Abstract
The hepatitis delta virus (HDV) ribozyme is an RNA motif embedded in human pathogenic HDV RNA. Previous experimental studies have established that the active-site nucleotide C75 is essential for self-cleavage of the ribozyme, although its exact catalytic role in the process remains debated. Structural data from X-ray crystallography generally indicate that C75 acts as the general base that initiates catalysis by deprotonating the 2′-OH nucleophile at the cleavage site, while a hydrated magnesium ion likely protonates the 5′-oxygen leaving group. In contrast, some mechanistic studies support the role of C75 acting as general acid and thus being protonated before the reaction. We report combined quantum chemical/molecular mechanical calculations for the C75 general base pathway, utilizing the available structural data for the wild type HDV genomic ribozyme as a starting point. Several starting configurations differing in magnesium ion placement were considered and both one-dimensional and two-dimensional potential energy surface scans were used to explore plausible reaction paths. Our calculations show that C75 is readily capable of acting as the general base, in concert with the hydrated magnesium ion as the general acid. We identify a most likely position for the magnesium ion, which also suggests it acts as a Lewis acid. The calculated energy barrier of the proposed mechanism, ~20 kcal/mol, would lower the reaction barrier by ~15 kcal/mol compared to the uncatalyzed reaction and is in good agreement with experimental data.
Keywords: catalysis, RNA, MPW1K, CCSD(T), ONIOM
Introduction
The ribozyme embedded in the genomic and antigenomic RNAs of hepatitis delta virus (HDV) (Figure 1) is a representative of a group of naturally occurring, small, non-protein coding RNAs that catalyze site-specific self-cleavage of their own backbones (Figure 2).1–3 The HDV ribozyme is unusual in that it is the only known ribozyme from a human pathogen, boding well for potential applications in human gene therapy. It has also recently been found to reside in the intron of the human CPEB3 gene, giving rise to the speculation that HDV may have evolved from the human transcriptome.4 Moreover, the HDV ribozyme was the first catalytic RNA motif for which structural and biochemical data suggested participation of a specific side chain, cytosine 75 (C75), in reaction chemistry.1–3 Two main models were proposed. In the first model C75 acts as the general base that deprotonates the cleavage site 2′-OH, activating it as a nucleophile that attacks the adjacent 3′,5′-phosphodiester backbone (Figure 2A).5 According to the second model, C75 instead protonates the 5′-oxygen leaving group, thereby generating the 2′,3′-cyclic and 5′-OH termini of the reaction products (Figure 2B).6 In these models a hydrated Mg2+ ion is proposed to provide the complementary general acid and base functionality, respectively. The notion that RNA side chains may be chemical participants in RNA-catalyzed reactions has inspired several similar proposals for other ribozymes,7–10 even though not all such proposals could be substantiated.11 Clearly, studies of HDV ribozyme catalysis have considerably heightened our appreciation of the chemical capabilities of RNA, but much remains to be learnt about the catalytic processes involved.
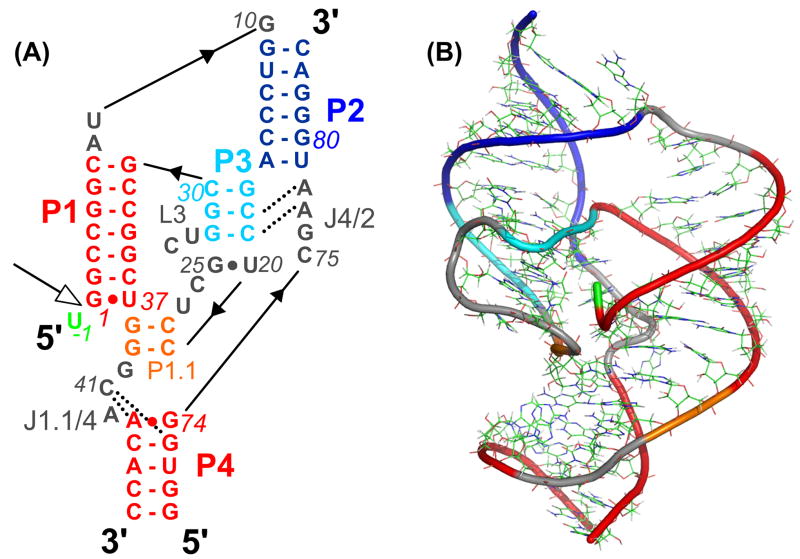
Figure 1.
The structure of the precursor form of HDV ribozyme: A) Sequence and secondary structure of the HDV ribozyme genomic form. The scissile phosphate between U-1 and G1 is highlighted by an open arrow (panel taken from ref. 13). B) Three-dimensional structures of the HDV ribozyme. The colors of the structural elements correspond to those in A.
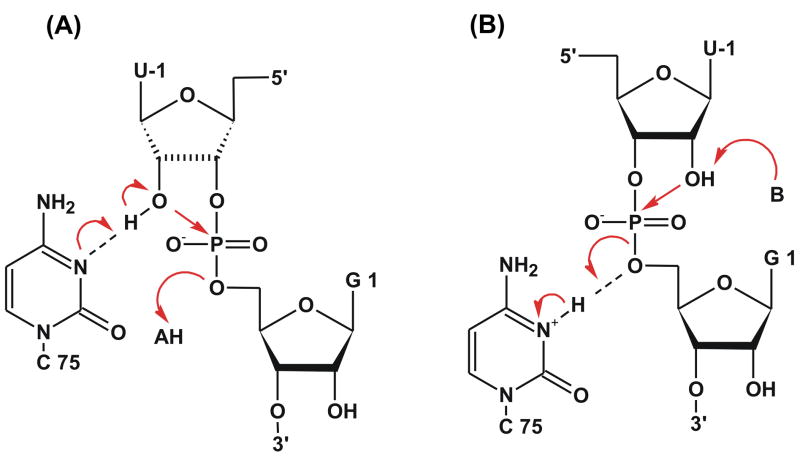
Figure 2.
Two proposed general acid-base reaction mechanisms for HDV ribozyme: A) C75 general base mechanism - the U-1(O′2) nucleophile is deprotonated by nucleobase C75 acting as a general base. The leaving G1(O′5) alcoholate group is protonated by a general acid (e.g., a water molecule coordinated to the Mg2+ ion), B) General acid mechanism - the leaving G1(O′5) alcoholate group is protonated by nucleobase C75 acting as a general acid and the (U-1)O′2 nucleophile is deprotonated by a general base.
Despite intense investigation, the self-cleavage mechanism of the HDV ribozyme is not fully understood. Been and coworkers suggested that C75 acts as the general base in the reaction,5 based on evidences that the rate of the reaction catalyzed by the wild-type C75, which has an apparent pKa of ~6.1, increases with pH; the activity diminishes following shifts in apparent pKa associated with C75A substitution; and activity is completely lost upon C75U substitution, however, it can be partially rescued by the presence of external imidazole. This view was subsequently supported by X-ray crystallographic studies of reaction precursors of the cis-acting genomic ribozyme,12 and by extensive molecular dynamics (MD) simulations based on these structures,13,14 because an active site architecture consistent with the general base mechanism formed spontaneously in the simulations. In contrast, no stable structural arrangement with protonated C75H+ supporting the general acid mechanism has been obtained as yet, despite extensive MD simulations.13,14 However, X-ray structure of C75 wild type was obtained with low resolution while considerably better X-ray structures were obtained only for the sequence with an inactive C75U substitution. The molecular dynamic simulations are obviously influenced by the starting X-ray geometries.
By contrast, Bevilacqua and coworkers observed an inverted rate-pH profile for genomic form in high monovalent salt concentrations, in the absence of Mg2+, prompting the proposal that C75 instead acts as general acid.6 Subsequent studies by both Been and Bevilacqua suggested that rather than C75 a critical base quartet adjacent to the catalytic core involving a protonated C41 gives rise or at least contributes to the inverted rate-pH profile in the absence of structurally stabilizing Mg2+.15–17 The product crystal structure lacking the 5′-leader sequence shows a hydrogen bond between C75 and the 5′-OH terminus generated during the reaction.18 This hydrogen bond is frequently interpreted as a plausible remnant of proton transfer from a C75H+ general acid to the 5′-oxygen leaving group (e.g. refs. 6,15). Interestingly, the general base mechanism was proposed by Doudna et al. in their original study presenting the product crystal structure.18 The most explicit support for C75 acting as the general acid has come from a recent site-specific chemical modification study of a trans-acting form of the HDV antigenomic ribozyme with external substrate. This work showed that C75 played a limited role in catalysis when sulfur was employed as the 5′-leaving group, quite unambiguously supporting the C75 general acid hypothesis.19 However, while such trans-acting ribozymes are structural and mechanistic analogs of the naturally occurring cis-acting forms, they exhibit an amplified conformational change upon cleavage20–22 relative to that observed in the cis-acting ribozyme.12,23 In addition, they are about an order of magnitude catalytically slower due to the long-range impact of the strand scission(s) required to sever the substrate from the ribozyme. Therefore, there is a possibility that the use of an external substrate with site-specific modifications may alter the balance between general base and general acid reaction pathways, if these are competing.24
Both the C75 general base and general acid mechanisms require the local pKa of C75 to be significantly shifted. This has been suggested by continuum solvent calculations utilizing the available ground-state crystal structures.25 However, the calculations did not take into account the fact that the open, highly negatively charged catalytic pocket of the HDV ribozyme is likely continuously occupied by divalent or monovalent cations.13,14,26 The ions probably neutralize the electrostatic potential of the catalytic core to a significant extent, and are known to compete with C75 protonation.6 Most examples of protonated nucleobases in structured nucleic acids show the excess proton to be involved in very specific planar hydrogen bonding interactions.27–35 Such protonated bases not only neutralize the negative electrostatic potentials but also bring clear structural gains which cannot be achieved with neutral bases and cations.27–35 Unless the excess proton is involved in such tight H-bonding interaction, or binding of ions is sterically disfavored as in case of the Hairpin ribozyme,36 protonation does not appear to be a common way to respond to negative potentials in nucleic acids. The catalytic pocket of HDV ribozyme is clearly open for extensive interactions with ions. No pKa shift of C75 was observed in equilibrium NMR experiments,37 but a recent Raman crystallography approach revealed a significant pKa shift toward neutrality that is anticooperatively coupled with Mg2+ binding.38 The unique structural context of C75 thus appears to perturb its pKa towards optimal for general acid-base catalysis at physiologic pH, but these experiments do not provide evidence that C75 acts as either general base or general acid and do not provide structural insights.
In light of the conflicting experimental evidence and the need to better understand the nature of the transition state computational approaches have been employed in several recent studies. In particular, two studies have described the reaction process by quantum mechanical (QM) methods.39,40 Gauld and coworkers compared the uncatalyzed reaction with the C75 general acid mechanism by applying high-quality density functional theory (DFT) to a small-molecule model of the active site, lacking the remainder of the ribozyme.39 However, while the uncatalyzed reaction can be considered a highly suitable reference point for the transition state energetics, both the C75 proton affinity and hydrated Mg2+ ion basicity are likely influenced by the HDV ribozyme structure outside the narrow boundaries employed in these calculations. In addition, the structure of the underlying active site did not correspond to any published conformation observed by either X-ray crystallography or MD simulation, limiting the utility of the otherwise very interesting calculations.12–14,18,26 Liu, Guo and coworkers used estimates for the effective concentrations of the reactive C75 adopting general base and acid roles using the near-attack-conformations concept and classical molecular dynamic simulations. They also used quantum chemical DFT calculations of a truncated active site model to compare the C75 general base and acid mechanisms. They concluded that the acid mechanism is energetically favored.40 However, while the reactive state corresponding to the C75 general base mechanism was significantly populated in MD simulations,13,40 making it possible to estimate that the Gibbs Energy correction that should be applied to account for its proportion was only a few kcal/mol, the C75 general acid reactive state was rare (0.01%), corresponding to only a 0.1-ps occurrence in a 1-ns MD simulation.40 Earlier, we have reported much longer 12-ns simulation with protonated C75H+13 but we have never observed formation of a suitable geometry. Such a negligible concentration combined with the limited MD sampling suggests that the reactive population and corresponding correction term could not be accurately estimated. In addition, the small size of the truncated model system used in DFT calculations again did not take into account the important structural context of the ribozyme’s active site, and the active site model employed did not fit the ribozyme fold observed in crystallographic studies. One problem of truncated active site models used by Liu et al. and Gauld et al. is that the C75H+ base is oriented differently than in the X-ray structures and MD, i.e., the direction from N1 to N4 of the base is antiparallel to the sugar-phosphate backbone direction from scissile phosphate to G1 sugar, instead of the usual parallel orientation. Therefore there is a clear need to address the mechanism using computational methods that incorporate the structural context of the HDV ribozyme.
In this study, we present results of a thorough theoretical study that utilizes a hybrid quantum mechanical/molecular mechanical (QM/MM) treatment41 to evaluate the plausibility of the C75 general base mechanism of the HDV genomic ribozyme. We describe the mechanism using combined QM/MM methodology in which the active site (QM region) containing ~80 atoms is described by a hybrid density functional in the context of the complete, AMBER force field-treated HDV ribozyme fold. We thus overcome the limitations associated with the use of small active site models. Our calculations show that the C75 general base mechanism, in concert with a hydrated magnesium ion as general and Lewis acid, is a viable catalytic strategy for the HDV ribozyme.
Methods
(a) Structure Preparation
The HDV genomic ribozyme crystal structures of C75U inactivated precursor and lower resolution C75 structures are consistent with the possibility that C75 acts as the general base, although the U-1(O2′) does not form a hydrogen (H-)bond with U75(N3) in C75U mutant structures and the U-1(O2′) atom was not identified in the wild type C75 structure. Formation of the U-1(O′2-H′2) … C75(N3) H-bond is assumed to be essential to initiate the C75 general base mechanism. However, the appropriate geometry can be readily obtained by rotation of the U-1 residue.12 Indeed, the U-1(O2′) … C75(N3) H-bond was formed spontaneously with a ~10% population in the course of multiple 10-ns scale explicit solvent MD simulations.13 Therefore, it was convenient to use a specific snapshot from the previously published molecular dynamics (MD) simulations of the HDV genomic ribozyme precursor with canonical C75 and protonated C41+ in the presence of Na+ counter ions to prepare all starting structures described below.13 The starting structure (see Supplementary Materials for coordinates) was chosen based on a geometrical arrangement achieving one of the best U-1(O′2-H′2) and C75(N3) H-bonds among those formed during MD simulation. Subsequently, it was necessary to place an Mg2+ ion in an appropriate position for the reaction. However, the crystallographic positions of Mg2+ ions are perturbed by the C75U mutation and obscured by the limited resolution.12 Furthermore, MD simulations are not able to properly sample the dynamics of divalent ions due to current force field limitations.14 Thus, we considered a range of Mg2+ ion positions in the active site, in order to reduce the uncertainty from sampling limitations. Initially, two Na+ ions present in the active site of the initial structure were replaced by two water molecules and the complete system was minimized by molecular mechanics using the AMBER force field (parm99).42,43 The positions of water oxygen atoms in the active site were chosen as potential positions for the Mg2+ ion. Subsequently, each water oxygen atom within the first three solvation spheres of the reaction center was replaced by the Mg2+ ion. This approach produced 13 diverse Mg2+ ion positions that were then subjected to QM/MM minimization (see below), covering a wide range of relaxed Mg2+ positions.
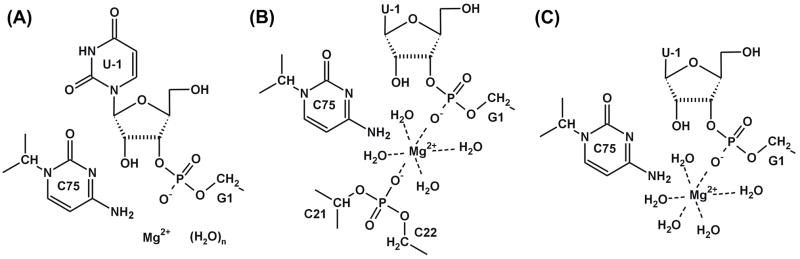
In these optimizations, the quantum (QM) region included all active site water molecules from the first and second solvation shells of the Mg2+ ion (Figure 3A), i.e., 13 to 25 water molecules for each Mg2+ ion position. All systems underwent geometric optimization and only two plausible positions of the Mg2+ ion were identified: (i) one in which the Mg2+ ion possesses a double inner-shell coordination with the C21(O2P) and scissile G1(O1P) phosphates (Figure 3B), and (ii) one in which it has a single inner-shell coordination to G1(O1P) (Figure 3C). Both of these Mg2+ positions are located in the same active site region, where metal ions have been consistently identified in crystal structures of HDV ribozyme mutants.12 Furthermore, both positions are consistent with the notion that Mg2+ simultaneously plays two catalytic roles: (i) acting as the general acid catalyst by increasing the acidity of a coordinated water molecule that forms an H-bond to G1(O′5), and (ii) exerting an electrophilic (Lewis acid) effect that activates the scissile phosphate for nucleophilic attack through direct coordination to G1(O1P). We did not consider other geometries in which the Mg2+ ion was located too far from the reaction center to participate.
Figure 3.
Schemes of three quantum regions used in QM/MM calculations. A) The large quantum region was used for a rapid pre-optimization of the HDV ribozyme active site with an Mg2+ ion in 13 different positions. The number of water molecules ranged from 13 to 25 depending on the position of the Mg2+ ion. B) The quantum region used to study the reaction energy profile for reaction coordinates including the Mg2+ ion with double inner-shell arrangements (PD and HD structures). C) The quantum region used to study the reaction energy profile for the reaction coordinate including the Mg2+ ion with single inner-shell arrangement (HS).
After QM/MM minimization, the Mg2+ ion in the double inner-shell position (Figure 3B) was pentacoordinated and its first coordination shell consisted of two phosphates (those of G1 and C21) and three water molecules. The sixth position of the octahedral coordination sphere was partially occupied by the C75(N4) nitrogen atom, sterically preventing coordination of a water molecule to this position. Since the coordination of a nitrogen atom from the exocyclic amino group to Mg2+ ion is unusual,44 a short MD simulation (see Supplementary Materials for details) was subsequently performed to better relax the solvation shell of the Mg2+ ion in the double inner-shell arrangement. Thus, three starting structures were eventually chosen for detailed QM/MM calculations (see Supplementary Materials for their coordinates): (i) the structure with a hexacoordinated Mg2+ ion and a single inner-shell bond to G1(O1P) – denoted the Hexacoordinated Single inner-shell structure (HS); (ii) a snapshot from the MD simulation (taken at 78 ps), in which the Mg2+ ion was pentacoordinated (to two phosphates, from G1 and C21, and three water molecules) – denoted the Pentacoordinated Double inner-shell structure (PD); and (iii) a snapshot from the MD simulation (taken at 125 ps) with a hexacoordinated Mg2+ ion (to two phosphates, from G1 and C21, and four water molecules) – denoted the Hexacoordinated Double inner-shell structure (HD) (see Figure 4). The HD structure also differs from the PD and HS structures in the U-1 ribose conformation, since its 2′,3′-cyclic phosphate attains the exo conformation in the HD product state compared to an endo conformation in the PD and HS product structures.
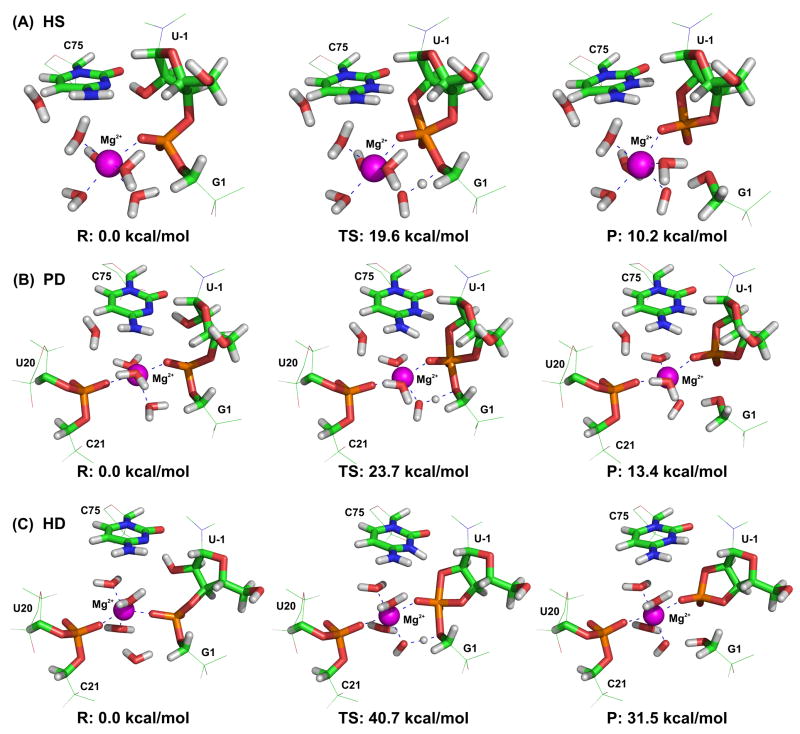
Figure 4.
Structures of HDV ribozyme active sites and the corresponding MPW1K/6-31+G(d,p):AMBER(parm99) energies in precursor (R), transition (TS), and product (P) states of the following reaction profiles with different Mg2+ ion positions and coordinations: A) the structure including a hexacoordinated Mg2+ ion with a single inner-shell bond to the G1(O1P) oxygen of the scissile phosphate (denoted HS in the text), B) the structure including a pentacoordinated Mg2+ ion with double inner-shell contacts to phosphates from C21 and G1 (denoted PD), and C) the structure including a hexacoordinated Mg2+ ion with double inner-shell bonds to phosphates from C21 and G1 (denoted HD). The quantum region and part of the molecular mechanical model neighborhood are shown in sticks and wire representation, respectively.
(b) QM/MM calculations
A two-layer ONIOM scheme45 implemented in Gaussian0346 was used for all QM/MM calculations. The low level (MM region) was handled by the AMBER molecular mechanical force field parm99,43 while the high level (QM region) was described by quantum mechanical method using electronic embedding. Hydrogen atoms were added to the dangling bonds at the interface between QM and MM regions. In all QM/MM calculations, the ribozyme was immersed in a water drop with a ~10 Å thick layer of water molecules surrounding the RNA molecule. The initial positions of counter ions were taken from the MD snapshot from which the starting structures were prepared. The ~2 Å thick layer of waters on the surface of the drop and counter ions outside the drop were fixed during all QM/MM calculations to prevent any changes in energy caused by H-bond network reorganization at the water-vacuum interface. The whole system contained ~13,000 atoms, of which ~4,500 were fixed.
Three different QM regions were used (Figure 3). Firstly, a large quantum region was subjected to QM/MM calculations (Figure 3A) in conjunction with a rapid, albeit less accurate HF/STO-3G method to pre-optimize 13 structures differing in Mg2+ ion placement (see above for details). The HF/STO-3G method was deemed sufficiently accurate for this initial screening. The BLYP/cc-pVDZ method in combination with smaller quantum regions was applied to explore reaction coordinates of the HD, PD and HS structures (Figure 3B and C). The BLYP functional47,48 with the density fitting approximation49,50 was chosen as a compromise providing sufficient geometrical accuracy at affordable computational cost. Since the energy barriers calculated at the BLYP level are likely to be underestimated, mainly due to the self-interaction error of the BLYP functional, the energy values were finally recalculated by the more accurate MPW1K DFT method51,52 as described below. The reaction profile was explored by shortening the distance between G1(O′5) and G1(P) to proceed from the product state back to the precursor. The forward reaction from precursor to product was studied by a potential energy surface scan in two dimensions, including shortening of the distance between U-1(O′2) and G1(P) and the distance between C75(N3) and U-1(H′2). The scan was performed in 0.1-Å steps and all remaining degrees of freedom were relaxed at each point (except for the fixed water molecules at the surface of the water drop, as mentioned in the previous paragraph). Finally, single-point energies were calculated for the BLYP/cc-pVDZ minima and saddle-point geometries corresponding to product, precursor and transition states, respectively, using the MPW1K hybrid functional51,52 with a 6-31+G(d,p) basis set, which is expected to provide sufficiently accurate energetics for the reaction barriers.
A similar ONIOM-QM/MM protocol was used in our recent study of the catalytic mechanism of haloalkane dehalogenase.53 In this case, the ONIOM-QM/MM approach (with mechanical embedding) was compared with the well established and widely used54–57 Car-Parrinello QM/MM (CP-QM/MM) method using a fully Hamiltonian coupling scheme.58,59 Our study showed that both approaches yield the same reaction mechanism, but the QM/MM scheme with mechanical embedding overestimated the reaction barrier because of the missing electronic coupling between the QM and MM regions. Therefore, in the present study, ONIOM-QM/MM with electronic embedding was employed to obtain reliable indications of both the reaction mechanism and energetics.
(c) Reference calculations and discussion of the limitations
The quality of the description of this reaction provided by the MPW1K functional was verified, and Gibbs energy corrections were estimated using the uncatalyzed reaction in a water environment, modeled as follows. The starting geometries of reactant, transition states, intermediates, and products of the uncatalyzed reaction, taken from the literature 39 were reoptimized in water, with implicit solvation model (IEFPCM(εr=78.4)/B3LYP/6-31G(d,p)). The frequencies were calculated at the same level for each optimized structure to estimate corrections to the Gibbs energy at temperature 298.15 K and pressure 1 atm. The differences between gas phase B3LYP/6-31G(d,p) energies and IEFPCM(εr=78.4)/B3LYP/6-31G(d,p) energies were used to calculate the solvation contributions to the reaction energy profile. Subsequently, single-point calculations were performed at MP2/cc-pVTZ and MP2/cc-pVQZ levels to estimate the MP2/complete basis set limit (CBS) energies,60,61 and at MP2/cc-pVDZ and CCSD(T)/cc-pVDZ to estimate a CCSD(T) correction.62 CCSD(T) stands for the Coupled Cluster method with noniterative triple electron excitations, which is currently the most accurate QM method that can be applied to such systems. The CCSD(T)/CBS method was obviously not affordable for any QM/MM calculations. The extrapolation scheme for the CCSD(T)/CBS energies was taken from literature.63 The MPW1K/6-31+G(d,p) single-point gas phase SCF energies were calculated at the same geometries, and the differences between MPW1K/6-31+G(d,p) and CCSD(T)/CBS energies were used to estimate the mean unsigned error of the MPW1K functional used in the presented QM/MM study. The SCF energies at MPW1K/6-31+G(d,p), extrapolated CCSD(T)/CBS and the respective MPW1K functional errors, solvation energies, Gibbs energy corrections, and total Gibbs energies at the extrapolated CCSD(T)/CBS level are summarized in Table 1.
Table 1.
Summary of single point energies (kcal/mol) calculated for reactants, transition states, intermediates and product structures of reference uncatalyzed reaction (geometries were optimized at IEFPCM(εr=78.4)/B3LYP/6-31G(d,p) level, see Methods for details and Supporting information for structures labeled R, TS1, I1, TS2, I2, TS3, P, and P′ endo/exo) at MPW1K/6-31+G(d,p) and extrapolated CCSD(T)/CBS levels, corresponding errors of the MPW1K functional, solvation terms and the corrections to Gibbs energies at IEFPCM(εr=78.4)/B3LYP/6-31G(d,p) level, and extrapolated total Gibbs energies in water solution at the CCSD(T)/CBS level.
| endo | R | TS1 | I1 | TS2 | I2 | TS3 | P | P′ |
|---|---|---|---|---|---|---|---|---|
| MPW1K/6-31+G(d,p) | 0.0 | 30.2 | 27.5 | 36.8 | 30.4 | 36.8 | 2.7 | 17.8 |
| CCSD(T)/CBS | 0.0 | 29.6 | 27.9 | 37.2 | 30.4 | 37.6 | 4.4 | 19.6 |
| MPW1K errora | 0.0 | −0.6 | 0.4 | 0.4 | −0.1 | 0.0 | 1.7 | 2.2 |
| Solvation energyb | 0.0 | −2.7 | −7.8 | −8.4 | −7.2 | −0.9 | 0.1 | −12.9 |
| Gibbs energy correctionc | 0.0 | 0.8 | 1.2 | −0.1 | 1.9 | −2.4 | −0.5 | −13.2 |
| CCSD(T)/CBS Gibbs energy in water | 0.0 | 27.7 | 21.3 | 28.8 | 25.0 | 34.4 | 4.0 | −6.5 |
|
| ||||||||
| exo | R | TS1 | I1 | TS2 | I2 | TS3 | P | P′ |
|
| ||||||||
| MPW1K/6-31+G(d,p) | 0.0 | 27.9 | 21.2 | 30.6 | 25.5 | 37.2 | 2.7 | 17.8 |
| CCSD(T)/CBS | 0.0 | 27.6 | 21.6 | 30.5 | 26.1 | 38.1 | 4.4 | 19.6 |
| MPW1K errora | 0.0 | −0.7 | 0.4 | 0.1 | 0.5 | 0.9 | 1.7 | 2.2 |
| Solvation energyb | 0.0 | 0.2 | −3.1 | −3.0 | −3.8 | −1.6 | 0.1 | −12.9 |
| Gibbs energy correctionc | 0.0 | 0.5 | 1.0 | 0.1 | 1.3 | −0.2 | −0.5 | −13.2 |
| CCSD(T)/CBS Gibbs energy in water | 0.0 | 28.3 | 19.5 | 27.7 | 23.5 | 36.3 | 4.0 | −6.5 |
differences between CCSD(T)/CBS and MPW1K/6-31+G(d,p) energies.
solvation term calculated as the difference between IEFPCM(εr=78.4)/B3LYP/6-31G(d,p) and gas phase B3LYP/6-31G(d,p) SCF energies.
the corrections to Gibbs energies were calculated at the IEFPCM(εr=78.4)/B3LYP/6-31G(d,p) level.
The Gibbs energy profile of the reference reaction in water was calculated as a sum of extrapolated CCSD(T)/CBS gas phase energies, Gibbs energy corrections and solvation terms obtained at the IEFPCM(εr=78.4)/B3LYP/6-31G(d,p) level. The Gibbs energy barrier for the nucleophile attack and the intermediates’ rearrangement was found to be ~28–29 kcal/mol, while the disruption of the intermediate involving departure of the alcoholate ion and transport of a proton to the leaving alcoholate is the rate-limiting step of the uncatalyzed reaction, leading to Gibbs energy barriers of 36.3 kcal/mol and 34.4 kcal/mol for exo and endo conformations, respectively.
The results showed that the mean unsigned error of the MPW1K/6-31+G(d,p) method relative to CCSD(T)/CBS was 0.7 kcal/mol, and the maximum unsigned errors were 0.9 kcal/mol for the transition state and intermediates, and 1.9 kcal/mol for the products.
The calculated QM/MM energy barriers, as described above, do not account for the Gibbs energy correction term, which could be estimated, within harmonic approximation, via frequency calculations. However, such calculations are not feasible for the QM/MM system due to their enormous memory demands. Therefore, we roughly estimated this term using the uncatalyzed reaction model. The mean correction for entropy and zero-point vibrational energy of transition states was found to be −0.2 kcal/mol and the maximum unsigned value to be lower than 2.4 kcal/mol (see Table 1). These observations agree with previously published Gibbs energy corrections for this type of reaction, which are consistently smaller than 2.5 kcal/mol.64
There is yet another part of the reaction, the separation of the products after the chemical step, in which the Gibbs energy correction (namely the entropic term) significantly contributes to the reaction profile. The separation of the products lowers the Gibbs energy of the product state in the uncatalyzed reaction by −13.2 kcal/mol (see Table 1). Separation of the products does not affect the kinetics of the studied reaction and is beyond the scope of the presented QM/MM study. Nevertheless, product separation immediately follows the part of the reaction considered in this paper and the associated lowering by ~13 kcal/mol shifts the equilibrium toward the separated products.
The calculated barriers also do not take into consideration the correction that should be applied to take into account the reactive state population. However, it seems reasonable to assume that this contribution is rather small for the studied reaction, no more than a few kcal/mol; less than 1.4 kcal/mol assuming that the population of the reactive state is approximately 10%, in accordance with the occurrence of the U-1(O′2-H′2) … C75(N3) H-bond ~10% of the time during MD simulations.13 Generally, it is challenging to accurately assess the Gibbs energy correction term associated with the relative population of precursor reactive states. This would require exhaustive molecular dynamics simulations because systems with small populations of reactive states need very robust phase space sampling (i.e., long simulation timescales). Further, the calculated populations could be biased by force field inaccuracies, such as those for divalent ions or the flexible anionic sugar-phosphate backbone.
Results
We carried out extensive QM/MM calculations of the HDV genomic ribozyme, aiming to clarify whether its C75 nucleotide is capable of acting as the general base during the HDV ribozyme self-cleavage reaction, using structural data obtained for the cis-acting genomic precursor ribozyme by X-ray crystallography and refined by MD simulations.
Due to the absence of relevant structural data for the alternative scenario, in which C75 is protonated and acts as the general acid, an analogous investigation of the general acid mechanism would be more complex and will be attempted separately. However, to date we have not found any suitable structure for the C75H+ acting as the general acid in quite extensive MD simulations.13 Furthermore, after a 20-ns MD simulation in which the C75H+(N3) … G1(O′5) H-bond was restrained to make the geometry more suitable for the general acid mechanism, the contact between C75H+ and G1(O′5) disrupted immediately when the restraint was released and no such contact was re-established in a subsequent 20 ns unrestrained simulation (data not shown). In addition, pilot QM/MM calculations starting from the structure that appeared to be most suitable (but dynamically unstable) for the general acid mechanism obtained from the restrained MD simulation failed to provide a viable reaction path (data not shown). Thus, obtaining a plausible structure with C75H+ for the cis-acting genomic precursor ribozyme is not straightforward.
Mg2+ ions play a crucial role in the HDV ribozyme self-cleavage reaction. The requirement of Mg2+ ions can be both structural65 and/or catalytical.66 Initially, therefore, it was necessary to find likely positions for the Mg2+ ion in the active site, since it is generally appreciated that even subtle details of the active site arrangement can profoundly influence both the energetic profile and mechanism of an enzymatic reaction. Unfortunately, the position of the Mg2+ ion cannot be taken directly from the available X-ray structures because a direct inner-shell coordination of U75(O4) to Mg2+ observed in the C75U mutant precursor structure is unlikely to occur between C75(N4) and Mg2+ in the wild-type ribozyme.14 Thus, we prepared a set of 13 possible positions of the Mg2+ ion in the active site and screened them in preliminary QM/MM calculations (see Methods for details). This search identified three plausible positions for the Mg2+ ion with different solvation shells (Figure 4). These three structural arrangements, denoted HS, PD and HD (see Methods and Figure 4 for details), were further used to investigate the reaction profile of the C75 general base mechanism of HDV ribozyme self-cleavage.
Initially, the reaction profile was studied using a one-dimensional (1D) potential energy surface scan by shortening the distance between U-1(O′2) and G1(P), following the reaction pathway in the forward direction from precursor to producta. In these attempts, the U-1(H′2) hydrogen was either not transferred to C75 or it was transferred to C75(N3) only after reaching an unrealistically short U-1(O′2) - G1(P) distance with an unacceptably high energy barrier (cf. Figure 5, red pathway). Furthermore, no rupture of the bond between the G1(P) and G1(O′5) atoms was observed while scanning the reaction.
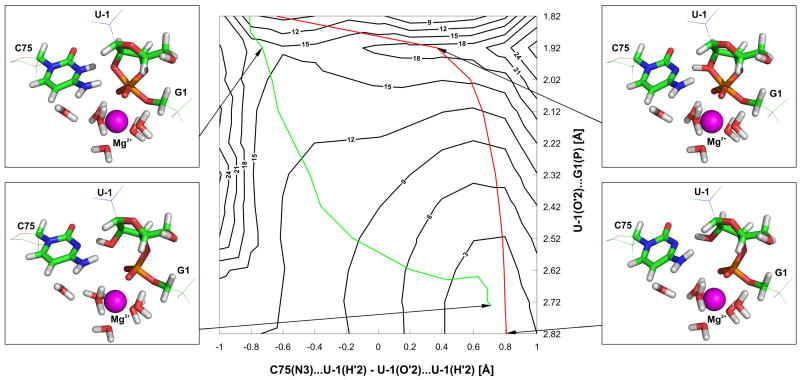
Figure 5.
Two-dimensional map of the potential energy surface (More O’Ferrall-Jencks diagram). The energy is presented as a function of the distance between U-1(O′2) and G1(P) (vertical axis) and the difference between two U-1(H′2)-C75(N3) and U-1(O′2-H′2) distances describing the proton transfer. The energy (kcal/mol) was calculated by the BLYP/cc-pVDZ:AMBER(parm99) method. The red line describes the reaction coordinate obtained by shortening the U-1(O′2)-G1(P) distance following the reaction from precursor to product, whilst the green line presents the reaction coordinate obtained by scanning the backward reaction from products to precursor via shortening the G1(O′5-P) distance (see text for details). The structures related to the corresponding parts of the surface are shown in small boxes.
To explain and overcome this problem, a two dimensional (2D) potential energy surface scan along both the U-1(O′2)-G1(P) and U-1(H′2)-C75(N3) distances was performed to investigate the shape and complexity of the energy surface of the HS system (Figure 5). These calculations revealed that simply shortening the distance between U-1(O′2) and G1(P) from the precursor state does not follow the realistic reaction path (i.e. the path via the lowest available saddle-point), and this was the reason for the unfeasibly high energy barriers discussed above. Although the potential energy surface implies that the realistic reaction path is easily accessible in range of U-1(O′2-H′2) vibration, the used geometry optimization algorithm is not able to relax geometry to this path due to the complexity of the potential energy surface. The shape of the potential energy surface further suggested that this problem could be overcome if the reaction coordinate was scanned in the opposite direction, i.e., from product to precursor.b The energy profiles along the reaction coordinate obtained by calculations in both directions (i.e., from product to precursor and vice versa) are presumably equivalent due to microscopic reversibility of the reaction studied.
Consequently, the reaction profiles of all three studied configurations were obtained from potential energy surface scans by shortening the distance between G1(O′5) and G1(P), following the reaction in the direction from product to precursor. Similar structural changes along the reaction coordinate were observed in all calculations. A spontaneous proton transfer from the G1(O′5-H′5) hydroxyl group to the hydroxide ion coordinated to Mg2+ occurred simultaneously with the shortening of the distance between the G1(O′5) and G1(P) atoms. We observed that the U-1(O′2) - G1(P) bond was disrupted after the G1(O′5) - G1(P) distance reached RO-P ≈ 2 Å, and the leaving alcoholate group of U-1(O′2) acted as the acceptor of the C75H+(H3) proton from the protonated C75H+ nucleobase (Figure 5). According to our calculations there is no stable intermediate along the calculated reaction pathway. This is in agreement with some other theoretical studies of HDV ribozyme cleavage39,40 and Hammerhead ribozyme cleavage.64 On the other hand, there are theoretical studies proposing stable intermediates in ribozyme reactions, e.g., a study of the HDV ribozyme general acid cleavage40 and another study of the Hammerhead ribozyme cleavage.66 The stable intermediate was also localized for the noncatalyzed reaction (39,67 and this study).
The calculated reaction barriers (activation energies) for the HS and PD systems were 19.6 kcal/mol and 23.7 kcal/mol, respectively. The activation barrier is presumably higher in the PD system due to the coordination of two negatively charged phosphates (from C21 and G1) to the Mg2+ ion compared to the single coordinated scissile phosphate of G1 in the HS structure, and the consequent reduction in the electrophilic (Lewis acid) catalytic power of the Mg2+ ion. On the other hand, a considerably higher barrier (40.7 kcal/mol) was found for the HD system (Figure 4).
The reaction rate constant of the HDV ribozyme self-cleavage was estimated to be equal kcat ≈ 52 min−1 by Tanner et al. 68 and subsequently measured directly by Brown et al. 69 (kcat = 60 min−1). Using the Eyring equation,70 the experimental Gibbs energy barrier can be estimated from the available rate constant (kcat = 60 min−1) to be 18.2 kcal/mol. A direct comparison of the calculated and experimental activation barriers is not straightforward because the Gibbs energy correction is not directly included in our QM/MM calculations and the calculated barriers are not corrected for the population of reactive precursor state. However, using our model reference reaction we estimated that the Gibbs energy correction for the transition state is lower than 2.5 kcal/mol and the correction for the reactive state population is ~1.4 kcal/mol (see Methods). These values, together with the estimated mean unsigned error of the MPW1K functional (~0.7 kcal/mol) lead to a qualified estimate of the upper limit of uncertainty for our calculations of ~5 kcal/mol. Taking these considerations into account, we can safely conclude that the reaction barriers of the HS system, and even the PD system, are in good agreement with the experimentally observed value, and well within the accuracy typically achieved with QM/MM methods for enzymes.71–74 Furthermore, the calculated barriers are in the typical range of many enzymatic reactions (10–20 kcal/mol),75 they are consistent with the calculated barriers of the hammerhead ribozyme (20.8 kcal/mol,64 19.3 kcal/mol66), and they are significantly smaller than the estimated barrier of the uncatalyzed reaction (34–36 kcal/mol; see Methods and refs. 39,67). These observations support the conclusion that we identified a plausible mechanistic scenario for the catalysis of HDV ribozyme self-cleavage.
For the sake of completeness, it should be mentioned that the calculated energies of the product states (i.e. the structures immediately after the cleavage reaction) are +10.2 kcal/mol and +13.4 kcal/mol above the precursor state for the HS and PD systems, respectively. At first sight these findings indicate that a thermodynamic penalty may be associated with the spontaneous reaction from precursor to products. However, these calculated product states are not the final experimentally observable product states since other processes occur after they have been reached. More specifically, product dissociation following the calculated part of the reaction pathway reduces the Gibbs energy of the product state by an estimated ~13 kcal/mol (see Methods). In addition, other processes such as neutralization of the protonated C75H+ and Mg2+-coordinated hydroxide ion are likely to follow, which are expected to further decrease the Gibbs energy and shift the equilibrium toward the product state, in favor of the HS and/or PD reaction pathways.
By contrast, a comparison of the experimental catalytic and uncatalyzed barriers (18.2 kcal/mol and 34–36 kcal/mol, respectively) with the reaction barrier of the HD system (40.7 kcal/mol) suggests that the self-cleavage reaction is unlikely to follow this route.
Our calculated reaction barriers and total reaction energy differences are related to the relative conformations of the scissile phosphate and C75 nucleobase (Table 2). The H-bond between the C75(N4–H4) exocyclic amino group and the G1(O1P) oxygen of the scissile phosphate was observed in the transition and product states of the PD and HS systems (those with lower reaction barriers). By contrast, this interaction was missing in the HD system because the waters of the hexacoordinate Mg2+ solvation shell filled the space around the G1(O1P) oxygen, resulting in an unfavorable conformation of the C75 nucleobase in which C75(N4–H4) points away from the scissile phosphate (Figure 4C). The C75(N4–H4) … G1(O1P) H-bond is essential since it decreases the reaction barrier by stabilizing the transition state in two ways: (i) by polarizing the scissile phosphate, thus making it more susceptible to nucleophilic attack; and (ii) by supporting sp2 hybridization of the exocyclic amino group, thus stabilizing protonation of C75 in the transition and product states.
Table 2.
The distances (in Å) between C75(N4) and G1(O1P) for various structures. Precursor, transition, and product states are labeled R, TS, and P, respectively. The labels PD, HD, and HS stand for different structural arrangements in the HDV ribozyme active site (see text and Figure 4).
| R | TS | P | |
|---|---|---|---|
| PD | 2.95 | 2.76 | 2.82 |
| HD | 4.69 | 3.40 | 3.76 |
| HS | 3.64 | 3.13 | 2.93 |
Discussion and Conclusions
QM/MM calculations were used to study the possibility that C75 may act as the general base in the HDV ribozyme self-cleavage reaction. In this mechanistic scenario, C75(N3) accepts a proton from the U-1(O′2-H′2) nucleophile. Our results also highlight two roles for the Mg2+ ion in the reaction mechanism: acting as both a general (Brønsted) and a Lewis acid. QM/MM analysis of the C75 general base role in catalysis is simplified by the fact that this mechanism is consistent with the available ground-state structural data obtained from X-ray crystallography and MD simulations. The QM/MM calculations were capable of identifying a plausible reaction path with C75 acting as the general base.
Currently no structural data are available that correspond to the alternative mechanism, in which C75 is protonated and acts as the general acid. This makes investigations of the putative C75H+ mechanistic role challenging. It should be noted that rather extensive MD simulations13 with the protonated C75H+ were not able to generate a suitable micro-environment for the catalytic center with a protonated C75H+, although even ~10–20 ns long simulations utilizing the available X-ray structures as the starting point may be still too short. Hence, some more complex structural rearrangements, as suggested for instance for the minimal hammerhead ribozyme by MD and QM/MM calculations,76 would be required to facilitate the C75H+ general acid mechanism for the cis-acting genomic HDV ribozyme.
The calculated activation barriers are sensitive to the Mg2+ ion’s position, conformation of its solvation sphere, and active site structural arrangements. This means that the self-cleavage reaction in the presence of Mg2+ should be preferably described by an ensemble of transition states taking into account relative populations of precursor conformations preceding each transition state, rather than by a single reaction coordinate. This obviously limits the reliability of the calculations. Since the reaction profile is highly sensitive to details of the active site’s structural arrangement the sampling must clearly be sufficiently extensive to ensure that adequately reliable estimates of the relative proportions of possible conformations are obtained. Efficient sampling could be accomplished by large scale QM/MM molecular dynamics, but this would require a considerably cheaper description of the QM core. However, if the QM/MM description has insufficient quality the results can also be biased. In our particular case we decided to utilize a high-quality method (in terms of the level of calculations and size of the QM region) while attempting to overcome the sampling problem by considering a range of starting structures. More specifically, we studied the reaction profiles of three different starting geometries chosen from 13 initial structures. The selected structures differed primarily in the positions and coordination spheres of the critical Mg2+ ion. It was found that the HS structure (in which the Mg2+ ion is hexacoordinated with a single inner-shell contact to the G1(O1P) oxygen of the scissile phosphate) provides the lowest reaction barrier, 19.6 kcal/mol, which is in good agreement with the experimental estimate of 18.2 kcal/mol.6,19 In addition, the structural arrangement of the Mg2+ ion in the HS structure is commonly observed in RNA structures.44
Interestingly, the PD structure (with pentacoordinated Mg2+ ion and the double inner-shell coordination sphere) also provides a feasible energy barrier, of 23.7 kcal/mol, indicating that it could provide another plausible route to the reaction products. However, the validity of the pentacoordinated solvation sphere of the Mg2+ ion is disputable since Mg2+ prefers hexacoordination,77 although it is possible that the Mg2+ coordination number may be reduced following the first-sphere binding of two highly negatively charged first-shell ligands, as in the PD structure. The reasonable barrier observed for the PD structure indicates that coupled catalytic functions of the hydrated Mg2+ ion and C75 are sufficient to catalyze the HDV ribozyme self-cleavage reaction effectively, regardless of the exact Mg2+ ion coordination.
The highest energy reaction barrier (40.7 kcal/mol) was computed for the HD structure (with hexacoordinated Mg2+ ion and double inner-shell Mg2+ coordination). This barrier is similar to that in the uncatalyzed reaction, because the hexacoordinated solvation shell of Mg2+ sterically shifts the position of C75 and thus abrogates its stabilizing effect. Thus, this geometry cannot lead to catalytic activity.
From a structural perspective the Mg2+ ions in both the PD and HD structures are coordinated by the two phosphates of C21 and G1 in trans orientation. Such Mg2+ coordination is rarely observed in RNA, in contrast to the common Mg2+ coordination of the HS structure.44
Three structural requirements were identified as critical for C75 general base catalysis in our calculations: (i) the catalytic water has increased acidity due to its coordination to the Mg2+ ion and so acts as an acid that passes its proton to the leaving alcoholate G1(O′5); (ii) the Mg2+ ion acts also as an electrophilic (Lewis acid) catalyst as it activates the scissile phosphate through direct coordination to the G1(O1P) atom and the resultant electron pull makes the phosphate more susceptible to nucleophilic attack, and (iii) the nucleobase C75 stabilizes the transition state via two H-bonds to the scissile phosphate, i.e. C75(N4-H4) … G1(O1P) and C75(N3)-U-1(H′2) … G1(O′2). Regarding the point (ii) above, it is fair to admit that such a role requiring inner-shell coordination of Mg2+ ion in cis-acting HDV ribozyme could be controversial. For example, Nishikawa et al. provided biochemical evidence that the thio-substitution of one of both non-bridging oxygens of scissile phosphate in trans-acting HDV ribozyme has no significant effect to the cleavage rate, and thus suggested that these non-bridging oxygens do not coordinate the Mg2+ ion directly in trans-acting HDV ribozyme.78 On the other hand, the X-ray structure of inactivated C75U precursor indicates (despites its limited resolution) inner shell binding to U75(O4). It cannot be ruled that, when the C75 is present, the divalent ion can replace the U75(O4) by a phosphate oxygen, as observed in our earlier simulations.13 Obviously, the computational analysis of the Mg2+ binding has major limitations, and especially the force fields are not sufficiently accurate for divalent ions.79,80 Thus, our capability to independently predict Mg2+ binding by simulations is not satisfactory and all literature attempts should be viewed in this context. In summary, while we found a plausible path with inner shell coordination in our calculations, we cannot rule out that there exist also suitable outer shell binding patterns, especially for the trans-acting ribozyme.
The stabilization of the transition state by the C75 nucleobase has two catalytic effects: (i) making the scissile phosphate more susceptible to nucleophilic attack due to polarization by the C75(N4-H4) … G1(O1P) H-bond reducing its electron density, and (ii) activating the nucleophile U-1(O′2-H′2) by the presence of a general base C75(N3). The nucleobase C75 is geometrically well suited for effective catalysis because it pulls electron density from the scissile phosphate through the H-bond between its exocyclic amino group C75(N4) and the G1(O1P) oxygen and pushes it through conjugation of its aromatic ring to the lone electron pair of the C75(N3) nitrogen, and thus toward the proton from the U-1(O′2-H′2) nucleophile.
A similar structural motif that stabilizes the transition state with the C75H+ bound to the scissile phosphate through two H-bonds (C75H+(N4-H4) … G1(O1P) and C75H+(N3-H3) … G1(O′5)) was observed in previous DFT calculations of the general acid mechanism using the truncated model of the HDV ribozyme active site.39,40 However, the orientation of C75 (and the overall active site arrangement) in these small-molecule models of the active site did not fit any available crystal structures. Superposing the sugar phosphate backbone from these active site models39,40 on the complete HDV ribozyme structure, we found that these two H-bonds positioned the C75 sugar too close to G1. Therefore, these models do not satisfy the steric requirements of the X-ray structures, in which the C75 sugar resides on the opposite side of the active site. This finding rather limits the biological relevance of the results from the model calculations39,40 and highlights the importance of considering the context of the complete ribozyme fold. These requirements are fulfilled in our QM/MM calculations, in which the sugar-phosphate backbone steric constraints of the ribozyme active site are respected and all proximal and distal functional groups contributing to the catalysis are included in the quantum region.
We show that under these conditions the C75 general base mechanism is chemically feasible. However, we do not conclude that the general base mechanism is the dominant mode of HDV ribozyme self-cleavage. The exact conclusions of this study are that the local and global arrangements suggested by the C75U and lower resolution C75 precursor X-ray structures of the cis-acting HDV genomic ribozyme indicate that the general base mechanism is readily available, while the general acid mechanism is much less structurally consistent. The general base mechanism is also supported by the fold of the ribozyme over the uncatalyzed reaction, which was not detected in previous QM studies based on small-molecule models of the active site. However, we cannot yet compare the C75 general base mechanism with the general acid mechanism (in which C75 is protonated and acts as the general acid) energetically, due to the paucity of suitable starting structures that would support this mechanism. Thus, we cannot comment on which of these two mechanisms is more feasible. It appears that the general acid mechanism could be associated with some local structural rearrangements compared to presently available structures. However, modeling these rearrangements would not be trivial, as attempts to obtain such geometries via classical MD simulations starting from the available X-ray structures (and assuming C75H+) were not successful.13,14 Nevertheless, it seems reasonable to assume that in principle the cytosine should be able to act both as the general base and the general acid in the cleavage reaction. Therefore, we suggest a possibility that the HDV ribozyme could use multiple, competing micro-strategies depending on the circumstances. This possibly could best reconcile the available experimental data.
Supplementary Material
The content of Supporting Information includes full citations for references 42 and 46, the setup for molecular dynamics simulations, and the geometries from the model study of the uncatalyzed reaction in water and the geometries from QM/MM calculations.
Acknowledgments
This study was supported by grants LC512, LC06030, MSM0021622413 and MSM6198959216 from the Ministry of Education of the Czech Republic, and grants IAA400040802 and 1QS500040581 from the Grant Agency of the Academy of Sciences of the Czech Republic. This work was also supported by the Academy of Sciences of the Czech Republic, grants no. AV0Z50040507, AV0Z40550506 and AV0Z50040702, and NIH grant GM62357 (to NGW). We thank Petr Juretka (Olomouc, CZ) for his advice regarding CCSD(T)/CBS calculations.
Footnotes
The term product state refers here to a structure of the HDV ribozyme immediately after the cleavage 3′ to U-1, when 2′,3′-uridincyclophosphate, a hydroxide ion coordinated to Mg2+, and a protonated C75 reside in the active site. The active site likely regenerates after the reaction and such a process is expected to include proton transfer from C75H+ to the hydroxide ion via a water network, as well as dissociation of the 2′,3′-uridincyclophosphate from the active site.
This process (the reversed scan) is not equivalent or related to the ligation reaction, because the product is not the same as the reactant of the ligation. It should be considered as a standard computational technique to explore the path of the cleavage reaction.
References
- 1.Fedor MJ, Williamson JR. Nat Rev Mol Cell Biol. 2005;6:399. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Lorsch JR. Nat Struct Mol Biol. 2005;12:395. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- 3.Been MD. Curr Top Microbiol Immunol. 2006;307:47. doi: 10.1007/3-540-29802-9_3. [DOI] [PubMed] [Google Scholar]
- 4.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. Science. 2006;313:1788. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 5.Perrotta AT, Shih I, Been MD. Science. 1999;286:123. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 6.Nakano S, Chadalavada DM, Bevilacqua PC. Science. 2000;287:1493. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 7.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Science. 2000;289:920. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 8.Muth GW, Ortoleva-Donnelly L, Strobel SA. Science. 2000;289:947. doi: 10.1126/science.289.5481.947. [DOI] [PubMed] [Google Scholar]
- 9.Pinard R, Hampel KJ, Heckman JE, Lambert D, Chan PA, Major F, Burke JM. EMBO J. 2001;20:6434. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafontaine DA, Norman DG, Lilley DM. Biochimie. 2002;84:889. doi: 10.1016/s0300-9084(02)01406-2. [DOI] [PubMed] [Google Scholar]
- 11.Beringer M, Rodnina MV. Mol Cell. 2007;26:311. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Ke AL, Zhou KH, Ding F, Cate JHD, Doudna JA. Nature. 2004;429:201. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 13.Krasovska MV, Sefcikova J, Spackova N, Sponer J, Walter NG. J Mol Biol. 2005;351:731. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Krasovska MV, Sefcikova J, Reblova K, Schneider B, Walter NG, Sponer J. Biophys J. 2006;91:626. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano S, Bevilacqua PC. Biochemistry. 2007;46:3001. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- 16.Wadkins TS, Shih I, Perrotta AT, Been MD. J Mol Biol. 2001;305:1045. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- 17.Perrotta AT, Wadkins TS, Been MD. RNA-Publ RNA Soc. 2006;12:1282. doi: 10.1261/rna.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferre-D’Amare AR, Zhou K, Doudna JA. Nature. 1998;395:567. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 19.Das SR, Piccirilli JA. Nature Chem Biol. 2005;1:45. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 20.Jeong S, Sefcikova J, Tinsley RA, Rueda D, Walter NG. Biochemistry. 2003;42:7727. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]
- 21.Pereira MJ, Harris DA, Rueda D, Walter NG. Biochemistry. 2002;41:730. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 22.Tinsley RA, Harris DA, Walter NG. Biochemistry. 2004;43:8935. doi: 10.1021/bi049471e. [DOI] [PubMed] [Google Scholar]
- 23.Harris DA, Tinsley RA, Walter NG. J Mol Biol. 2004;341:389. doi: 10.1016/j.jmb.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 24.Tinsley RA, Walter NG. Biol Chem. 2007;388:705. doi: 10.1515/BC.2007.088. [DOI] [PubMed] [Google Scholar]
- 25.Tang CL, Alexov E, Pyle AM, Honig B. J Mol Biol. 2007;366:1475. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Ke A, Ding F, Batchelor JD, Doudna JA. Structure. 2007;15:281. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Gao H, Gaffney BL, Jones RA. J Am Chem Soc. 1991;113:5486. [Google Scholar]
- 28.Doronina SO, Behr JP. Chem Soc Rev. 1997;26:63. [Google Scholar]
- 29.Soliva R, Laughton CA, Luque FJ, Orozco M. J Am Chem Soc. 1998;120:11226. [Google Scholar]
- 30.Nixon PL, Giedroc DP. J Mol Biol. 2000;296:659. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- 31.Csaszar K, Spackova N, Stefl R, Sponer J, Leontis NB. J Mol Biol. 2001;313:1073. doi: 10.1006/jmbi.2001.5100. [DOI] [PubMed] [Google Scholar]
- 32.Ferre-D’Amare AR, Doudna JA. J Mol Biol. 2000;295:541. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 33.Gehring K, Leroy JL, Gueron M. Nature. 1993;363:561. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 34.Kang CH, Berger I, Lockshin C, Ratliff R, Moyzis R, Rich A. Proc Natl Acad Sci USA. 1994;91:11636. doi: 10.1073/pnas.91.24.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spackova N, Berger I, Egli M, Sponer J. J Am Chem Soc. 1998;120:6147. [Google Scholar]
- 36.Rhodes MM, Reblova K, Sponer J, Walter NG. Proc Natl Acad Sci USA. 2006;103:13380. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luptak A, Ferre-D’Amare AR, Zhou K, Zilm KW, Doudna JA. J Am Chem Soc. 2001;123:8447. doi: 10.1021/ja016091x. [DOI] [PubMed] [Google Scholar]
- 38.Gong B, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. J Am Chem Soc. 2007;129:13335. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 39.Liu HN, Robinet JJ, Ananvoranich S, Gauld JW. J Phys Chem B. 2007;111:439. doi: 10.1021/jp064292n. [DOI] [PubMed] [Google Scholar]
- 40.Wei K, Liu L, Cheng YH, Fu Y, Guo QX. J Phys Chem B. 2007;111:1514. doi: 10.1021/jp070120u. [DOI] [PubMed] [Google Scholar]
- 41.Warshel A, Levitt M. J Mol Biol. 1976;103:227. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 42.Case DA, et al. AMBER 8. University of California; San Francisco: 2004. See Supporting information for complete citation. [Google Scholar]
- 43.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179. [Google Scholar]
- 44.Klein DJ, Moore PB, Steitz TA. RNA- Publ RNA Soc. 2004;10:1366. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svensson M, Humbel S, Froese RDJ, Matsubara T, Sieber S, Morokuma K. J Phys Chem. 1996;100:19357. [Google Scholar]
- 46.Frisch MJ, et al. Gaussian 03. Gaussian, Inc.: Pittsburgh; 2003. See Supporting information for complete citation. [Google Scholar]
- 47.Becke AD. Phys Rev A. 1988;38:3098. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 48.Lee CT, Yang WT, Parr RG. Phys Rev B. 1988;37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 49.Dunlap BI. J Chem Phys. 1983;78:3140. [Google Scholar]
- 50.Dunlap BI. J Mol Struct. 2000;529:37. [Google Scholar]
- 51.Lynch BJ, Fast PL, Harris M, Truhlar DG. J Phys Chem A. 2000;104:4811. [Google Scholar]
- 52.Lynch BJ, Truhlar DG. J Phys Chem A. 2001;105:2936. [Google Scholar]
- 53.Otyepka M, Banas P, Magistrato A, Carloni P, Damborsky J. Proteins. 2008;70:707. doi: 10.1002/prot.21523. [DOI] [PubMed] [Google Scholar]
- 54.Carloni P, Rothlisberger U, Parrinello M. Acc Chem Res. 2002;35:455. doi: 10.1021/ar010018u. [DOI] [PubMed] [Google Scholar]
- 55.Piana S, Carloni P. Proteins. 2000;39:26. doi: 10.1002/(sici)1097-0134(20000401)39:1<26::aid-prot3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 56.Piana S, Sebastiani D, Carloni P, Parrinello M. J Am Chem Soc. 2001;123:8730. doi: 10.1021/ja003145e. [DOI] [PubMed] [Google Scholar]
- 57.Piana S, Bucher D, Carloni P, Rothlisberger U. J Phys Chem B. 2004;108:11139. [Google Scholar]
- 58.Laio A, VandeVondele J, Rothlisberger U. J Chem Phys. 2002;116:6941. [Google Scholar]
- 59.Laio A, Gervasio FL, VandeVondele J, Sulpizi M, Rothlisberger U. J Phys Chem B. 2004;108:7963. [Google Scholar]
- 60.Halkier A, Helgaker T, Jorgensen P, Klopper W, Olsen J. Chem Phys Lett. 1999;302:437. [Google Scholar]
- 61.Halkier A, Helgaker T, Jorgensen P, Klopper W, Koch H, Olsen J, Wilson AK. Chem Phys Lett. 1998;286:243. [Google Scholar]
- 62.Jurecka P, Hobza P. Chem Phys Lett. 2002;365:89. [Google Scholar]
- 63.Jurecka P, Hobza P. J Am Chem Soc. 2003;125:15608. doi: 10.1021/ja036611j. [DOI] [PubMed] [Google Scholar]
- 64.Torres RA, Himo F, Bruice TC, Noodleman L, Lovell T. J Am Chem Soc. 2003;125:9861. doi: 10.1021/ja021451h. [DOI] [PubMed] [Google Scholar]
- 65.Bevilacqua PC, Yajima R. Curr Opin Chem Biol. 2006;10:455. doi: 10.1016/j.cbpa.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 66.Leclerc F, Karplus M. J Phys Chem B. 2006;110:3395. doi: 10.1021/jp053835a. [DOI] [PubMed] [Google Scholar]
- 67.Lopez X, Dejaegere A, Leclerc F, York DM, Karplus M. J Phys Chem B. 2006;110:11525. doi: 10.1021/jp0603942. [DOI] [PubMed] [Google Scholar]
- 68.Tanner NK, Schaff S, Thill G, Petitkoskas E, Craindenoyelle AM, Westhof E. Curr Biol. 1994;4:488. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- 69.Brown TS, Chadalavada DM, Bevilacqua PC. J Mol Biol. 2004;341:695. doi: 10.1016/j.jmb.2004.05.071. [DOI] [PubMed] [Google Scholar]
- 70.Eyring H. J Chem Phys. 1935;3:107. [Google Scholar]
- 71.Kwiecien RA, Khavrutskii IV, Musaev DG, Morokuma K, Banerjee R, Paneth P. J Am Chem Soc. 2006;128:1287. doi: 10.1021/ja056333j. [DOI] [PubMed] [Google Scholar]
- 72.Lundberg M, Morokuma K. J Phys Chem B. 2007;111:9380. doi: 10.1021/jp071878g. [DOI] [PubMed] [Google Scholar]
- 73.Prabhakar R, Morokuma K, Musaev DG. J Comput Chem. 2005;26:443. doi: 10.1002/jcc.20179. [DOI] [PubMed] [Google Scholar]
- 74.Prabhakar R, Vreven T, Frisch MJ, Morokuma K, Musaev DG. J Phys Chem B. 2006;110:13608. doi: 10.1021/jp0619181. [DOI] [PubMed] [Google Scholar]
- 75.Warshel A, Sharma PK, Kato M, Xiang Y, Liu HB, Olsson MHM. Chem Rev (Washington, DC) 2006;106:3210. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 76.Radhakrishnan R. Biophys J. 2007;93:2391. doi: 10.1529/biophysj.107.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bock CW, Kaufman A, Glusker JP. Inorg Chem. 1994;33:419. [Google Scholar]
- 78.Fauzi H, Kawakami J, Nishikawa F, Nishikawa S. Nucleic Acids Res. 1997;25:3124. doi: 10.1093/nar/25.15.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sponer J, Sabat M, Gorb L, Leszczynski J, Lippert B, Hobza P. J Phys Chem B. 2000;104:7535. [Google Scholar]
- 80.Gresh N, Sponer JE, Spackova N, Leszczynski J, Sponer J. J Phys Chem B. 2003;107:8669. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The content of Supporting Information includes full citations for references 42 and 46, the setup for molecular dynamics simulations, and the geometries from the model study of the uncatalyzed reaction in water and the geometries from QM/MM calculations.