartate in the fourth position found in PDB protein structures (control peptides). Differences between cleavage sites and control peptides have associated p-values < 0.0001 by chi-square at all positions. (B) Sequence logo representations of secondary structure at each position of all known P4-P4′ caspase cleavage sites and each position of the control peptides described above, where L = loop, A = α-helix, and B = β-sheet. Differences between cleavage sites and control peptides have associated p-values < 0.0001 by chi-square at all positions, except for P2 with 0.0014 and P1 with 0.0007. (C) Distribution of secondary structure motifs for P4-P4′ caspase cleavage sites and for the control peptides described above. (D) Localization of caspase cleavage sites in substrates relative to functional domain boundaries annotated in Pfam compared to localization of all eight residue sequences containing aspartate in the fourth position found in the human Swiss-Prot database (control peptides). Differences between cleavage sites and control peptides have associated p-values < 0.0001 by chi-square in all cases.
Analysis of the location of cleavage sites in caspase substrates annotated in the Pfam database (Finn et al., 2006) indicates that 46% of them are located within an annotated functional domain, 38% are located between annotated domains, and 16% are located at protein termini, either before the first annotated domain or after the last (Figure 4D). This distribution is relatively similar to the distribution of a reference control of all octapeptide sequences in the human Swiss-Prot database containing an aspartate residue at the fourth position. Thus caspases do not exhibit a strong preference for cleavage of substrates either inside or outside functional domains. Caspase cleavage sites are also evenly distributed over the length of protein substrates (data not shown).
Analysis of protein-protein interactions between caspase substrates
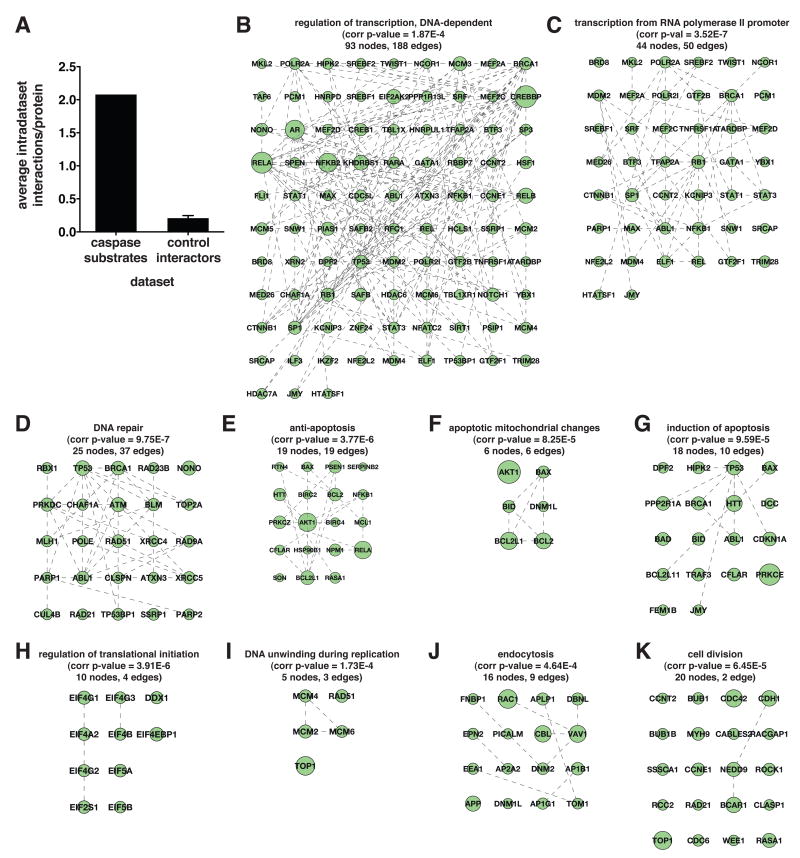
Upon inspection of the entire dataset of caspase substrates, we noted a number of instances where multiple proteins along a single biochemical pathway, or in a single protein complex, are targeted by caspases. For a more systematic analysis of this property, we utilized data from three different protein interaction databases (HPRD, IntAct, and MINT) for creation of a network of caspase substrate protein interactors (Chatr-aryamontri et al., 2007; Kerrien et al., 2007; Mishra et al., 2006). This network is made up solely of the substrates identified in our studies and reported human caspase substrates (Lüthi and Martin, 2007), but excludes the caspases themselves (binary interactions constituting this network are listed in Supplemental Table 9). A total of 415 interactors and 1253 interactions were found among the merged human caspase substrate dataset of 602 proteins, for an average of 2.1 intradataset interactions per caspase substrate. Ten datasets of 602 randomly chosen proteins from the protein interaction databases had an average of 0.2 intradataset interactions per protein. This indicates a tenfold enrichment in protein interactions between caspase substrates relative to randomly interacting proteins (Figure 5A) (a detailed description of this analysis is found in Supplemental Experimental Procedures).
Figure 5.
Network analysis of protein interactions between caspase substrates, including those identified in this work and previously reported human caspase substrates (Lüthi and Martin, 2007). (A) Enrichment in protein-protein interactions between the 602 total caspase substrates relative to an equally sized reference control set of protein interactors randomly selected from protein interaction databases. Data for the control set are represented as mean ± SD (n=10). (B–K) Caspase substrate protein interaction subnetworks encompassing substrates annotated to overrepresentated GO biological process terms relative to the entire human GO annotation. Substrates are labeled using gene symbols. Corrected p-values, number of nodes, and number of edges are indicated in each case.
To find protein networks and complexes that are preferentially targeted by caspases during apoptosis, we used the BiNGO (Maere et al., 2005) plugin of Cytoscape (Shannon et al., 2003) to find GO biological process terms that are overrepresented relative to the complete human GO annotation. We then focused on the three deepest levels of the GO hierarchy of overrepresented terms to find the ten most informative GO terms and the substrates annotated to those terms. This analysis yielded subnetworks of substrates involved in regulation of transcription, transcription from RNA polymerase II promoter, DNA repair, anti-apoptosis, induction of apoptosis, apoptotic mitochondrial changes, regulation of translational initiation, DNA unwinding during replication, endocytosis, and cell division (Figure 5B–5K). The regulation of transcription GO term yielded the densest subnetwork, with 188 edges among 93 nodes (Figure 5B). In sharp contrast to the other nine GO terms, the cell division GO term barely yielded a network at all, with only 2 edges among 20 nodes (Figure 5K).
The N-CoR/SMRT complex is a target of caspase proteolysis during apoptosis
To analyze whether multiple cleavages along a pathway or in a complex occur at physiologically relevant rates, we focused on the portion of the regulation of transcription subnetwork representing N-CoR/SMRT transcriptional corepressor complex components and interactors (Figure 6A and 6B). This complex is involved in the recruitment of histone deacetylase activity to chromatin. Our studies identified N-CoR/SMRT complex resident components N-CoR and TBLR1 (Karagianni and Wong, 2007), as well as additional N-CoR/SMRT complex interactors HDAC7 (Fischle et al., 2001), MINT/SHARP/SPEN (Shi et al., 2001), and RBBP7/RbAp46 (Takezawa et al., 2007) as caspase substrates (MS/MS spectra of N-terminal peptides corresponding to cleavage sites in these proteins are included as Supplemental Figures 7–14). We probed for cleavage of these proteins during etoposide-induced apoptosis in Jurkat cells by immunoblot in order to qualitatively determine extent of proteolysis in each case.
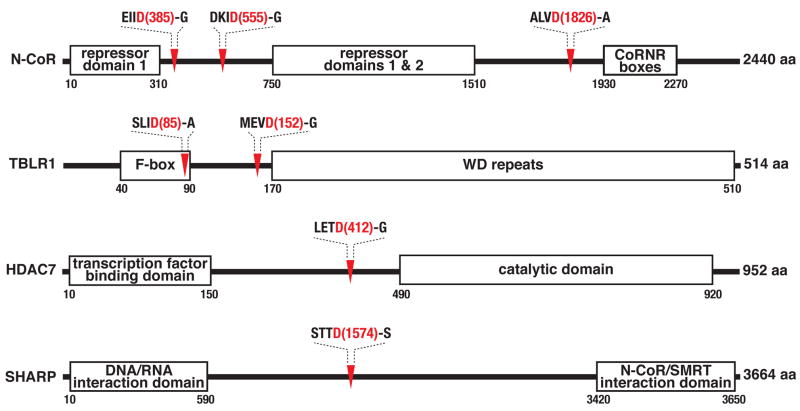
N-CoR, TBLR1, HDAC7, and SHARP were all fully cleaved at rates similar to those observed for hallmark substrates procaspase-3 and DFF45 (Figure 6C and 6D). This proteolysis also tracked reasonably well with the time course for DNA fragmentation. In contrast, only partial proteolysis of RBBP7 was observed, suggesting it to be a possible bystander substrate (Figure 6D). Although not detected in our proteomic studies, we predicted the N-CoR homolog SMRT (Karagianni and Wong, 2007) to also be a caspase substrate based on high sequence similarity to N-CoR cleavage sites. Indeed, SMRT was fully cleaved during etoposide-induced apoptosis in Jurkat cells (Figure 6E). The previously identified caspase substrate HDAC3 (Escaffit et al., 2007), another N-CoR/SMRT complex component (Karagianni and Wong, 2007), was also fully cleaved. Organization of functional domains in these proteins indicates that proteolytic processing at the cleavage sites identified in our studies likely results in inactivation of protein function by virtue of separating functional domains from one another (Figure 7).
Figure 7.
Caspases cleave resident N-CoR/SMRT complex components and visiting interactors at regions leading to separation of functional domains. Functional domain organization and candidate caspase cleavage sites in SMRT and TBL1 are similar to those indicated for the respective homologs, N-CoR and TBL1R.
DISCUSSION
One of the most striking findings of this study is that caspase substrates as a whole tend to physically interact with one or more other caspase substrates, either in protein complexes or networks. We interpret this as an indication that caspases target a limited set of biological pathways to elicit programmed cell death, as opposed to indiscriminately targeting the entire cellular proteome. These data also suggest that caspases target protein complexes that are hubs for cell viability in essential processes such as transcription, and that targeting of multiple components in each complex is required for a full commitment to apoptosis. In this regard, it is notable that active caspases are dimeric, which is rare for proteases. A dimer is well equipped for semi-processive activity consistent with targeting multiple components of protein complexes. Another reported example of targeted proteolysis of a protein complex is the cleavage of SET, HMG-2, and Ape1, three components of the SET complex, by the cytotoxic lymphocyte protease granzyme A (Lieberman and Fan, 2003). Interestingly, granzyme A is also dimeric.
The discovery that several components of the N-CoR/SMRT transcriptional corepressor complex are targets of caspase proteolysis presents a remarkable example of multiple cleavages in a single protein complex or pathway during apoptosis. Six proteins that are part of, or interact with, the N-CoR/SMRT complex are fully cleaved during etoposide-induced apoptosis in Jurkat cells, including the corepressors N-CoR and SMRT themselves. This finding was made possible by our large-scale discovery-oriented proteomic approach, as opposed to a more typical focused hypothesis-driven approach. Inactivation of the N-CoR/SMRT complex during apoptosis may achieve a result similar to the effect of HDAC inhibitors, with decreased histone deacetylation leading to transcriptional upregulation of proapoptotic genes (Bolden et al., 2006). Interestingly, HDAC 7 has recently been implicated as a physiological substrate of caspase-8, with its proteolytic inactivation leading to upregulation of Nur77 (Scott et al., 2008).
Our studies indicate that a change in function of proteins targeted by caspases during apoptosis must be rationalized by one or occasionally a few cuts per protein. We have found that caspase cleavages occur inside functional domains and between functional domains at approximately equal frequencies. In either case, relatively stable products must be produced following cleavage of the substrates since we detected them. Stability of these products is also consistent with the strict P1′ glycine, serine, and alanine specificity we observe for the cellular caspase-like activity, which creates fragments conforming to the N-end rule (Varshavsky, 1992). In addition to functional disruption of the substrate protein, such cleavages may result in products that function as dominant negatives. For example, in the case of the N-CoR and SMRT corepressors, the C-terminal cleavage products contain the CoRNR boxes known to interact with nuclear receptors (Hu and Lazar, 1999). These proteolysis products could thus inhibit interaction between N-CoR/SMRT and nuclear receptors.
By globally identifying caspase-like cleavage sites in the proteome of apoptotic cells, this work presents a large-scale substrate specificity profile of caspase processing of endogenous proteins in intact cells. Importantly, this profile is influenced not only by the primary structure of cleavage sites, but also solvent accessibility, secondary and higher order protein structure, and possibly post-translational modifications of substrates (Tözsér et al., 2003). Our finding that caspases often target proteins in complexes underscores the value of studying determinants of proteolysis under physiologically relevant conditions. Although the aggregate substrate specificity of the caspase-like activity observed during etoposide-induced apoptosis in Jurkat cells is most similar to the known substrate specificity of executioner caspases, substrate specificity studies of caspases using peptides do not fully account for the observed cellular specificity (Schilling and Overall, 2008; Stennicke et al., 2000; Thornberry et al., 1997). Peptide-centric approaches are best suited for determination of optimal protease substrate specificity, invaluable in development of sensitive synthetic substrates or potent inhibitors. In contrast, a protein-centric methods such as the one presented here is best suited for characterization of endogenous proteolysis in biological samples.
This work indicates that the widely used primary structural determinants of caspase in vitro substrate specificity are insufficient to predict physiological caspase cleavage sites. However, the cellular cleavage sites we have identified experimentally double the number of annotated caspase cleavage sites, significantly expanding a dataset that can be used to train algorithms for predicting cleavage sites. Indeed, a proof of principle is provided by an accurate prediction of caspase cleavage sites by our preliminary HMMs. In addition to demonstrating that caspase cleavage sites are most commonly found in solvent accessible loop regions, as shown for other proteases (Hubbard et al., 1991), our analysis also indicates that a number of cleavage sites appear in partially solvent inaccessible regions and α-helices. This information could also be incorporated into predictive algorithms. Finally, based on our protein interaction analysis, predictive algorithms may also benefit from scoring that considers physical interactions of candidate substrates with other caspase substrates.
Common approaches for the study of proteolysis in complex mixtures employ gel electrophoresis and mass spectrometry for analysis of proteins in cells, typically identifying tens of protein substrates at a time (Machuy et al., 2005). These approaches do not usually identify specific cleavage sites. In contrast, modern proteomic methods using positive enrichment of phosphorylated, glycosylated, or ubiquitinated polypeptides can lead to the identification of hundreds or thousands of post-translationally modified sites on proteins (Collins et al., 2007; Peng et al., 2003; Vosseller et al., 2006). However, selective capture of the products of proteolysis is not facile. Gevaert et al. and McDonald et al. have reported methods for negative selection of N-terminal peptides, while Timmer et al. have reported an approach for positive selection of N-terminal peptides (Gevaert et al., 2003; McDonald et al., 2005; Timmer et al., 2007). These chemical approaches require two consecutive and quasi-orthogonal derivatization steps, the first to block lysine ε-amines and the second to label terminal α-amines. We believe the success of our method is based on the advantage of achieving great selectivity for α-amines in a single labeling step through use of the enzyme subtiligase.
The incomplete overlap between cleavage sites and protein substrates identified in our separate experiments is not uncommon for tandem mass spectrometric analysis of complex mixtures, in which analysis of many species, whether peptidic or not, precludes complete sampling (Elias et al., 2005). The number of caspase substrates we have identified is thus likely smaller than the total number of caspase substrates in apoptotic Jurkat cells. We identified 50 of approximately 361 previously reported human caspase substrates (Supplemental Figure 3C), 48 of approximately 227 previously reported caspase substrates for which cleavage sites are known (Supplemental Figure 3D), and 50 of approximately 307 previously reported human caspase cleavage sites (Supplemental Figure 3E) (Lüthi and Martin, 2007). In addition to incomplete proteomic sampling, three additional factors likely account for the modest overlap with previously identified substrates.
First, only cleavage sites corresponding to N-terminal semi-tryptic peptides 7 to 40 residues in length (without the additional SY dipeptide label) are generally identified in our studies. With collision-induced dissociation (CID), fragmentation of peptides below this range generally does not provide enough information for unambiguous matching to databases, and most peptides above this range do not fragment efficiently. Approximately half of the 307 previously reported human caspase cleavage sites result in N-terminal semi-tryptic peptides that fall outside this range and would not be identified using our current method (Supplemental Figure 15). Second, each analytical method, whether global or focused, will have its own associated biases and limitations. For example, a limitation of the subtiligase labeling method is that the enzyme cannot access protein N-termini that are buried or occluded. Third, we employed a single apoptotic inducer in a single cell line, using a single analysis method, whereas the previously reported set of substrates comes from a multitude of studies using varied inducers, cell types, and methods.
Although the dataset of substrates we have identified is not comprehensive, it doubles the number of known cleavage sites in human targets of caspase-like proteolysis in apoptosis. The study of apoptotic pathways has important ramifications for identification of pathways that are critical for cellular homeostasis, and for development of potential anti-cancer therapeutics. A number of caspase targets are active or established drug targets for treating cancer, including topoisomerase II, Bcl-2, Hdm2, MEK1, and Akt, to name a few. Thus, it is possible that the list of substrates we have identified includes new candidate chemotherapeutic targets. The products of caspase proteolysis may also serve as useful biomarkers for assessment of chemotherapeutic efficacy, as demonstrated in the case of cytokeratin-18 for breast cancer (Olofsson et al., 2007). Along with MS-based quantitation, the technology we describe should enable global analysis of the apoptotic phenotype as a function of time, cellular context, and type of induction. Finally, the technology should also be broadly applicable for global sequencing of proteolytic cleavage sites in other biological settings.
EXPERIMENTAL PROCEDURES
Preparation of subtiligase and peptide ester substrate
Subtiligase was recombinantly expressed in B. subtilis and purified essentially as previously described (Abrahmsén et al., 1991). The biotinylated peptide glycolate ester was synthesized by solid-phase peptide synthesis as described for other subtiligase substrates (Braisted et al., 1997). See Supplemental Experimental Procedures for more detail.
Cell culture, induction of apoptosis, and cell lysate preparation
Jurkat clone E6-1 (ATCC) cells at a density of 1 × 106 cells/ml were treated with etoposide (50 μM) for 0 or 12 hours prior to harvesting. Detergent lysates were prepared at a typical concentration of 2 × 108 cells/ml (approximately 20 mg/ml) using buffered 1.0% Triton X-100 in the presence of protease inhibitors. See Supplemental Experimental Procedures for more detail.
Sample biotinylation, denaturation, reduction, alkylation, and gel filtration
Cell lysates were biotinylated by treatment with subtiligase (1 μM), biotinylated peptide ester substrate (1 mM), and DTT (2 mM). Ligation reactions were typically left to proceed at room temperature for 60 minutes. Samples were then denatured, reduced, alkylated, and subjected to gel filtration for removal of hydrolyzed peptide ester substrate. See Supplemental Experimental Procedures for more detail.
Trypsinization, capture of biotinylated peptides, and recovery of biotinylated peptides
Filtered samples were subjected to solution digestion with sequencing grade modified trypsin (Promega). Biotinylated N-terminal peptides were captured from trypsinized samples using NeutrAvidin agarose (Pierce). Captured peptides were recovered by treatment of agarose resin with recombinant TEV protease (1 μM). See Supplemental Experimental Procedures for more detail.
LC/MS/MS
N-terminal peptide samples were analyzed by one-dimensional reversed-phase LC/MS/MS or two-dimensional strong cation exchange/reversed-phase LC/MS/MS. In the latter case, samples were fractionated by offline strong cation exchange chromatography with a 60 minute gradient on a 2.1 × 200 mm PolySULFOETHYL Aspartamide column at a flow rate of 0.3 ml/min. Reversed-phase chromatography of unfractionated or fractionated samples was carried out with a 60 minute gradient on a 75 μm × 15 cm C18 column at a flow rate of 350 nl/min. The capillary column was coupled to a QSTAR Pulsar, QSTAR XL, or QSTAR Elite mass spectrometer (Applied Biosystems). For each acquired MS spectrum, either the single or the two most intense multiply charged peaks were selected for generation of subsequent CID spectra. A dynamic exclusion window of 3 minutes was applied. CID spectra not included as supplemental data will be made available upon request. See Supplemental Experimental Procedures for more detail.
Interpretation of MS/MS spectra
Data were analyzed using Analyst QS software, and MS/MS centroid peak lists were generated using the Mascot.dll script. Data were searched against the Swiss-Prot human database (March 2008 release) using Protein Prospector 5.0 (University of California, San Francisco). Peptide tolerances in MS and MS/MS modes were 100 ppm and 300 ppm, respectively. The digest protease specified was trypsin, allowing for two missed cleavages and non-specific cleavage at N-termini. An N-terminal SY modification and cysteine carbamidomethylation were specified as a fixed modifications, and methionine oxidation was specified as a variable modification. Peptides with scores ≥ 22 and expectation values ≤ 0.05 were considered positively identified. False discovery rates for peptide identifications were estimated using a target-decoy strategy. See Supplemental Experimental Procedures for more detail.
Bioinformatics
Cleavage site prediction was performed with HMMer version 2.3.2 (Eddy, 1998). The training and testing substrate sets were obtained by a jacknife procedure from data for the cleavage sites identified in our work and those previously reported in the literature (Lüthi and Martin, 2007). Solvent accessible surface area and secondary structure analysis of cleavage sites was carried out on a set of experimentally determined structures from the Protein Data Bank (Berman et al., 2002) and “good quality” comparative models from ModBase (Pieper et al., 2006). Domain analysis was performed using domain assignments from the Pfam database, (Finn et al., 2006). Protein-protein interaction analysis was carried out using data from the HPRD (Mishra et al., 2006), IntAct (Kerrien et al., 2007), and MINT (Chatr-aryamontri et al., 2007) databases. See Supplemental Experimental Procedures for more detail.
DNA fragmentation
Fragmentation of whole cell DNA was analyzed by agarose gel electrophoresis with the Apoptotic DNA Ladder Kit (Roche).
Immunoblotting
Jurkat cells at a density of 1 × 106 cells/ml were treated with etoposide (50 μM) for 0, 2, 4, 8, 12, and 24 hours prior to harvesting. Whole cell lysates were prepared at a concentration of 2 × 107 cells/ml using buffered 1.0% SDS in the presence of protease inhibitors and sonication. Lysates were normalized to a protein concentration of approximately 2 mg/ml prior to analysis by SDS-PAGE and Western blot. See Supplemental Experimental Procedures for more detail and a list of utilized antibodies.
Supplementary Material
Acknowledgments
We are grateful to members of the Wells, Burlingame, and Sali labs for useful discussions. This work was supported by NIH F32 GM074458 (SM), NIH R01 GM081051 (JAW), the Hartwell Foundation (JAW), NIH NCRR 01614 (ALB), the Adelson Medical Research Foundation (ALB), NIH R01 GM54762 (AS), NIH P01 GM71790 (AS), NIH U54 GM074945 (AS), the Sandler Family Supporting Foundation (AS), Hewlett-Packard (AS), NetApps (AS), IBM (AS), and Intel (AS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahmsén L, Tom J, Burnier J, Butcher KA, Kossiakoff A, Wells JA. Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry. 1991;30:4151–4159. doi: 10.1021/bi00231a007. [DOI] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nature reviews Drug discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Braisted AC, Judice JK, Wells JA. Synthesis of proteins by subtiligase. Methods Enzymol. 1997;289:298–313. doi: 10.1016/s0076-6879(97)89053-2. [DOI] [PubMed] [Google Scholar]
- Brown JL, Roberts WK. Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J Biol Chem. 1976;251:1009–1014. [PubMed] [Google Scholar]
- Chang TK, Jackson DY, Burnier JP, Wells JA. Subtiligase: a tool for semisynthesis of proteins. Proc Natl Acad Sci U S A. 1994;91:12544–12548. doi: 10.1073/pnas.91.26.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Yu L, Choudhary JS. Analysis of protein phosphorylation on a proteome-scale. Proteomics. 2007;7:2751–2768. doi: 10.1002/pmic.200700145. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Elias J, Haas W, Faherty B, Gygi S. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- Escaffit F, Vaute O, Chevillard-Briet M, Segui B, Takami Y, Nakayama T, Trouche D. Cleavage and cytoplasmic relocalization of histone deacetylase 3 are important for apoptosis progression. Mol Cell Biol. 2007;27:554–567. doi: 10.1128/MCB.00869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- Fulda S, Meyer E, Friesen C, Susin SA, Kroemer G, Debatin KM. Cell type specific involvement of death receptor and mitochondrial pathways in drug-induced apoptosis. Oncogene. 2001;20:1063–1075. doi: 10.1038/sj.onc.1204141. [DOI] [PubMed] [Google Scholar]
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J Mol Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Karagianni P, Wong J. HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26:5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp Cell Res. 2000;256:42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct--open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman J, Fan Z. Nuclear war: the granzyme A-bomb. Curr Opin Immunol. 2003;15:553–559. doi: 10.1016/s0952-7915(03)00108-0. [DOI] [PubMed] [Google Scholar]
- Lüthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- Machuy N, Thiede B, Rajalingam K, Dimmler C, Thieck O, Meyer TF, Rudel T. A global approach combining proteome analysis and phenotypic screening with RNA interference yields novel apoptosis regulators. Mol Cell Proteomics. 2005;4:44–55. doi: 10.1074/mcp.M400089-MCP200. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- McDonald L, Robertson DH, Hurst JL, Beynon RJ. Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nat Methods. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson MH, Ueno T, Pan Y, Xu R, Cai F, van der Kuip H, Muerdter TE, Sonnenberg M, Aulitzk WE, Schwarz S, et al. Cytokeratin-18 is a useful serum biomarker for early determination of response of breast carcinomas to chemotherapy. Clin Cancer Res. 2007:3198–3206. doi: 10.1158/1078-0432.CCR-07-0009. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem Biophys Res Commun. 1968;32:898–902. doi: 10.1016/0006-291x(68)90326-4. [DOI] [PubMed] [Google Scholar]
- Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- Scott FL, Fuchs GJ, Boyd SE, Denault JB, Hawkins CJ, Dequiedt F, Salvesen GS. Caspase-8 cleaves histone deacetylase 7 and abolishes its transcription repressor function. J Biol Chem. 2008;283:19499–19510. doi: 10.1074/jbc.M800331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes & Development. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J. 2000;350 Pt 2:563–568. [PMC free article] [PubMed] [Google Scholar]
- Stepczynska A, Lauber K, Engels IH, Janssen O, Kabelitz D, Wesselborg S, Schulze-Osthoff K. Staurosporine and conventional anticancer drugs induce overlapping, yet distinct pathways of apoptosis and caspase activation. Oncogene. 2001;20:1193–1202. doi: 10.1038/sj.onc.1204221. [DOI] [PubMed] [Google Scholar]
- Takezawa S, Yokoyama A, Okada M, Fujiki R, Iriyama A, Yanagi Y, Ito H, Takada I, Kishimoto M, Miyajima A, et al. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 2007;26:764–774. doi: 10.1038/sj.emboj.7601548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault JB, Ma Y, Dummitt B, Chang YH, Mast AE, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tözsér J, Bagossi P, Zahuczky G, Specht SI, Majerova E, Copeland TD. Effect of caspase cleavage-site phosphorylation on proteolysis. Biochem J. 2003;372:137–143. doi: 10.1042/BJ20021901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, et al. O-Linked N-Acetylglucosamine Proteomics of Postsynaptic Density Preparations Using Lectin Weak Affinity Chromatography and Mass Spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- Wiegand UK, Corbach S, Prescott AR, Savill J, Spruce BA. The trigger to cell death determines the efficiency with which dying cells are cleared by neighbours. Cell Death Differ. 2001;8:734–746. doi: 10.1038/sj.cdd.4400867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.