Abstract
DNA containing unmethylated CpG dinucleotides (CpG DNA) is a potent activator of innate and acquired immune responses. Although the sequence-specific immunostimulatory activity of CpG DNA has been extensively explored, little information is available about the importance of the stereochemical properties of CpG DNA. In this study, Y-shaped oligodeoxynucleotides (Y-ODNs) were prepared using three ODNs with the halves of each ODN being partially complementary to a half of the other two ODNs. Y-ODN induced greater amounts of tumour necrosis factor-α and interleukin-6 from RAW264.7 macrophage-like cells than did conventional single-stranded ODN (ssODN) or double-stranded ODN (dsODN). The Y-ODN was less stable in serum than dsODN, but greater amounts of Y-ODN were taken up by macrophage-like cells compared with dsODN. A newly designed Y-ODN containing three potent CpG motifs generated significantly higher levels of cytokines compared with dsODN containing the identical sequences. These results indicate that the Y-shaped form of ODN is a novel, reproducible and reliable approach to enhancing the immunostimulatory activity of ODNs.
Keywords: CpG motif, immunostimulatory activity, oligodeoxynucleotides, Toll-like receptor 9, Y-shape formation
Introduction
Bacterial DNA contains unmethylated CpG dinucleotides (CpG motifs) that induce a potent immunostimulatory response upon recognition by the Toll-like receptor 9 (TLR9) expressed on dendritic cells, B cells and macrophages.1,2 Synthetic oligodeoxynucleotides (ODNs) containing CpG motifs (CpG ODNs) can mimic the immunostimulatory activity of bacterial DNA and exhibit similar immune responses.3,4 Once activated by bacterial DNA or CpG ODN, i.e. CpG DNA, immune cells secrete various cytokines, including interleukin-6 (IL-6), IL-12, interferon-α/β (IFN-α/β), IFN-γ and tumour necrosis factor-α (TNF-α), and increase the expression of various costimulatory molecules.5,6 Thus, CpG DNA can induce T helper type 1 cytokine production; this promotes a cytotoxic T-lymphocyte response with enhanced immunoglobulin production, which has been used in the treatment of a broad spectrum of diseases, including cancer, viral and bacterial infections, allergic diseases and inflammatory disorders.1,7–9
The immunostimulatory activity of DNA has been extensively investigated using ODNs with varying base combinations, and several rules have been proposed. The activity of CpG DNA depends on the flanking sequences, and an ODN containing a GACGTT hexameric nucleotide motif strongly stimulates the immune system in rodents.10,11 Other parameters have also been reported to be important for immunostimulatory activity of DNA, e.g. TpC dinucleotide on the 5′ end, pyrimidine-rich on the 3′ side of the motif and the presence of two or three CpG motifs in a sequence.1 Thus, the optimal sequence of CpG ODN for activating mouse or human immune cells was elucidated by examining many possible base combinations. Although a few reports have shown that aggregation of ODNs increases their immunostimulatory activity,12,13 the stereochemical effects of CpG ODN on immunostimulatory activity have hardly been explored.
DNA possesses many desirable chemical and physical properties as a polymeric material and much progress has been made in DNA computing 14,15 and DNA nanotechnology.16–19 Recently, Li et al. established a reproducible method for constructing dendrimer-like DNA, by connecting Y-shaped DNA (Y-DNA) composed of three ODNs.20 This unique-structured DNA has been applied to various experimental settings, including nanobarcodes.21 However, the biological and immunological characteristics of such structured DNA preparations have not been examined. Their unique structure may be recognized differently by immune cells.
In the present study, Y-ODN was prepared using three ODNs with the halves of each ODN being partially complementary to a half of the other two ODNs. Then, the immunostimulatory activity of Y-ODN was examined using RAW264.7, a mouse macrophage-like cell line. Here, we show that newly designed Y-ODN containing three potent CpG motifs can be a powerful immunostimulatory compound through increased uptake by immune cells.
Materials and methods
Chemicals
Dulbecco's modified Eagle's minimum essential medium (DMEM), RPMI-1640 medium and phosphate-buffered saline (PBS) were obtained from Nissui Pharmaceutical (Tokyo, Japan). Fetal bovine serum (FBS) was obtained from MP Biomedicals (Eschwege, Germany). Opti-modified Eagle's medium (Opti-MEM) was purchased from Invitrogen (Carlsbad, CA). DNase I and 20-base-pair (bp) DNA ladder were purchased from Takara Bio (Otsu, Japan). Polymyxin B sulphate salt was purchased from Sigma Chemical Co. (St Louis, MO). All other chemicals were of the highest grade available and were used without further purification.
Cell cultures
RAW264.7 macrophage-like cells were grown in RPMI-1640 medium supplemented with 10% heat-inactivated FBS, 0·15% NaHCO3, 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mm l-glutamine at 37° in humidified air containing 5% CO2. Cells were then plated on 24-well culture plates at a density of 5 × 105 cells/ml and cultured for 24 hr. B16-BL6/Luc, a clone of murine melanoma B16-BL622 that stably expresses the firefly luciferase gene,23 was grown in 5% CO2 in humidified air at 37° with DMEM supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine. They were then plated on 24-well culture plates at a density of 2 × 104 cells/ml and cultured for 8 hr.
Oligodeoxynucleotides
Phosphodiester ODNs (Table 1) were purchased from Invitrogen. For cellular uptake studies, Y0a labelled with fluorescein at the 5′ end was used for all fluorescein-labelled ODN preparations.
Table 1.
Oligodeoxynucleotide (ODN) sequences used for preparation of single stranded-, double stranded- and Y-shaped ODNs
| Name | Sequence (5′→3′) |
|---|---|
| Y0a | TGACTGGATCCGCATGACATTCGCCGTAAG |
| Y0b | TGACCTTACGGCGAATGACCGAATCAGCCT |
| Y0c | TGACAGGCTGATTCGGTTCATGCGGATCCA |
| r.Y0a | TGACCTTACGGCGAATGTCATGCGGATCCA |
| r.Y0b | TGACAGGCTGATTCGGTCATTCGCCGTAAG |
| r.Y0c | TGACTGGATCCGCATGAACCGAATCAGCCT |
| Y0a(CpG) | TGACGACGTTCGCATGACATTCGCCGTAAG |
| Y0b(CpG) | TGACCTTACGGCGAATGACCGAATCAGCCT |
| Y0c(CpG) | TGACAGGCTGATTCGGTTCATGCGAACGTC |
| Y0a(CpG3) | TCGACGTTTCCGCATGACATTCGCCGAACG |
| Y0b(CpG3) | TCGACGTTCGGCGAATGACCGAATCAAACG |
| Y0c(CpG3) | TCGACGTTTGATTCGGTTCATGCGGAAACG |
| r.Y0a(CpG3) | TCGACGTTCGGCGAATGTCATGCGGAAACG |
| r.Y0b(CpG3) | TCGACGTTTGATTCGGTCATTCGCCGAACG |
| r.Y0c(CpG3) | TCGACGTTTCCGCATGAACCGAATCAAACG |
All ODNs have a phosphodiester backbone. The ‘r.’ indicates that the sequence is complementary to the ODN with four base overhangs at both 5′ ends, and each of these complementary ODNs was used to obtain double-stranded ODN. The potent immunostimulatory CpG motif (GACGTT) is underlined.
Preparation of Y-ODN and double-stranded ODN
Y-ODN was prepared by mixing equimolar amounts of three 30-base ODNs as reported previously.20 In brief, three ODNs dissolved in an annealing buffer [10 mm Tris–HCl, pH 8·0, 1 mm ethylenediaminetetraacetic acid (EDTA) and 50 mm NaCl] were mixed in sterile Milli-Q water at a final concentration of 0·5 mm for each ODN. Mixtures were incubated at 95° for 5 min, 65° for 2 min, 62° for 1 min, then slowly cooled to 4°. Formation of Y-ODN was confirmed by 21% polyacrylamide gel electrophoresis (PAGE) at 200 V for 1·5–2 hr. Double-stranded ODN (dsODN) with four base overhangs at both 5′ ends was prepared by addition of a complementary ODN (r.Y0a, Table 1) to Y0a ODN.
Dynamic light-scattering analysis
The apparent hydrodynamic sizes of Y-ODN, Y-ODN(CpG), Y-ODN(CpG3), single-stranded ODN (ssODN; Y0a) and dsODN were measured by laser light scattering using a Malvern Zetasizer 3000HS (Malvern Instruments, Malvern, UK) equipped with a helium-neon laser (633 nm).
Cytokine release from RAW264.7 cells
RAW264.7 cells were washed three times with 0·5 ml PBS before use. Then, ssODN, one of three kinds of dsODN, Y-ODN, Y-ODN(CpG) or Y-ODN(CpG3) diluted in 0·5 ml Opti-MEM was added to cells. The cells were incubated for 8 hr (TNF-α) or 24 hr (IL-6), and the supernatants were collected and stored at −80° until use. To exclude the effect of contaminated lipopolysaccharide (LPS) on cytokine release, polymyxin B, an inhibitor of LPS, was added to samples at a final concentration of 50 μg/ml. The levels of TNF-α and IL-6 in supernatants were determined by enzyme-linked immunosorbent assay using OptEIA™ sets (Pharmingen, San Diego, CA).
Stability of Y-ODN and dsODN in serum
The Y-ODN and dsODN (10 μg/100 μl) were incubated with 50% non-heat inactivated FBS at 37°. After 0, 2, 4, 8, or 24 hr of incubation, the reaction was terminated by adding 2 μl 0·5 m EDTA solution per 10 μl of samples. ODNs were extracted with phenol/chloroform/isoamyl alcohol, and the extracts were run on a 21% polyacrylamide gel and stained with ethidium bromide. Before the extraction, a fixed amount of dsODN of 45 bp was added to each sample, and the intensity of the band on gels was used to validate the efficiency of the extraction step. The amount of remaining ODNs was estimated by a Cool Saver (ATTO, Tokyo, Japan).
Uptake of ODNs in RAW264.7 cells
Fluorescein-labelled Y0a ODN was used for the preparation of fluorescein-labelled (F-) ssODN, dsODN and Y-ODN. RAW264.7 cells were plated on 96-well culture plates at a density of 5 × 105 cells/ml and cultured for 24 hr. Cells were washed three times with 100 μl PBS, incubated with F-ODN for 1 hr at 37° or 4°, harvested, and washed three times with 100 μl PBS. Then, the intensity of fluorescence of the cells was analysed by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA) using CellQuest software (version 3.1; BD Biosciences).
Growth inhibition of B16-BL6/Luc cells
RAW264.7 cells (5 × 105 cells/ml) were mixed with each ODN (10 or 20 μg/ml) and incubated for 8 hr. Then, the conditioned medium was added to B16-BL6/Luc cells, and the cells were cultured for 48 hr. The number of B16-BL6/Luc cells was determined by measuring the luciferase activity of cell lysates in a luminometer (Lumat LB 9507, EG & G Berthold, Bad Wildbad, Germany) as previously reported.23
Statistical analysis
Differences were statistically evaluated by one-way analysis of variance (anova) followed by the Fisher's protected least significant difference (PLSD) test for multiple comparisons. A P-value of < 0·05 was considered to be statistically significant.
Results
Physicochemical properties of Y-ODN
Equimolar amounts of three ODNs (Y0a, Y0b and Y0c) were hybridized to obtain Y-ODN, the putative structure of which is shown in Fig. 1(a). Figure 1(b) shows the gel electrophoresis of the DNA preparations. As reported in a previous study,20 Y-ODN showed a single band, the mobility of which was less than that of ssODN or dsODN, supporting the assembly of all three ODNs to form Y-ODN. Similarly, newly designed Y-ODN containing a potent CpG motif, Y-ODN(CpG), was prepared with Y0a(CpG), Y0b(CpG) (identical to Y0b) and Y0c(CpG). The Y-ODN containing three potent CpG motifs, Y-ODN(CpG3), was also prepared with Y0a(CpG3), Y0b(CpG3) and Y0c(CpG3). Polyacrylamide gel electrophoresis of the newly designed Y-ODN(CpG) and Y-ODN(CpG3) showed one major band with a mobility similar to that of Y-ODN (Fig. 1b, lanes 5 and 6, respectively), suggesting a Y-shape formation of these ODNs containing CpG motifs. The apparent sizes were estimated to be 7·02 ± 0·22, 7·07 ± 0·40 and 7·09 ± 0·24 nm for Y-ODN, Y-ODN(CpG) and Y-ODN(CpG3), respectively. Similarly, the sizes of ss-(Y0a) and dsODN were estimated to be 3·61 ± 0·68 and 6·86 ± 1·34 nm, respectively.
Figure 1.
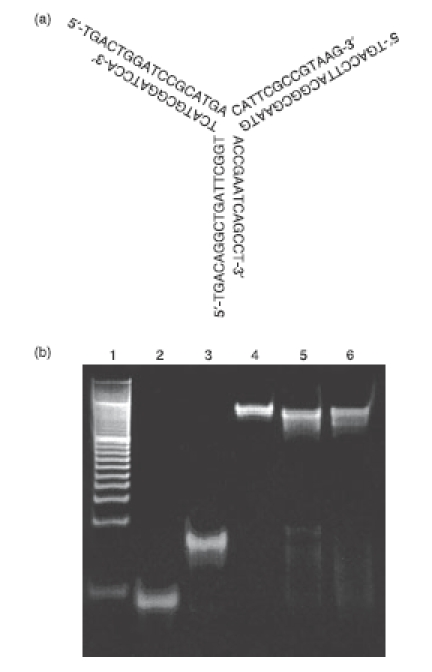
Formation of Y-shaped oligodeoxynucleotide (Y-ODN). (a) Putative structure of Y-ODN, which was prepared with three ODNs (Y0a, Y0b, Y0c) with the halves of each ODN being partially complementary to a half of the other two ODNs. (b) Polacrylamide gel electrophoresis analysis of single-stranded (ss), double-stranded (ds) and Y-ODN. Each ODN was run on 21% polyacrylamide gel at 200 V for 2 hr. Lane 1, 20-bp DNA ladder (Takara Bio); lane 2, ssODN; lane 3, dsODN; lane 4, Y-ODN; lane 5, Y-ODN(CpG); lane 6, Y-ODN(CpG3).
Cytokine release from RAW264.7 cells by ODNs
Y0a, one of the three ODNs consisting of the Y-ODN, was used as ssODN. A complementary ODN (r.Y0a) to Y0a was hybridized to obtain dsODN with four base overhangs at both 5′ ends. Similarly, three kinds of dsODN were prepared by designing complementary ODNs to Y0b and Y0c. The mixture of these three kinds of dsODNs (dsODN × 3) contained exactly the same bases in the same structural configuration of four base overhangs at both 5′ ends as Y-ODN, representing a good control to evaluate the effects of the Y-shape formation on the immunostimulatory activity of ODNs. Figure 2(a) shows the TNF-α concentration in the culture media of RAW264.7 cells. The addition of ssODN, dsODN or dsODN × 3 to RAW264.7 cells induced only weak secretion of TNF-α at concentrations of 2 and 6 μg/ml. Increasing the concentration of these ODNs to 18 μg/ml slightly increased TNF-α secretion to levels of up to 400 pg/ml. In marked contrast, large amounts of TNF-α were secreted from RAW264.7 cells after the addition of Y-ODN and the amounts varied in a concentration-dependent manner. Figure 2(b) shows the IL-6 concentration in the culture media. Again, Y-ODN induced significantly greater amounts of IL-6 secretion from cells than the other ODNs at all concentrations examined. These results indicate that Y-ODN has a stronger immunostimulatory activity than conventional ssODN or dsODN, even though there are no potent immunostimulatory CpG motifs in the sequence.
Figure 2.
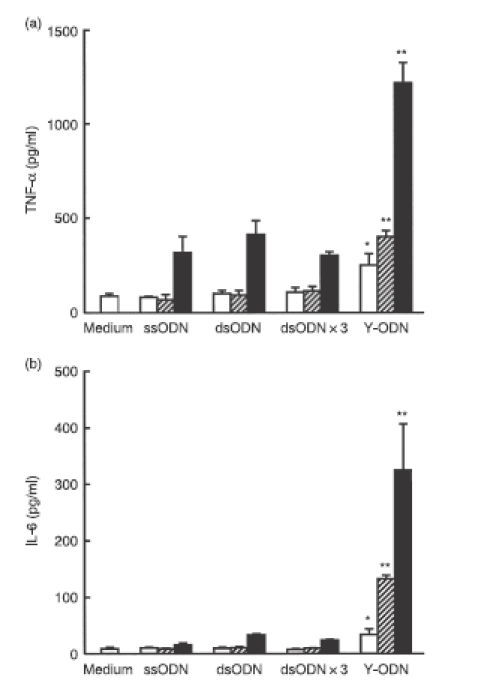
Secretion of cytokines from RAW264.7 cells after addition of oligodeoxynucleotide (ODN). Concentrations of (a) tumour necrosis factor-α (TNF-α) and (b) interleukin-6 (IL-6) in culture media were measured at 8 hr (TNF-α) or 24 hr (IL-6) after addition of each ODN to RAW264.7 cells at varying concentrations: (open bars) 2 μg/ml; (hatched bars) 6 μg/ml; (closed bars) 18 μg/ml. Results are expressed as the mean ± SD of three determinations. The experiment shown was a representative of three experiments with similar results. *P < 0·05, **P < 0·01, significantly different from single-stranded ODN (ss-ODN), double-stranded ODN (ds-ODN) and dsODN × 3 at the same concentration.
Stability of Y-ODN
Because the stability of ODN would affect their biological activity, the stability of dsODN and Y-ODN in 50% non-heat inactivated FBS solution was examined. Each ODN extracted was subjected to a PAGE analysis (Fig. 3a). The bands for 45-bp ODN, which was added to the mixtures just before extraction of ODNs, confirmed that the extraction efficiency of ODNs was almost identical in all samples. Both dsODN and Y-ODN were degraded with time in the FBS solution. A densitometric analysis of gel bands was performed in four identical experiments and the remaining amounts of dsODN and Y-ODN were plotted against the incubation time (Fig. 3b). Y-ODN showed a similar profile of degradation to dsODN, at least for the first 4 hr. Thereafter, it tended to be degraded more quickly than dsODN. Similar results were obtained when these ODNs were added to a solution containing DNase I (data not shown). These findings indicate that Y-ODN is degraded at a similar or slightly faster rate than dsODN under the conditions examined.
Figure 3.
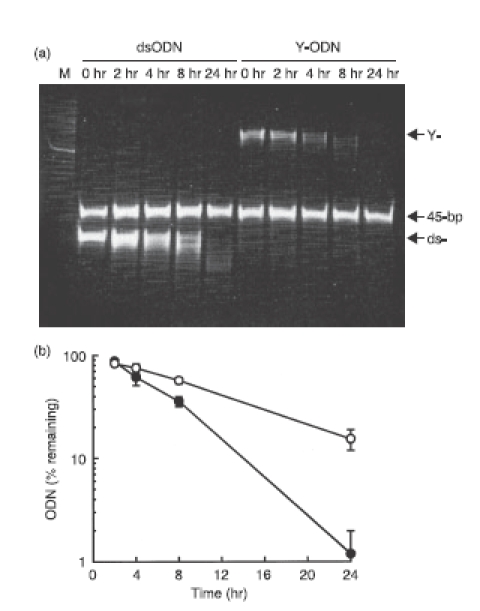
Stability of double-stranded oligodeoxynucleotide (dsODN) and Y-shape ODN (Y-ODN) in 50% non-heat inactivated fetal bovine serum (FBS). (a) The dsODN and Y-ODN were incubated in 50% non-heat inactivated FBS at 37° for the indicated times and the reaction was terminated by adding ethylenediaminetetraacetic acid. ODNs extracted were run on 21% polyacrylamide gel at 200 V for 2 hr and stained with ethidium bromide. ‘M’ represents 20-bp DNA ladder (Takara Bio). (b) The amounts of ODNs on the gel were estimated by Cool Saver. The remaining amounts of dsODN (○) and Y-ODN (•) were plotted against the incubation time. Results are expressed as the mean ± SD of four determinations.
Cellular uptake of ODNs in RAW264.7 cells
To investigate whether the enhanced immunostimulatory activity of Y-ODN is mediated by an increased cellular uptake, the uptake of F-dsODN, F-dsODN × 3 and F-Y-ODN was examined in RAW264.7 cells. The uptake of these F-ODNs was greater at 37° than at 4° (data not shown) in all cases examined. A fluorescence-activated cell sorting (FACS) analysis was performed in three identical experiments and the mean fluorescence intensity (MFI) was measured at each concentration of ODN. MFI (37°–4°) was plotted against the concentration of ODN (Fig. 4). The MFI of cells treated with F-Y-ODN was significantly (P < 0·05) greater than the MFI of those treated with other F-ODN preparations, suggesting that an increased cellular uptake of Y-ODN contributes to the enhanced immunostimulatory activity of Y-ODN.
Figure 4.
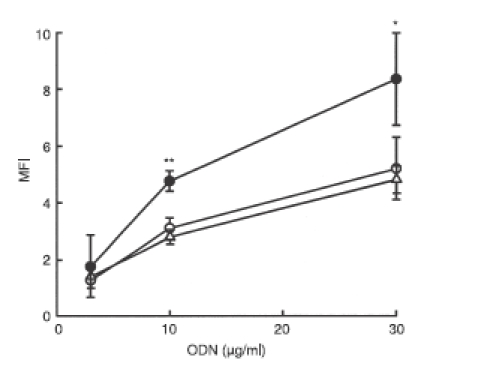
Uptake of fluorescein-labelled oligodeoxynucleotide (ODN) in RAW264.7 cells. Cells were incubated with fluorescein-labelled double-stranded ODN (dsODN) (○), dsODN × 3 (△) or Y-ODN (•) for 1 hr at 4° or 37°, and the amounts of ODN associated with cells were measured by flow cytometry. The mean fluorescence intensity (MFI) was plotted against the concentration of ODN. Results are expressed as the mean ± SD of three determinations. *P < 0·05, **P < 0·01, significantly different from dsODN and dsODN × 3.
Cytokine release from RAW264.7 cells by CpG ODNs
Activation of RAW264.7 cells by the newly designed Y-ODN(CpG) and Y-ODN(CpG3) was examined under the same conditions as above. Y0a(CpG3), an ODN containing a potent CpG motif, was selected as ssODN with a CpG motif, ssODN(CpG3). Then, dsODN(CpG3) and dsODN(CpG3) × 3 were prepared as described above. Again, dsODN(CpG3) × 3 had the same sequence as Y-ODN(CpG3). Contrary to ODNs with no potent immunostimulatory CpG motifs, any preparation of ODNs containing CpG motifs showed a marked secretion of TNF-α in a concentration-dependent manner (Fig. 5a). Approximately six-fold higher amounts of TNF-α were released from RAW264.7 cells treated with Y-ODN(CpG3) compared with those treated with dsODN(CpG3) × 3. The IL-6 concentration in the culture media was also measured, and similar results were obtained (Fig. 5b). All ODN preparations contained trace amounts of LPS, up to 2·5 EU/mg DNA, when measured by a Limulus test (Wako, Tokyo, Japan). To exclude the effects of contaminated LPS on cytokine release, polymyxin B was added to ssODN and Y-ODN(CpG3). The addition of polymyxin B to Y-ODN(CpG3) slightly reduced the level of TNF-α released from RAW264.7 cells (Table 2). However, the level was much greater than those obtained with the medium + LPS/polymyxin B or ssODN + LPS/polymyxin B, suggesting that contaminated LPS has little effect on the ODN-mediated cytokine release from RAW264.7 cells. These results suggest that the immunostimulatory activity of the CpG motif-containing ODNs can be significantly increased by the Y-shape formation. In addition, increasing the number of potent CpG motifs in Y-ODN is a useful approach to increasing the immunostimulatory activity of Y-ODN.
Figure 5.
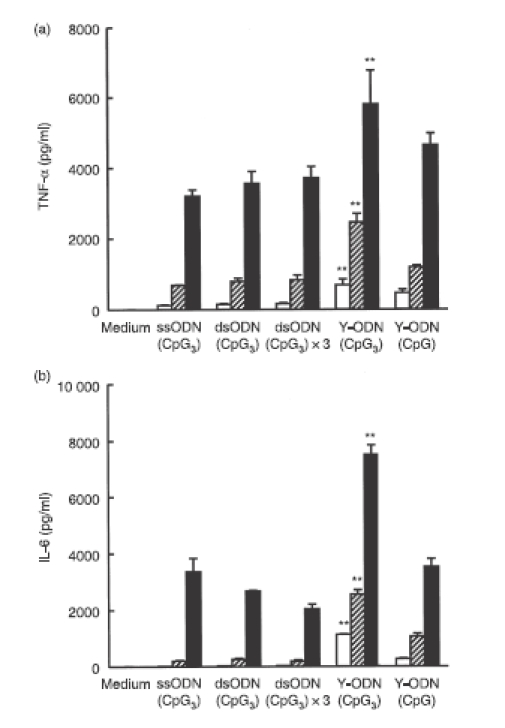
Secretion of cytokines from RAW264.7 cells after addition of CpG oligodeoxynucleotides (ODNs). Concentrations of (a) tumour necrosis factor-α (TNF-α) and (b) interleukin-6 (IL-6) in culture media were measured at 8 hr (TNF-α) or 24 hr (IL-6) after addition of each ODN to RAW264.7 cells at various concentrations: 2 μg/ml (open bars); 6 μg/ml (hatched bars); 18 μg/ml (closed bars). Results are expressed as the mean ± SD of three determinations. The experiment shown was representative of three experiments with similar results. **P < 0·01, significantly different from double-stranded ODN(CpG3) 3 at the same concentration.
Table 2.
Effect of polymyxin B on tumour ncrosis factor-α (TNF-α) release from RAW264.7 cells after addition of oligodeoxynucleotides (ODNs) with or without lipopolysaccharide (LPS)
| Compound | TNF-α (pg/ml) | |
|---|---|---|
| Control | + polymyxin B | |
| Medium | 123 ± 11 | 394 ± 21 |
| Medium + LPS | 28 900 ± 700 | 448 ± 16 |
| ssODN | 278 ± 30 | 495 ± 23 |
| ssODN + LPS | 30 700 ± 300 | 557 ± 16 |
| Y-ODN(CpG3) | 31 200 ± 700 | 27 400 ± 700 |
| Y-ODN(CpG3) + LPS | 34 900 ± 1000 | 27 700 ± 1200 |
Polymyxin B, an inhibitor of LPS, was added to medium (Opti-MEM), single-stranded (ss) ODN or Y-ODN(CpG3) at a final concentration of 50 μg/ml. LPS was added to samples at a final concentration of 1 ng/ml. Each sample was added to cells, and the TNF-α concentration in supernatants was measured after an 8-hr incubation. Results are expressed as the mean ± SD of three determinations.
Effect of conditioned medium of ODN-treated RAW264.7 cells on proliferation of B16-BL6/Luc cells
To examine whether Y-ODN(CpG3) is effective in inhibiting the proliferation of tumour cells, the conditioned medium of ODN-treated RAW264.7 cells was added to B16-BL6/Luc cells. The conditioned medium of Y-ODN(CpG3)-treated RAW264.7 cells significantly inhibited the proliferation of tumour cells compared with the media of other ODNs (Fig. 6).
Figure 6.
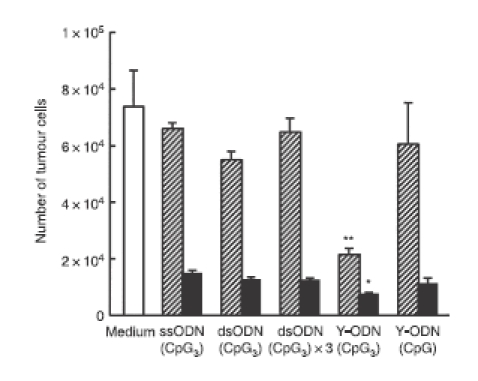
Growth inhibition of tumour cells by conditioned medium of RAW264.7 cells treated with oligodeoxynucleotides (ODN). Conditioned medium of ODN-treated RAW264.7 cells was added to B16-BL6/Luc cells, and the cells were cultured for additional 48 hr. Then, the number of cells was determined by measuring the luciferase activity of cell lysates. Results are expressed as the mean ± SD of three determinations. *P < 0·05, **P < 0·01, significantly different from other conditions.
Discussion
CpG DNA activates immune cells expressing TLR91,2 and induces the cells to release a broad repertoire of chemokines and cytokines, including IL-6, IL-12, IFN-α/β, and TNF-α,5,6 when it is sorted to endosomes after internalization via endocytosis.24 CpG dinucleotide flanked by two purine bases on the 5′ side and two pyrimidine bases on the 3′ side, such as GACGTT, efficiently activates the murine immune system, whereas the optimal motif for humans is GTCGTT. Although the optimal sequences of CpG ODN for activating mouse or human immune cells have been identified by examining many possible base combinations, little is known about the structural requirements of CpG ODN for the recognition of TLR9 and following the activation of immune cells. Phosphodiester ODN is rapidly degraded by nucleases in body fluids, such as serum, and intracellular compartments, which greatly limits the use of such ODNs as immunostimulatory agents. However, a recent study has shown that polyguanosine at the 3′ end of ODNs can increase the immunostimulatory activity of phosphodiester ODN.12 Furthermore, it has been reported that the immunostimulatory effect was enhanced by using self-stabilized CpG DNA.25–27 In the present study, we prepared Y-shaped ODNs, with or without immunostimulatory CpG motifs, and found that the Y-shape formation increased the immunostimulatory responses of ODNs in RAW264.7 macrophage-line cells, even though the stability was not increased by the Y-shape formation.
Y-shaped ODN (Y-ODN), the element of dendrimer-like preparations of ODN, was prepared using three 30-base ODNs. As reported in a previous paper,20 Y-ODN showed a single band on polyacrylamide gels, the mobility of which was less than that of the bands for ssODN or dsODN (Fig. 1b). The addition of Y-ODN to RAW264.7 cells resulted in a significant secretion of both TNF-α and IL-6 in a DNA concentration-dependent manner, even though it had no potent immunostimulatory CpG motifs. DNA sequences other than potent immunostimulatory CpG motifs may trigger weak, but detectable, cytokine responses, and these could be involved in the immunostimulatory responses to ODNs without any potent CpG motifs. Although all ODN preparations contained trace amounts of LPS, up to 2·5 EU/mg DNA, this level of LPS induced only a little TNF-α secretion from RAW264.7 cells (data not shown). Furthermore, the addition of polymyxin B, an inhibitor of LPS, had little effect on the release of TNF-α from the Y-ODN(CpG3)-treated RAW264.7 cells (Table 2). These results strongly support the idea that DNA is responsible for the cytokine response of cells to all ODN preparations under these experimental conditions, even though no potent CpG motifs were included in the sequences.
The RAW264.7 cells and other TLR9-positive cells secrete inflammatory cytokines on recognition of CpG DNA. TLR9 localizes in the intracellular compartments, such as the endoplasmic reticulum, and translocates to the lysosomal compartment when CpG DNA is taken up by the cells.28 Therefore, CpG DNA should be transferred to such subcellular compartments to induce cytokine production. The level of cytokine release from TLR9-positive cells would be a function of variables, including the stability of DNA and the amount of DNA taken up by cells. Therefore, we examined whether the Y-shape formation of DNA increased the stability to nucleases and/or the cellular uptake. In the present study, we found that Y-ODN was less stable than dsODN. Y-ODNs have three terminals in one unit of the structure, whereas conventional dsODNs have two. This difference may explain the high susceptibility of Y-ODN to nuclease-mediated degradation.
In marked contrast, the Y-shape formation significantly increased the uptake of ODNs in RAW264.7 cells (Fig. 3b). A previous study demonstrated that increasing the length of ODN increases its endocytic uptake when ODNs with a length of 250 bp or less are used.29 Because one unit of dsODN and Y-ODN contains 60 and 90 bases, respectively, this difference may be responsible for the greater uptake of Y-ODN compared with that of dsODN. However, the high immunostimulatory activity of Y-ODN could not be simply attributed to the increased uptake of Y-ODN because the increase in the level of cytokine release (three- to six-fold) by the Y-shape formation was greater than the increase in the uptake (about two-fold). Therefore, other factors, such as the affinity of ODN for TLR9 and intracellular localization, may also contribute to the increased immunostimulatory activity of Y-ODN.
To construct a new Y-ODN preparation with a potent immunostimulatory CpG motif, the GACGTT sequence, the most potent one in rodents, was inserted close to the 5′-terminal of Y0a, one of the components of Y-ODN, based on the information that TLR9 reads the CpG from the 5′-end of DNA.30 Although the ssODN and dsODN with the CpG motifs were effective in inducing cytokines, such as TNF-α and IL-6, when added to RAW264.7 cells, the Y-ODN containing CpG motifs induced greater amounts of cytokines than these conventional CpG ODN preparations. Because all ODN preparations, i.e. ssODN(CpG3), dsODN(CpG3), dsODN(CpG3) × 3 and Y-ODN(CpG3), have identical numbers of potent CpG motifs, the Y-shape formation of ODN significantly increases the efficiency of cytokine production by CpG DNA. Furthermore, Y-ODN(CpG3), which contained three potent CpG motifs in one unit, was much more effective in inducing TNF-α and IL-6 compared with Y-ODN(CpG) containing only one potent CpG motif. In accordance with the levels of cytokines, the conditioned medium of Y-ODN(CpG3)-treated RAW264.7 cells showed a greater inhibitory effect on the growth of melanoma cells than other ODNs. Previous studies have demonstrated that CpG ODNs are a very strong activator of TLR9-positive cells, such as macrophages, B cells and dendritic cells, and have been considered as therapeutic agents against cancer, and infectious and allergic diseases.1,31 As shown in the present study, cytokines possessing anti-proliferative activity of tumour cells, such as TNF-α, are induced by CpG ODNs. Therefore, cytokines released from CpG ODN-treated RAW264.7 cells would contribute to the growth inhibition of melanoma cells. These results indicate that highly potent immunostimulatory ODNs can be designed by increasing the number of CpG motifs in the sequences of Y-ODN.
The involvement of TLR9 in CpG DNA-mediated immune activation has been reported using TLR9−/− mice or cells isolated from those mice. Our preliminary experiments using TLR9−/− and wild-type mice suggested that Y-ODN(CpG3) induces TNF-α production in a TLR9-dependent manner, although the level obtained was lower than those obtained with DNA–cationic liposome complexes. The involvement of TLR9 in the immune activation by Y-ODNs was therefore evident, but it should be further investigated in future experiments.
In conclusion, the Y-shape formation of ODN has been shown to be effective in inducing greater amounts of cytokines, such as TNF-α and IL-6, in macrophage-like, TLR9-positive cells than conventional ssODN or dsODN. These enhanced immunostimulatory effects of Y-ODN are, at least partly, associated with an increase in the uptake by TLR9-positive cells, but not with stabilization of ODN. CpG DNA has been explored as a therapeutic agent for cancer, asthma, allergy, and infectious diseases and as an adjuvant in immunotherapy, but it generally requires phosphorothioate or other chemical modification. Such modification may have disadvantages associated with systemic toxicity, such as a transient anticoagulant effect, activation of complement cascade, and inhibition of basic fibroblast growth factor binding to surface receptors, because of non-specific protein binding.32 The findings of the present study provide a novel strategy for the development of potent immunostimulatory CpG ODN preparations free from such modification problems.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and by a grant from the Ministry of Health, Labour and Welfare, Japan.
References
- 1.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–60. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 2.Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 3.Tokunaga T, Yamamoto H, Shimada S, et al. Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG. I. Isolation, physicochemical characterization, and antitumor activity. J Natl Cancer Inst. 1984;72:955–62. [PubMed] [Google Scholar]
- 4.Hartmann G, Weiner GJ, Krieg AM. CpG DNA: a potent signal for growth, activation, and maturation of human dendritic cells. Proc Natl Acad Sci U S A. 1999;96:9305–10. doi: 10.1073/pnas.96.16.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klinman DM, Yi AK, Beaucage SL, Conover J, Krieg AM. CpG motifs present in bacterial DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon γ. Proc Natl Acad Sci U S A. 1996;93:2879–83. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparwasser T, Miethke T, Lipford G, Erdmann A, Hacker H, Heeg K, Wagner H. Macrophages sense pathogens via DNA motifs: induction of tumor necrosis factor-α-mediated shock. Eur J Immunol. 1997;27:1671–9. doi: 10.1002/eji.1830270712. [DOI] [PubMed] [Google Scholar]
- 7.Sato Y, Roman M, Tighe H, et al. Immunostimulatory DNA sequences necessary for effective intradermal gene immunization. Science. 1996;273:352–4. doi: 10.1126/science.273.5273.352. [DOI] [PubMed] [Google Scholar]
- 8.Dalpke A, Zimmermann S, Heeg K. Immunopharmacology of CpG DNA. Biol Chem. 2002;383:1491–500. doi: 10.1515/BC.2002.171. [DOI] [PubMed] [Google Scholar]
- 9.Kandimalla ER, Yu D, Agrawal S. Towards optimal design of second-generation immunomodulatory oligonucleotides. Curr Opin Mol Ther. 2002;4:122–9. [PubMed] [Google Scholar]
- 10.Yi AK, Chang M, Peckham DW, Krieg AM, Ashman RF. CpG oligodeoxyribonucleotides rescue mature spleen B cells from spontaneous apoptosis and promote cell cycle entry. J Immunol. 1998;160:5898–906. [PubMed] [Google Scholar]
- 11.Rankin R, Pontarollo R, Ioannou X, Krieg AM, Hecker R, Babiuk LA, van Drunen Little-van den Hurk S. CpG motif identification for veterinary and laboratory species demonstrates that sequence recognition is highly conserved. Antisense Nucleic Acid Drug Dev. 2001;11:333–40. doi: 10.1089/108729001753231713. [DOI] [PubMed] [Google Scholar]
- 12.Dalpke AH, Zimmermann S, Albrecht I, Heeg K. Phosphodiester CpG oligonucleotides as adjuvants: polyguanosine runs enhance cellular uptake and improve immunostimulative activity of phosphodiester CpG oligonucleotides in vitro and in vivo. Immunology. 2002;106:102–12. doi: 10.1046/j.1365-2567.2002.01410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CC, Lee J, Raz E, Corr M, Carson DA. Necessity of oligonucleotide aggregation for Toll-like receptor 9 activation. J Biol Chem. 2004;279:33071–8. doi: 10.1074/jbc.M311662200. [DOI] [PubMed] [Google Scholar]
- 14.Adleman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–4. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto K, Gouzu H, Komiya K, Kiga D, Yokoyama S, Yokomori T, Hagiya M. Molecular computation by DNA hairpin formation. Science. 2000;288:1223–6. doi: 10.1126/science.288.5469.1223. [DOI] [PubMed] [Google Scholar]
- 16.Yan H, Zhang X, Shen Z, Seeman NC. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002;415:62–5. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 17.Watson KJ, Park SJ, Im JH, Nguyen ST, Mirkin CA. DNA–block copolymer conjugates. J Am Chem Soc. 2001;123:5592–3. doi: 10.1021/ja0156845. [DOI] [PubMed] [Google Scholar]
- 18.Condon A. Designed DNA molecules: principles and applications of molecular nanotechnology. Nat Rev Genet. 2006;7:565–75. doi: 10.1038/nrg1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Tseng YD, Kwon SY, D’Espaux L, Bunch JS, Mceuen PL, Luo D. Controlled assembly of dendrimer-like DNA. Nat Mater. 2004;3:38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Cu YT, Luo D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat Biotechnol. 2005;23:885–9. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poste G, Doll J, Hart IR, Fidler IR. In vitro selection of murine B16 melanoma variants with enhanced tissue-invasive properties. Cancer Res. 1980;40:1636–44. [PubMed] [Google Scholar]
- 23.Hyoudou K, Nishikawa M, Umeyama Y, Kobayashi Y, Yamashita F, Hashida M. Inhibition of metastatic tumor growth in mouse lung by repeated administration of polyethylene glycol-conjugated catalase: quantitative analysis with firefly luciferase-expression melanoma cells. Clin Cancer Res. 2004;10:7685–91. doi: 10.1158/1078-0432.CCR-04-1020. [DOI] [PubMed] [Google Scholar]
- 24.Manzel L, Macfarlane DE. Lack of immune stimulation by immobilized CpG-oligonucleotide. Antisense Nucleic Acid Drug Dev. 1999;9:459–64. doi: 10.1089/oli.1.1999.9.459. [DOI] [PubMed] [Google Scholar]
- 25.Kandimalla ER, Bhagat L, Cong YP, Pandey RK, Yu D, Zhao Q, Agrawal Q. Secondary structures in CpG oligonucleotides affect immunostimulatory activity. Biochem Biophys Res Commun. 2003;306:948–53. doi: 10.1016/s0006-291x(03)01080-5. [DOI] [PubMed] [Google Scholar]
- 26.Cong YP, Song SS, Bhagat L, Pandey RK, Yu D, Kandimalla ER, Agrawal S. Self-stabilized CpG DNAs optimally activate human B cells and plasmacytoid dendritic cells. Biochem Biophys Res Commun. 2003;310:1133–9. doi: 10.1016/j.bbrc.2003.09.134. [DOI] [PubMed] [Google Scholar]
- 27.Shimosato T, Kimura T, Tohno M, et al. Strong immunostimulatory activity of AT-oligodeoxynucleotide requires a six-base loop with a self-stabilized 5′-C…G-3′ stem structure. Cell Microbiol. 2006;8:485–95. doi: 10.1111/j.1462-5822.2005.00640.x. [DOI] [PubMed] [Google Scholar]
- 28.Latz E, Schoenemeyer A, Visintin A, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–8. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 29.Roberts TL, Dunn JA, Terry TD, Jennings MP, Hume DA, Sweet MJ, Stacey KJ. Differences in macrophage activation by bacterial DNA and CpG-containing oligonucleotides. J Immunol. 2005;175:3569–76. doi: 10.4049/jimmunol.175.6.3569. [DOI] [PubMed] [Google Scholar]
- 30.Kandimalla ER, Bhagat L, Yu D, Cong Y, Tang J, Agrawal S. Conjugation of ligands at the 5′-end of CpG DNA affects immunostimulatory activity. Bioconjug Chem. 2002;13:966–74. doi: 10.1021/bc0200374. [DOI] [PubMed] [Google Scholar]
- 31.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–84. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 32.Henry SP, Giclas PC, Leeds J, Pangburn M, Auletta C, Levin AA, Kornbrust DJ. Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action. J Pharmacol Exp Ther. 1997;281:810–6. [PubMed] [Google Scholar]


