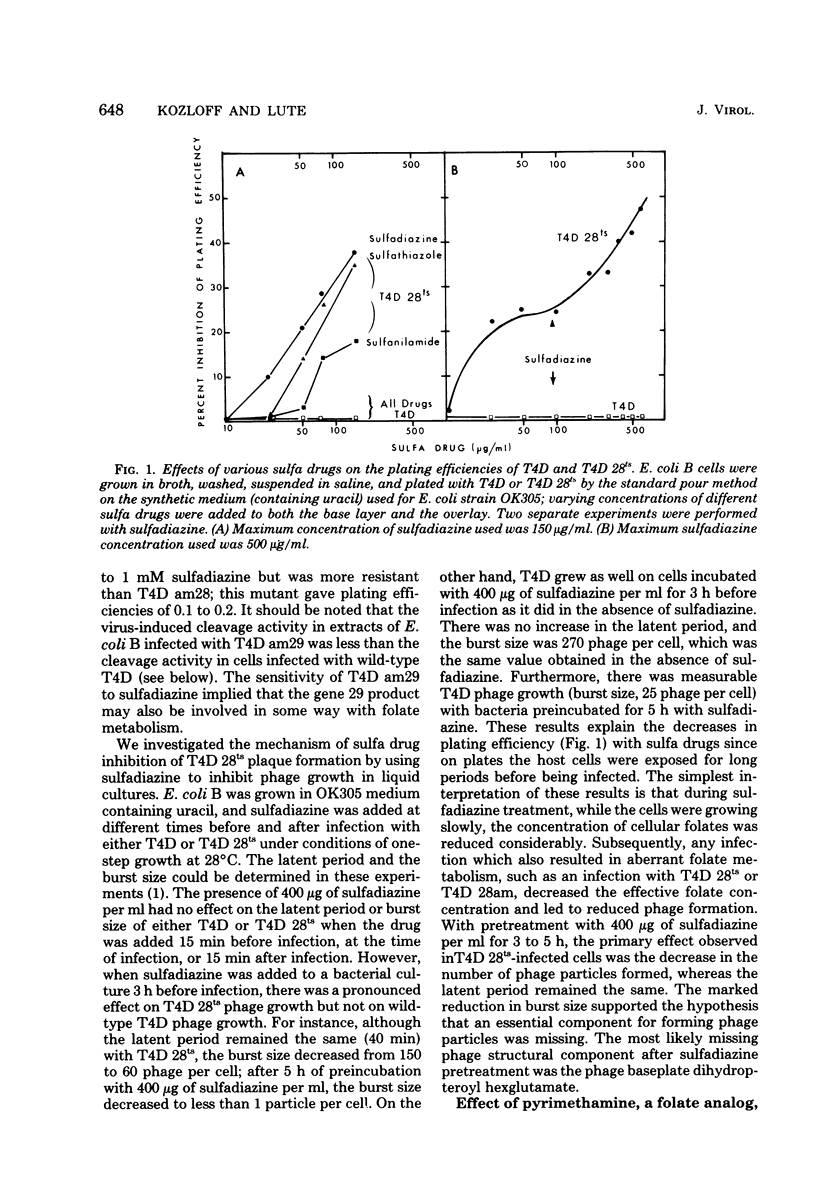
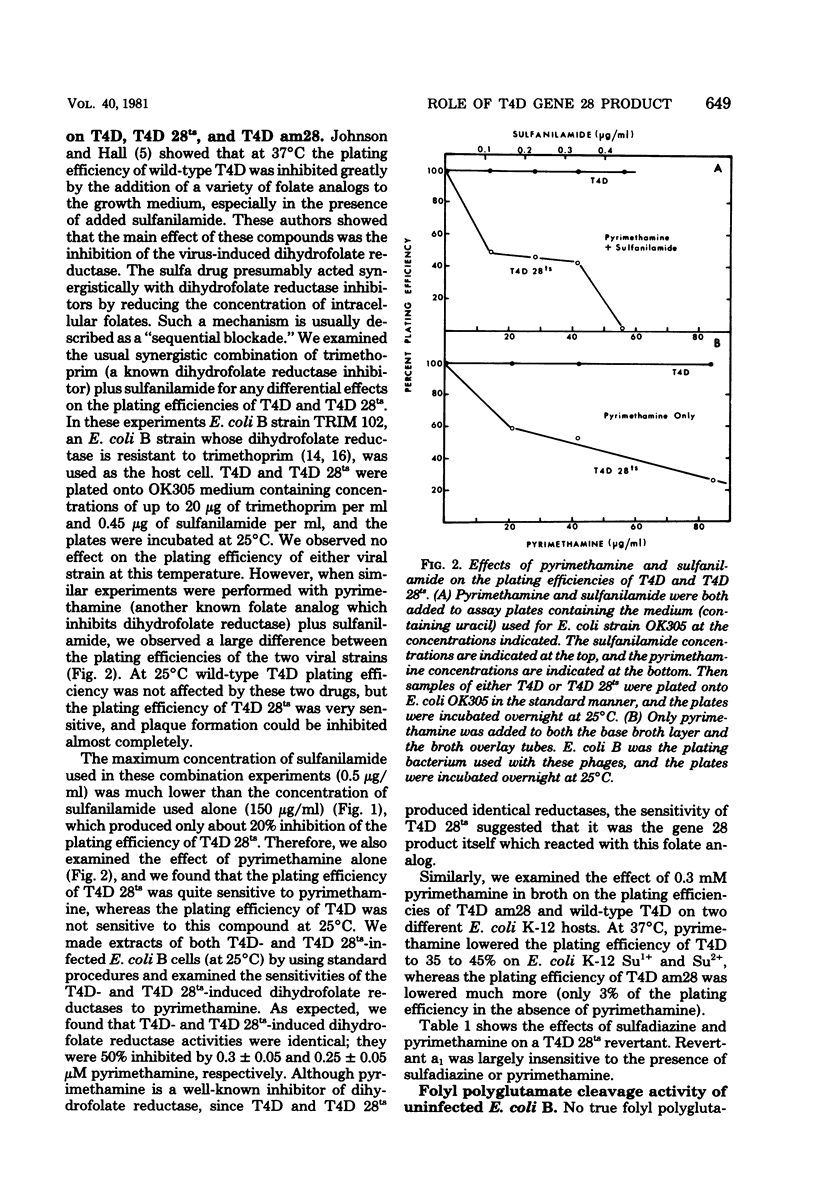
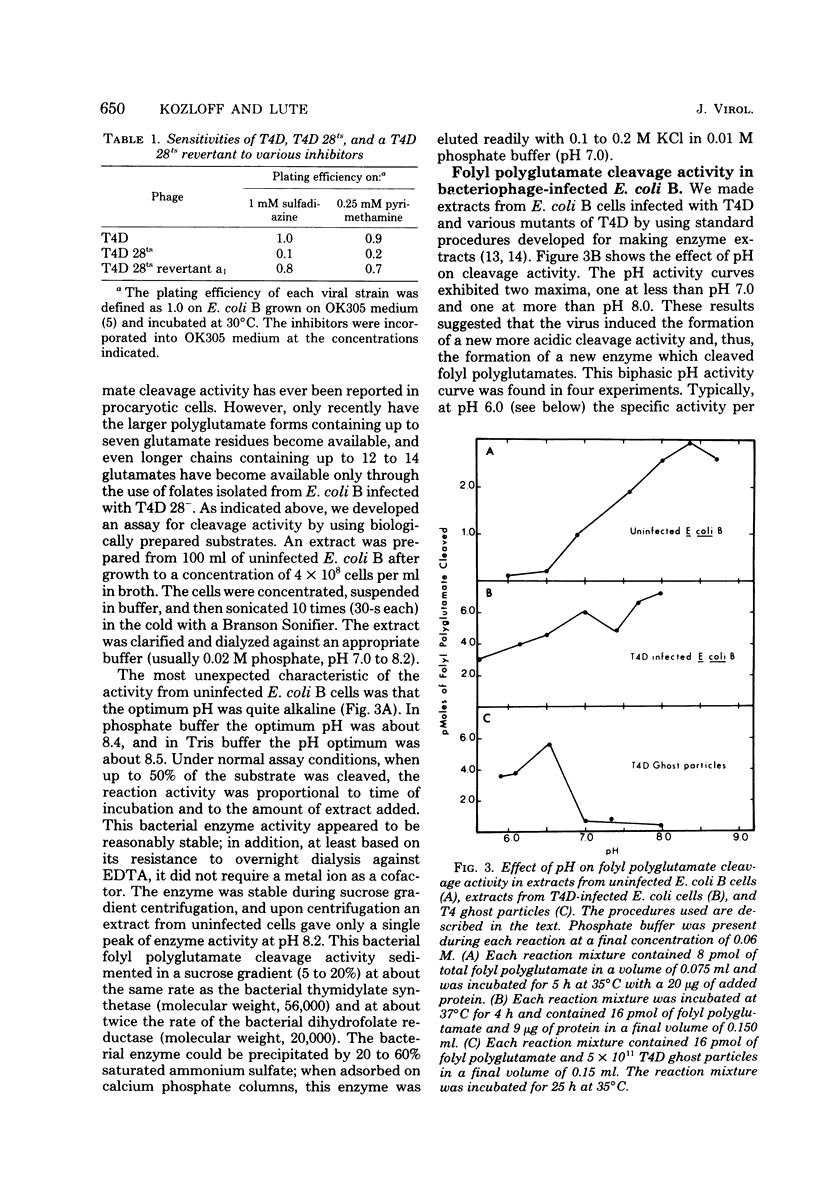
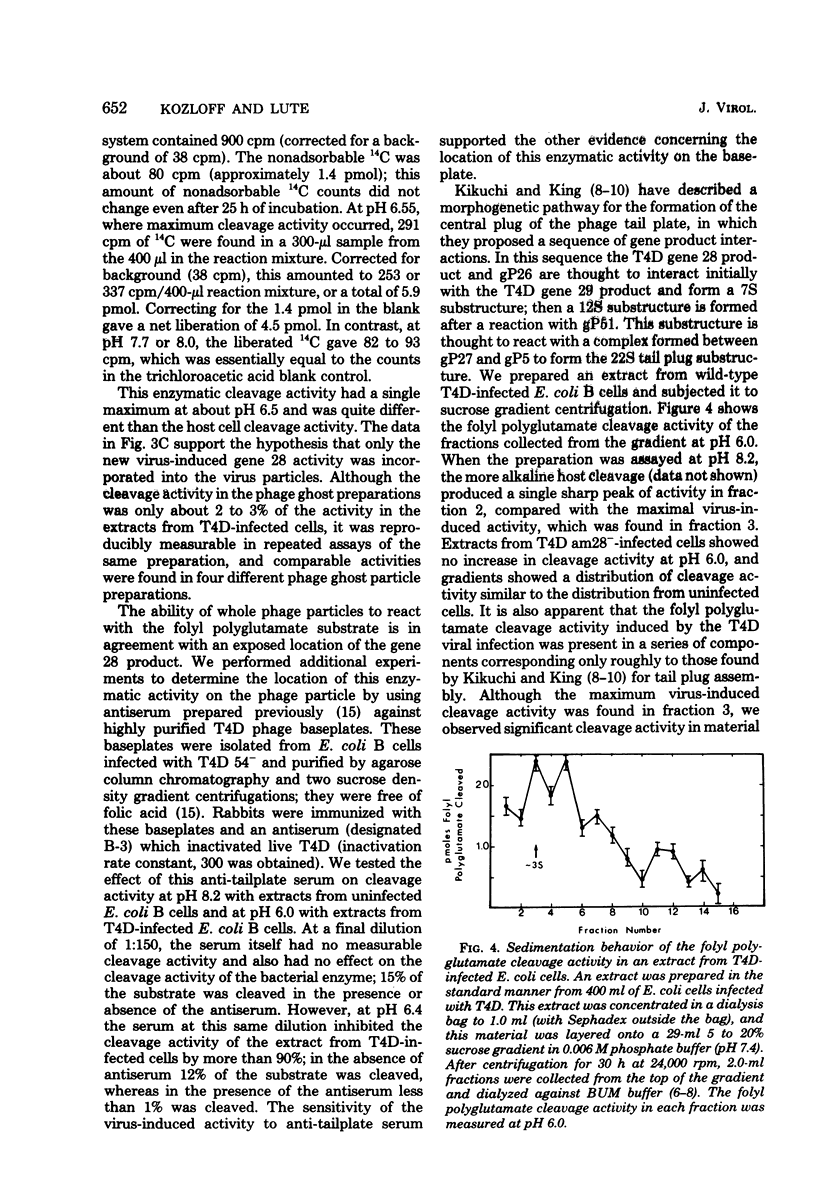
Abstract
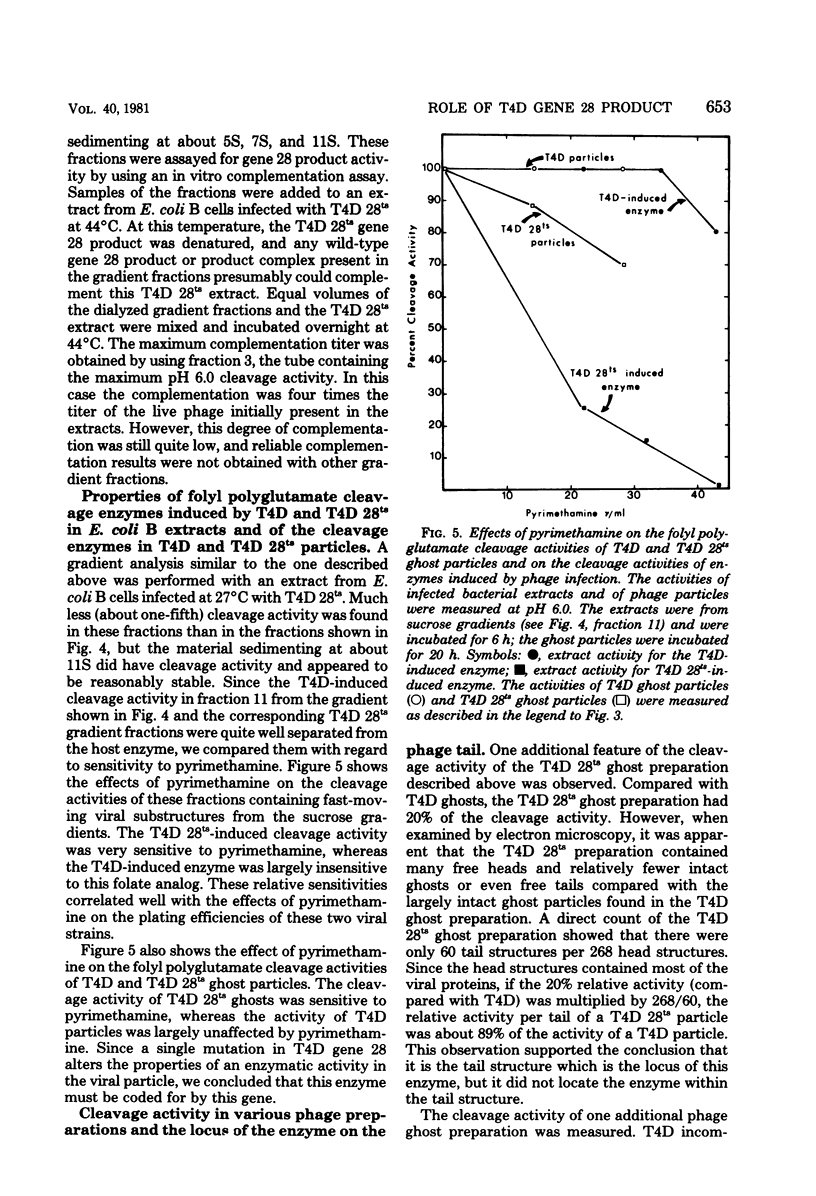
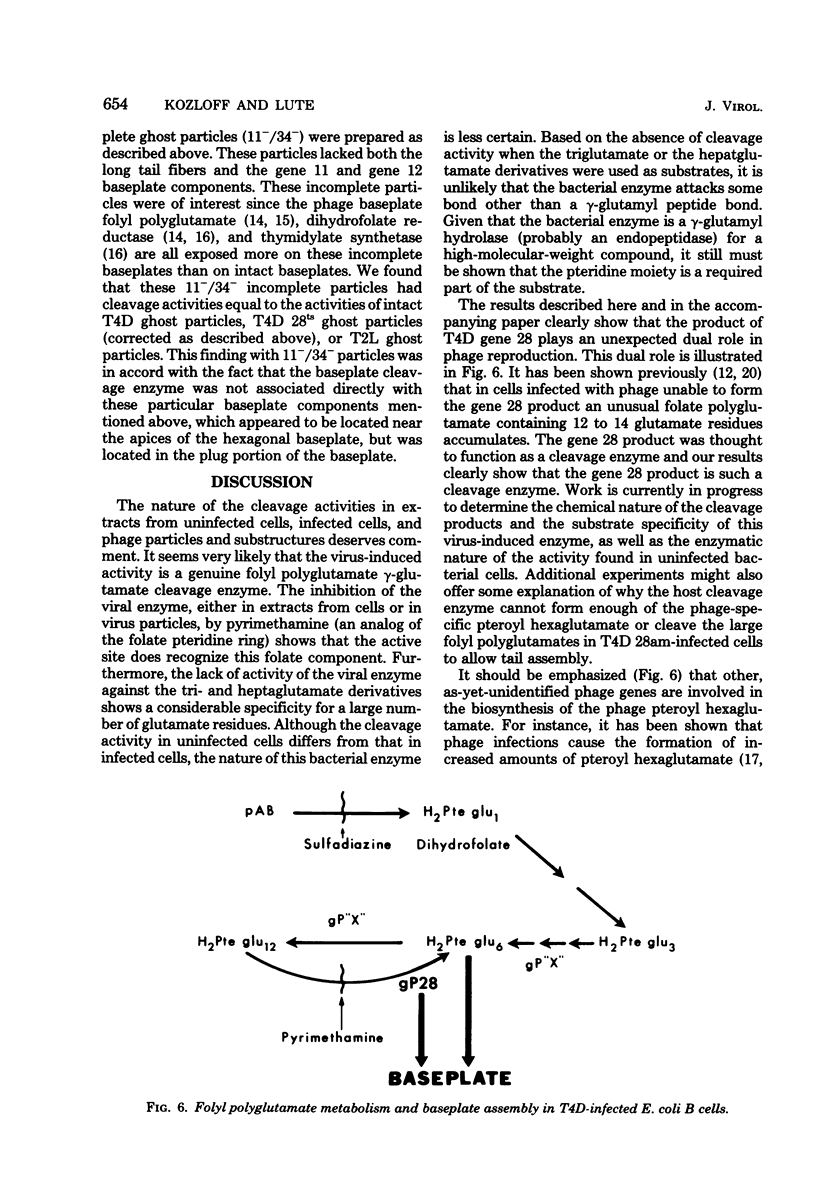
We investigated the role of the T4D bacteriophage gene 28 product in folate metabolism in infected Escherichia coli cells by using antifolate drugs and a newly devised assay for folyl polyglutamate cleavage activity. Preincubation of host E. coli cells with various sulfa drugs inhibited phage production by decreasing the burst size when the phage particles produced an altered gene 28 product (i.e., after infection under permissive conditions with T4D 28ts or T4D am28). In addition, we found that another folate analog, pyrimethamine, also inhibited T4D 28ts production and T4D 28am production, but this analog did not inhibit wild-type T4D production. A temperature-resistant revertant of T4D 28ts was not sensitive to either sulfa drugs or pyrimethamine. We developed an assay to measure the enzymatic cleavage of folyl polyglutamates. The high-molecular-weight folyl polyglutamate substrate was isolated from E. coli B cells infected with T4D am28 in the presence of labeled glutamic acid and was characterized as a folate compound containing 12 to 14 labeled glutamate residues. Extracts of uninfected bacteria liberated glutamate residues from this substrate with a pH optimum of 8.4 to 8.5. Extracts of bacteriophage T4D-infected E. coli B cells exhibited an additional new folyl polyglutamate cleavage activity with a pH optimum of about 6.4 to 6.5, which was clearly distinguished from the preexisting activity in the uninfected host cells. This new activity was induced in E. coli B cells by infection with wild-type T4D and T4D amber mutants 29−, 26−, 27−, 51−, and 10−, but it was not induced under nonpermissive conditions by T4D am28 or by T4D 28ts. Mutations in gene 28 affected the properties of the induced cleavage enzyme. Wild-type T4D-induced cleavage activity was not inhibited by pyrimethamine, whereas the T4D 28ts activity induced at a permissive temperature was inhibited by this folate analog. Folyl polyglutamate cleavage activity characteristic of the activity induced in host cells by wild-type T4D or by T4D gene 28 mutants was also found in highly purified preparations of these phage ghost particles. The T4D-induced cleavage activity could be inhibited by antiserum prepared against highly purified phage baseplates. We concluded that T4D infection induced the formation of a new folyl polyglutamate cleavage enzyme and that this enzyme was coded for by T4D gene 28. Furthermore, since this gene product was a baseplate tail plug component which had both its antigenic sites and its catalytic sites exposed on the phage particle, it was apparent that this enzyme formed part of the distal surface of the phage baseplate central tail plug.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baugh C. M., Braverman E., Nair M. G. The identification of poly-gamma-glutamyl chain lengths in bacterial folates. Biochemistry. 1974 Nov 19;13(24):4952–4957. doi: 10.1021/bi00721a012. [DOI] [PubMed] [Google Scholar]
- Baugh C. M., Krumdieck C. L. Naturally occurring folates. Ann N Y Acad Sci. 1971 Nov 30;186:7–28. [PubMed] [Google Scholar]
- Johnson J. R., Hall D. H. Isolation and characterization of mutants of bacteriophage T4 resistant to folate analogs. Virology. 1973 Jun;53(2):413–426. doi: 10.1016/0042-6822(73)90221-3. [DOI] [PubMed] [Google Scholar]
- Kao S. H., McClain W. H. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J Virol. 1980 Apr;34(1):95–103. doi: 10.1128/jvi.34.1.95-103.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S. H., McClain W. H. Roles of bacteriophage T4 gene 5 and gene s products in cell lysis. J Virol. 1980 Apr;34(1):104–107. doi: 10.1128/jvi.34.1.104-107.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J Mol Biol. 1975 Dec 25;99(4):645–672. doi: 10.1016/s0022-2836(75)80178-1. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. II. Mutants unable to form the central part of the baseplate. J Mol Biol. 1975 Dec 25;99(4):673–694. doi: 10.1016/s0022-2836(75)80179-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975 Dec 25;99(4):695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M. Bacteriophage tail components. IV. Pteroyl polyglutamate synthesis in T4D-infected Escherichia coli B. J Virol. 1973 May;11(5):630–636. doi: 10.1128/jvi.11.5.630-636.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M., Baugh C. M. Bacteriophage tail components. V. Complementation of T4D gene 28 - -infected bacterial extracts with pteroyl hexaglutamate. J Virol. 1973 May;11(5):637–641. doi: 10.1128/jvi.11.5.637-641.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M., Crosby L. K. Bacteriophage T4 virion baseplate thymidylate synthetase and dihydrofolate reductase. J Virol. 1977 Sep;23(3):637–644. doi: 10.1128/jvi.23.3.637-644.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M., Crosby L. K., Rao N., Chapman V. A., DeLong S. S. Bacteriophage tail components. I. Pteroyl polyglutamates in T-even bacteriophages. J Virol. 1970 Jun;5(6):726–739. doi: 10.1128/jvi.5.6.726-739.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M. Folic acid, a structural component of T4 bacteriophage. J Mol Biol. 1965 Jul;12(3):780–792. doi: 10.1016/s0022-2836(65)80327-8. [DOI] [PubMed] [Google Scholar]
- Kozloff L. M., Verses C., Lute M., Crosby L. K. Bacteriophage tail components. II. Dihydrofolate reductase in T4D bacteriophage. J Virol. 1970 Jun;5(6):740–753. doi: 10.1128/jvi.5.6.740-753.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Zorzopulos J. Dual functions of bacteriophage T4D gene 28 product: structural component of the viral tail baseplate central plug and cleavage enzyme for folyl polyglutamates. I. Identification of T4D gene 28 product in the tail plug. J Virol. 1981 Dec;40(3):635–644. doi: 10.1128/jvi.40.3.635-644.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumdieck C. L., Baugh C. M. Radioactive assay of folic acid polyglutamate conjugase(s). Anal Biochem. 1970 May;35(1):123–129. doi: 10.1016/0003-2697(70)90017-5. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kozloff L. M. Folate polyglutamates in T4D bacteriophage and T4D-infected Escherichia coli. Biochim Biophys Acta. 1978 May 3;540(2):313–319. doi: 10.1016/0304-4165(78)90144-7. [DOI] [PubMed] [Google Scholar]
- Silink M., Reddel R., Bethel M., Rowe P. B. Gamma-glutamyl hydrolase conjugase). Purification and properties of the bovine hepatic enzyme. J Biol Chem. 1975 Aug 10;250(15):5982–5994. [PubMed] [Google Scholar]
- VOLCANI B. E., MARGALITH P. A new species (Flavobacterium polyglutamicum) which hydrolyzes the gamma-L-glutamyl bond in polypeptides. J Bacteriol. 1957 Nov;74(5):646–655. doi: 10.1128/jb.74.5.646-655.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]


